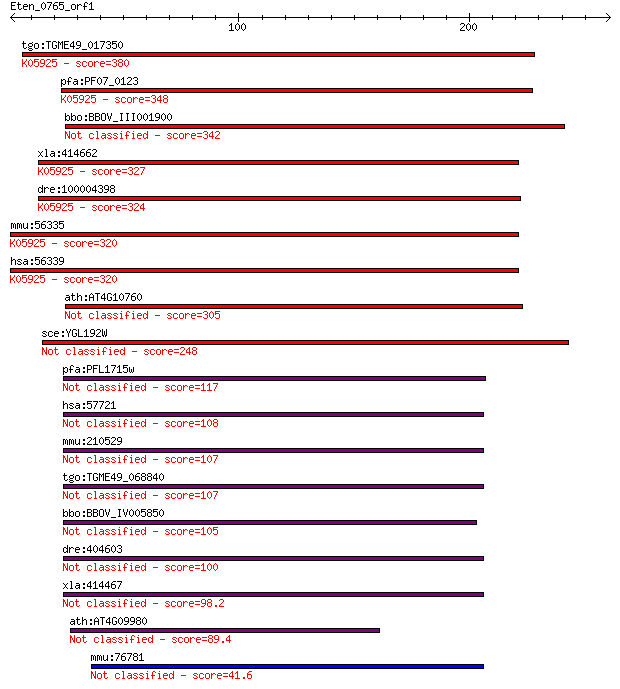
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0765_orf1
Length=260
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_017350 N6-adenosine-methyltransferase 70 kDa subuni... 380 3e-105
pfa:PF07_0123 mRNA (N6-adenosine)-methyltransferase, putative ... 348 2e-95
bbo:BBOV_III001900 17.m07187; MT-A70 family protein 342 8e-94
xla:414662 mettl3, MGC81069; methyltransferase like 3 (EC:2.1.... 327 3e-89
dre:100004398 mettl3, MGC77093, fj82g07, wu:fj82g07, zgc:77093... 324 3e-88
mmu:56335 Mettl3, 2310024F18Rik, M6A, Spo8; methyltransferase ... 320 3e-87
hsa:56339 METTL3, IME4, M6A, MGC4336, MT-A70, Spo8; methyltran... 320 3e-87
ath:AT4G10760 MTA; MTA (MRNA ADENOSINE METHYLASE); S-adenosylm... 305 1e-82
sce:YGL192W IME4, SPO8; Ime4p (EC:2.1.1.62) 248 1e-65
pfa:PFL1715w mRNA methyltransferase, putative 117 5e-26
hsa:57721 METTL14, KIAA1627; methyltransferase like 14 108 3e-23
mmu:210529 Mettl14, G430022H21Rik, mKIAA1627; methyltransferas... 107 4e-23
tgo:TGME49_068840 mRNA (2'-O-methyladenosine-N(6)-)-methyltran... 107 5e-23
bbo:BBOV_IV005850 23.m05775; hypothetical protein 105 1e-22
dre:404603 mettl14, MGC77296, zgc:77296; methyltransferase lik... 100 4e-21
xla:414467 mettl14; methyltransferase like 14 98.2
ath:AT4G09980 EMB1691 (EMBRYO DEFECTIVE 1691); S-adenosylmethi... 89.4 1e-17
mmu:76781 Mettl4, 2410198H06Rik, A730091E08Rik, AV296509, HsT6... 41.6 0.003
> tgo:TGME49_017350 N6-adenosine-methyltransferase 70 kDa subunit,
putative (EC:2.1.1.62); K05925 mRNA (2'-O-methyladenosine-N6-)-methyltransferase
[EC:2.1.1.62]
Length=819
Score = 380 bits (975), Expect = 3e-105, Method: Composition-based stats.
Identities = 180/225 (80%), Positives = 195/225 (86%), Gaps = 5/225 (2%)
Query 6 EEPLHPFSIG---PQCLTEKYPAQWIRCDIRTLDFSIFKPFVDVVMADPPWDIHMDLPYG 62
E P++IG TE YPAQWIRCDIRT DFSIF+ + VVMADPPWDIHMDLPYG
Sbjct 564 EASADPYAIGTISAGAYTE-YPAQWIRCDIRTFDFSIFRKLIRVVMADPPWDIHMDLPYG 622
Query 63 TMTDDEMRSLRVDLIQDEGLLFLWVTGRAMELARECLQLWGYRRVEEILWVKTNQLQRII 122
TMTD EMRSLRVDLIQ+EGLLFLWVTGRAMELARECLQLWGYRRVEEILWVKTNQLQRII
Sbjct 623 TMTDQEMRSLRVDLIQEEGLLFLWVTGRAMELARECLQLWGYRRVEEILWVKTNQLQRII 682
Query 123 RTGRTGHWLNHSKEHCLVGVKGNPKVNRNIDCDVIVSEVRQTSRKPDEIYRMIERMRPGT 182
RTGRTGHWLNHSKEHCLV VKGN NRNIDCDVIVSEVR+TSRKPDEIYRMIERM P +
Sbjct 683 RTGRTGHWLNHSKEHCLVAVKGNMAFNRNIDCDVIVSEVRETSRKPDEIYRMIERMAPDS 742
Query 183 LKIELFGRQHNIRNNWITLGNQLEGVKLEEPAELAQRYKAFVQQQ 227
LK+ELFGR HN+RNNWITLGNQL+GVK+E P L +RY AF +++
Sbjct 743 LKVELFGRMHNVRNNWITLGNQLKGVKIEHPL-LRERYNAFAERE 786
> pfa:PF07_0123 mRNA (N6-adenosine)-methyltransferase, putative
(EC:2.1.1.62); K05925 mRNA (2'-O-methyladenosine-N6-)-methyltransferase
[EC:2.1.1.62]
Length=760
Score = 348 bits (892), Expect = 2e-95, Method: Compositional matrix adjust.
Identities = 156/204 (76%), Positives = 174/204 (85%), Gaps = 1/204 (0%)
Query 23 YPAQWIRCDIRTLDFSIFKPFVDVVMADPPWDIHMDLPYGTMTDDEMRSLRVDLIQDEGL 82
Y QWIRCD+R D SIF +V VVMADPPWDIHMDLPYGTMTD+EM+ L V LIQDEG+
Sbjct 550 YGPQWIRCDLRNFDLSIFNKYVSVVMADPPWDIHMDLPYGTMTDNEMKLLPVQLIQDEGM 609
Query 83 LFLWVTGRAMELARECLQLWGYRRVEEILWVKTNQLQRIIRTGRTGHWLNHSKEHCLVGV 142
+FLWVTGRAMELARECLQ+WGY+RVEEILWVKTN LQRIIRTGRTGHWLNHSKEHCLVG+
Sbjct 610 IFLWVTGRAMELARECLQIWGYKRVEEILWVKTNHLQRIIRTGRTGHWLNHSKEHCLVGI 669
Query 143 KGNPKVNRNIDCDVIVSEVRQTSRKPDEIYRMIERMRPGTLKIELFGRQHNIRNNWITLG 202
KGNP +NRNIDC+VIVSEVR+TSRKPDEIY +IER+ P LKIELFGR HN R+NWITLG
Sbjct 670 KGNPIINRNIDCNVIVSEVRETSRKPDEIYSLIERLCPQNLKIELFGRPHNCRSNWITLG 729
Query 203 NQLEGVKLEEPAELAQRYKAFVQQ 226
NQL GV L P ++ +RY Q
Sbjct 730 NQLNGVVLHHP-QIKERYNKVATQ 752
> bbo:BBOV_III001900 17.m07187; MT-A70 family protein
Length=641
Score = 342 bits (877), Expect = 8e-94, Method: Compositional matrix adjust.
Identities = 149/216 (68%), Positives = 182/216 (84%), Gaps = 1/216 (0%)
Query 25 AQWIRCDIRTLDFSIFKPFVDVVMADPPWDIHMDLPYGTMTDDEMRSLRVDLIQDEGLLF 84
QWI CD+R LDFSIF PFV VVMADPPWDIHMDLPYGTM D EM+ L+V IQ+EGLLF
Sbjct 424 GQWICCDVRKLDFSIFNPFVSVVMADPPWDIHMDLPYGTMKDSEMKHLKVQNIQNEGLLF 483
Query 85 LWVTGRAMELARECLQLWGYRRVEEILWVKTNQLQRIIRTGRTGHWLNHSKEHCLVGVKG 144
LWVTGR +E+ REC++ WGYR+++EI+WVKTNQLQRIIRTGRTGHW+NHSKEHCL+G+KG
Sbjct 484 LWVTGRTLEVGRECMETWGYRQLDEIVWVKTNQLQRIIRTGRTGHWINHSKEHCLIGIKG 543
Query 145 NPKVNRNIDCDVIVSEVRQTSRKPDEIYRMIERMRPGTLKIELFGRQHNIRNNWITLGNQ 204
NP +NR +DCDV+VSEVR+TSRKPDEIY +IER+ PG+LK+E+FGR HN+RNNWITLGNQ
Sbjct 544 NPVINRFVDCDVLVSEVRETSRKPDEIYGLIERVAPGSLKLEIFGRSHNVRNNWITLGNQ 603
Query 205 LEGVKLEEPAELAQRYKAFVQQQQQQQQQQQQQPQQ 240
L+G KL P E+ +RY+A + Q Q+ ++ Q
Sbjct 604 LDGYKLTHP-EIKKRYEAHLAQSDNNTQETTEESNQ 638
> xla:414662 mettl3, MGC81069; methyltransferase like 3 (EC:2.1.1.62);
K05925 mRNA (2'-O-methyladenosine-N6-)-methyltransferase
[EC:2.1.1.62]
Length=573
Score = 327 bits (837), Expect = 3e-89, Method: Compositional matrix adjust.
Identities = 151/212 (71%), Positives = 176/212 (83%), Gaps = 5/212 (2%)
Query 13 SIGPQCLTEKYPAQWIRCDIRTLDFSIFKPFVDVVMADPPWDIHMDLPYGTMTDDEMRSL 72
++G + +PAQWIRCDIR LD SI F VVMADPPWDIHM+LPYGT+TDDEMR L
Sbjct 352 AVGDSSVGRLFPAQWIRCDIRYLDVSILGKF-SVVMADPPWDIHMELPYGTLTDDEMRKL 410
Query 73 RVDLIQDEGLLFLWVTGRAMELARECLQLWGYRRVEEILWVKTNQLQRIIRTGRTGHWLN 132
++ ++QD+G LFLWVTGRAMEL RECL+LWGY RV+EI+WVKTNQLQRIIRTGRTGHWLN
Sbjct 411 QIPVLQDDGFLFLWVTGRAMELGRECLKLWGYERVDEIIWVKTNQLQRIIRTGRTGHWLN 470
Query 133 HSKEHCLVGVKGNPK-VNRNIDCDVIVSEVRQTSRKPDEIYRMIERMRPGTLKIELFGRQ 191
H KEHCLVGVKG+P+ NR +DCDVIV+EVR TS KPDEIY MIER+ PGT KIELFGR
Sbjct 471 HGKEHCLVGVKGSPQGFNRGLDCDVIVAEVRSTSHKPDEIYGMIERLSPGTRKIELFGRP 530
Query 192 HNIRNNWITLGNQLEGVKLEEP---AELAQRY 220
HNI+ NWITLGNQL+G+ L +P A+ Q+Y
Sbjct 531 HNIQPNWITLGNQLDGIHLLDPDVVAQFKQKY 562
> dre:100004398 mettl3, MGC77093, fj82g07, wu:fj82g07, zgc:77093;
methyltransferase like 3 (EC:2.1.1.62); K05925 mRNA (2'-O-methyladenosine-N6-)-methyltransferase
[EC:2.1.1.62]
Length=584
Score = 324 bits (830), Expect = 3e-88, Method: Compositional matrix adjust.
Identities = 149/210 (70%), Positives = 174/210 (82%), Gaps = 3/210 (1%)
Query 13 SIGPQCLTEKYPAQWIRCDIRTLDFSIFKPFVDVVMADPPWDIHMDLPYGTMTDDEMRSL 72
++G + + +P+QWI CDIR LD SI F VVMADPPWDIHM+LPYGT+TDDEMR L
Sbjct 363 TVGDSNVGKLFPSQWICCDIRYLDVSILGKFA-VVMADPPWDIHMELPYGTLTDDEMRKL 421
Query 73 RVDLIQDEGLLFLWVTGRAMELARECLQLWGYRRVEEILWVKTNQLQRIIRTGRTGHWLN 132
+ ++QD+G LFLWVTGRAMEL RECL LWGY RV+EI+WVKTNQLQRIIRTGRTGHWLN
Sbjct 422 NIPILQDDGFLFLWVTGRAMELGRECLSLWGYDRVDEIIWVKTNQLQRIIRTGRTGHWLN 481
Query 133 HSKEHCLVGVKGNPK-VNRNIDCDVIVSEVRQTSRKPDEIYRMIERMRPGTLKIELFGRQ 191
H KEHCLVGVKGNP+ NR +DCDVIV+EVR TS KPDEIY MIER+ PGT KIELFGR
Sbjct 482 HGKEHCLVGVKGNPQGFNRGLDCDVIVAEVRSTSHKPDEIYGMIERLSPGTRKIELFGRP 541
Query 192 HNIRNNWITLGNQLEGVKLEEPAELAQRYK 221
HN++ NWITLGNQL+G+ L +P E+ R+K
Sbjct 542 HNVQPNWITLGNQLDGIHLLDP-EVVARFK 570
> mmu:56335 Mettl3, 2310024F18Rik, M6A, Spo8; methyltransferase
like 3 (EC:2.1.1.62); K05925 mRNA (2'-O-methyladenosine-N6-)-methyltransferase
[EC:2.1.1.62]
Length=580
Score = 320 bits (821), Expect = 3e-87, Method: Compositional matrix adjust.
Identities = 153/225 (68%), Positives = 176/225 (78%), Gaps = 6/225 (2%)
Query 1 EERPKEEPLHPFSIGPQCLTEK-YPAQWIRCDIRTLDFSIFKPFVDVVMADPPWDIHMDL 59
E P +E S+G ++ +P QWI CDIR LD SI F VVMADPPWDIHM+L
Sbjct 346 EHMPSQELALTQSVGGDSSADRLFPPQWICCDIRYLDVSILGKFA-VVMADPPWDIHMEL 404
Query 60 PYGTMTDDEMRSLRVDLIQDEGLLFLWVTGRAMELARECLQLWGYRRVEEILWVKTNQLQ 119
PYGT+TDDEMR L + ++QD+G LFLWVTGRAMEL RECL LWGY RV+EI+WVKTNQLQ
Sbjct 405 PYGTLTDDEMRRLNIPVLQDDGFLFLWVTGRAMELGRECLNLWGYERVDEIIWVKTNQLQ 464
Query 120 RIIRTGRTGHWLNHSKEHCLVGVKGNPK-VNRNIDCDVIVSEVRQTSRKPDEIYRMIERM 178
RIIRTGRTGHWLNH KEHCLVGVKGNP+ N+ +DCDVIV+EVR TS KPDEIY MIER+
Sbjct 465 RIIRTGRTGHWLNHGKEHCLVGVKGNPQGFNQGLDCDVIVAEVRSTSHKPDEIYGMIERL 524
Query 179 RPGTLKIELFGRQHNIRNNWITLGNQLEGVKLEEP---AELAQRY 220
PGT KIELFGR HN++ NWITLGNQL+G+ L +P A QRY
Sbjct 525 SPGTRKIELFGRPHNVQPNWITLGNQLDGIHLLDPDVVARFKQRY 569
> hsa:56339 METTL3, IME4, M6A, MGC4336, MT-A70, Spo8; methyltransferase
like 3 (EC:2.1.1.62); K05925 mRNA (2'-O-methyladenosine-N6-)-methyltransferase
[EC:2.1.1.62]
Length=580
Score = 320 bits (820), Expect = 3e-87, Method: Compositional matrix adjust.
Identities = 152/225 (67%), Positives = 176/225 (78%), Gaps = 6/225 (2%)
Query 1 EERPKEEPLHPFSIGPQCLTEK-YPAQWIRCDIRTLDFSIFKPFVDVVMADPPWDIHMDL 59
+ P +E S+G ++ +P QWI CDIR LD SI F VVMADPPWDIHM+L
Sbjct 346 DHTPSQELALTQSVGGDSSADRLFPPQWICCDIRYLDVSILGKFA-VVMADPPWDIHMEL 404
Query 60 PYGTMTDDEMRSLRVDLIQDEGLLFLWVTGRAMELARECLQLWGYRRVEEILWVKTNQLQ 119
PYGT+TDDEMR L + ++QD+G LFLWVTGRAMEL RECL LWGY RV+EI+WVKTNQLQ
Sbjct 405 PYGTLTDDEMRRLNIPVLQDDGFLFLWVTGRAMELGRECLNLWGYERVDEIIWVKTNQLQ 464
Query 120 RIIRTGRTGHWLNHSKEHCLVGVKGNPK-VNRNIDCDVIVSEVRQTSRKPDEIYRMIERM 178
RIIRTGRTGHWLNH KEHCLVGVKGNP+ N+ +DCDVIV+EVR TS KPDEIY MIER+
Sbjct 465 RIIRTGRTGHWLNHGKEHCLVGVKGNPQGFNQGLDCDVIVAEVRSTSHKPDEIYGMIERL 524
Query 179 RPGTLKIELFGRQHNIRNNWITLGNQLEGVKLEEP---AELAQRY 220
PGT KIELFGR HN++ NWITLGNQL+G+ L +P A QRY
Sbjct 525 SPGTRKIELFGRPHNVQPNWITLGNQLDGIHLLDPDVVARFKQRY 569
> ath:AT4G10760 MTA; MTA (MRNA ADENOSINE METHYLASE); S-adenosylmethionine-dependent
methyltransferase/ methyltransferase
Length=685
Score = 305 bits (781), Expect = 1e-82, Method: Compositional matrix adjust.
Identities = 141/198 (71%), Positives = 162/198 (81%), Gaps = 2/198 (1%)
Query 25 AQWIRCDIRTLDFSIFKPFVDVVMADPPWDIHMDLPYGTMTDDEMRSLRVDLIQDEGLLF 84
AQWI CDIR+ I F VVMADPPWDIHM+LPYGTM DDEMR+L V +Q +GL+F
Sbjct 458 AQWINCDIRSFRMDILGTF-GVVMADPPWDIHMELPYGTMADDEMRTLNVPSLQTDGLIF 516
Query 85 LWVTGRAMELARECLQLWGYRRVEEILWVKTNQLQRIIRTGRTGHWLNHSKEHCLVGVKG 144
LWVTGRAMEL RECL+LWGY+RVEEI+WVKTNQLQRIIRTGRTGHWLNHSKEHCLVG+KG
Sbjct 517 LWVTGRAMELGRECLELWGYKRVEEIIWVKTNQLQRIIRTGRTGHWLNHSKEHCLVGIKG 576
Query 145 NPKVNRNIDCDVIVSEVRQTSRKPDEIYRMIERMRPGTLKIELFGRQHNIRNNWITLGNQ 204
NP+VNRNID DVIV+EVR+TSRKPDE+Y M+ER+ P K+ELF R HN W++LGNQ
Sbjct 577 NPEVNRNIDTDVIVAEVRETSRKPDEMYAMLERIMPRARKLELFARMHNAHAGWLSLGNQ 636
Query 205 LEGVKLEEPAELAQRYKA 222
L GV+L L R+KA
Sbjct 637 LNGVRLINEG-LRARFKA 653
> sce:YGL192W IME4, SPO8; Ime4p (EC:2.1.1.62)
Length=600
Score = 248 bits (634), Expect = 1e-65, Method: Compositional matrix adjust.
Identities = 121/233 (51%), Positives = 162/233 (69%), Gaps = 8/233 (3%)
Query 15 GPQCLTEKYPAQWIRCDIRTLDFSIFKPFVDVVMADPPWDIHMDLPYGTMTDDEMRSLRV 74
C+ + PAQWIRCD+R DF + F VV+ADP W+IHM+LPYGT D E+ L +
Sbjct 314 SAHCIKKALPAQWIRCDVRKFDFRVLGKF-SVVIADPAWNIHMNLPYGTCNDIELLGLPL 372
Query 75 DLIQDEGLLFLWVTGRAMELARECLQLWGYRRVEEILWVKTNQLQRIIRTGRTGHWLNHS 134
+QDEG++FLWVTGRA+EL +E L WGY + E+ W+KTNQL R I TGRTGHWLNHS
Sbjct 373 HELQDEGIIFLWVTGRAIELGKESLNNWGYNVINEVSWIKTNQLGRTIVTGRTGHWLNHS 432
Query 135 KEHCLVGVKGNPK-VNRNIDCDVIVSEVRQTSRKPDEIYRMIERMRPGTL--KIELFGRQ 191
KEH LVG+KGNPK +N++ID D+IVS R+TSRKPDE+Y + ER+ GT K+E+FGR
Sbjct 433 KEHLLVGLKGNPKWINKHIDVDLIVSMTRETSRKPDELYGIAERL-AGTHARKLEIFGRD 491
Query 192 HNIRNNWITLGNQLEGVKLEEPAELAQRYKAFVQQQQQQQQQQQQQ--PQQPS 242
HN R W T+GNQL G + E ++ ++Y+ F++ + ++ +QPS
Sbjct 492 HNTRPGWFTIGNQLTGNCIYE-MDVERKYQEFMKSKTGTSHTGTKKIDKKQPS 543
> pfa:PFL1715w mRNA methyltransferase, putative
Length=646
Score = 117 bits (292), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 73/225 (32%), Positives = 114/225 (50%), Gaps = 42/225 (18%)
Query 24 PAQWIRCDIRTLDFSIFKPFVDVVMADPPW---------------DIHMDLPYGTMTDDE 68
PA++IRCD+RT D DV++ DPPW +I++D ++E
Sbjct 370 PARYIRCDLRTFDLGSLDTKFDVILIDPPWKEYYDRKIHNLHVLNNINLDQDLNNDMNNE 429
Query 69 ---------MRSLRVDLIQD-EGLLFLWVTGRAMELARECLQLWGYRRVEEILWVKTNQL 118
+ ++ ++ I + LF+W +E AR L WGYRR E+I W+KTN
Sbjct 430 KDKFWTLEDLANIEIEKIAEVPSFLFIWCGVTHLEDARVLLNKWGYRRCEDICWLKTNIN 489
Query 119 QRII------RTGRTGHWLNHSKEHCLVGVKGNPK-------VNRNIDCDVIVSEVRQ-- 163
++ +L + EHCLVG+KG + ++ N+D DVI++E +
Sbjct 490 EKNKKNKYLNEINNENSYLQRTTEHCLVGIKGAVRRSYDIHLIHANLDTDVIIAEETEQN 549
Query 164 --TSRKPDEIYRMIERMRPGTLKIELFGRQHNIRNNWITLGNQLE 206
+ KP+E+Y++IE+ G KIELFG NIRN W+TLG ++
Sbjct 550 IYDNNKPEELYKIIEKFCLGRRKIELFGTNRNIRNGWLTLGKHID 594
> hsa:57721 METTL14, KIAA1627; methyltransferase like 14
Length=456
Score = 108 bits (269), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 65/199 (32%), Positives = 102/199 (51%), Gaps = 17/199 (8%)
Query 24 PAQWIRCDIRTLDFSIFKPFVDVVMADPPWDIHMDLPYGTMTD-----DEMRSLRVDLIQ 78
P +++ DI D P DV++ +PP + + T + D++ L +D I
Sbjct 166 PPMYLQADIEAFDIRELTPKFDVILLEPPLEEYYRETGITANEKCWTWDDIMKLEIDEIA 225
Query 79 D-EGLLFLWV-TGRAMELARECLQLWGYRRVEEILWVKTNQLQ-RIIRTGRTGHWLNHSK 135
+FLW +G ++L R CL+ WGYRR E+I W+KTN+ +T +K
Sbjct 226 APRSFIFLWCGSGEGLDLGRVCLRKWGYRRCEDICWIKTNKNNPGKTKTLDPKAVFQRTK 285
Query 136 EHCLVGVKGNPK-------VNRNIDCDVIVSEVRQTSR--KPDEIYRMIERMRPGTLKIE 186
EHCL+G+KG K ++ N+D D+I++E + KP EI+ +IE G ++
Sbjct 286 EHCLMGIKGTVKRSTDGDFIHANVDIDLIITEEPEIGNIEKPVEIFHIIEHFCLGRRRLH 345
Query 187 LFGRQHNIRNNWITLGNQL 205
LFGR IR W+T+G L
Sbjct 346 LFGRDSTIRPGWLTVGPTL 364
> mmu:210529 Mettl14, G430022H21Rik, mKIAA1627; methyltransferase
like 14
Length=456
Score = 107 bits (268), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 65/199 (32%), Positives = 102/199 (51%), Gaps = 17/199 (8%)
Query 24 PAQWIRCDIRTLDFSIFKPFVDVVMADPPWDIHMDLPYGTMTD-----DEMRSLRVDLIQ 78
P +++ DI D P DV++ +PP + + T + D++ L +D I
Sbjct 166 PPMYLQADIEAFDIRELTPKFDVILLEPPLEEYYRETGITANEKCWTWDDIMKLEIDEIA 225
Query 79 D-EGLLFLWV-TGRAMELARECLQLWGYRRVEEILWVKTNQLQ-RIIRTGRTGHWLNHSK 135
+FLW +G ++L R CL+ WGYRR E+I W+KTN+ +T +K
Sbjct 226 APRSFIFLWCGSGEGLDLGRVCLRKWGYRRCEDICWIKTNKNNPGKTKTLDPKAVFQRTK 285
Query 136 EHCLVGVKGNPK-------VNRNIDCDVIVSEVRQTSR--KPDEIYRMIERMRPGTLKIE 186
EHCL+G+KG K ++ N+D D+I++E + KP EI+ +IE G ++
Sbjct 286 EHCLMGIKGTVKRSTDGDFIHANVDIDLIITEEPEIGNIEKPVEIFHIIEHFCLGRRRLH 345
Query 187 LFGRQHNIRNNWITLGNQL 205
LFGR IR W+T+G L
Sbjct 346 LFGRDSTIRPGWLTVGPTL 364
> tgo:TGME49_068840 mRNA (2'-O-methyladenosine-N(6)-)-methyltransferase,
putative (EC:2.1.1.62)
Length=525
Score = 107 bits (267), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 71/222 (31%), Positives = 107/222 (48%), Gaps = 40/222 (18%)
Query 24 PAQWIRCDIRTLDFSIFKPF-VDVVMADPPWDIHMD--LPYGTMTDD-------EMRSLR 73
P+ I+ ++ D+ I DV++ DPPW + D G +D EM L
Sbjct 247 PSLCIQANLHHFDWGILGGVKFDVILVDPPWQEYFDRCAAIGATNEDLTPWTLEEMLQLP 306
Query 74 VDLIQDE-GLLFLWVTGRAMELARECLQLWGYRRVEEILWVKTNQLQRIIRTGRTGHWLN 132
V+ I D FLW +E AR+ L WGYRR E+I W+KTN+ R + +N
Sbjct 307 VEKIGDTPSFCFLWCGVTHLEDARQLLHKWGYRRCEDICWLKTNKKAAQRRREQNAAHVN 366
Query 133 -------------------HSKEHCLVGVKGNPK-------VNRNIDCDVIVSEVRQ--- 163
+ EHCL+G+KG + ++ N+D D+++SE +
Sbjct 367 DVLDYKATQLVHDETSILQRTTEHCLMGIKGTVRRSQDSHFIHANLDTDILISEQEEEVG 426
Query 164 TSRKPDEIYRMIERMRPGTLKIELFGRQHNIRNNWITLGNQL 205
+RKP+E+Y +IER G +IELFGR N R W+T+G +
Sbjct 427 CTRKPEELYDIIERFCLGRRRIELFGRDWNRRAGWVTVGCEF 468
> bbo:BBOV_IV005850 23.m05775; hypothetical protein
Length=448
Score = 105 bits (263), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 70/231 (30%), Positives = 107/231 (46%), Gaps = 52/231 (22%)
Query 24 PAQWIRCDIRTLDFSIFKPFVDVVMADPPWDI----HMDLPYGTMTDDEMRSLRVDLIQD 79
PA++I D+RT DF + DVV+ +PPW + +G DD + + VD I D
Sbjct 153 PARYISADLRTFDFDSLRVLFDVVLINPPWRTPRMKALKQNFGWTLDDLIEHVPVDKIVD 212
Query 80 E-GLLFLWVTGRAMELARECLQLWGYRRVEEILWVKTNQL-----------QRIIRTGRT 127
F+W +++ AR L+ WG+R+ E+I W+KTN R G
Sbjct 213 AMSFCFIWCDYYSLDDARAALRHWGFRKCEDICWLKTNATWSNTNADSAFKMRNNAPGEH 272
Query 128 GHWLNHSKEHCLVGVKGNPK-------VNRNIDCDVIVSEVR------------------ 162
L+ + E CLVG++G + V+ N+D DVI++E R
Sbjct 273 QGLLHKTTERCLVGLRGPIRRNEDDYFVHSNLDTDVIIAEERDPRESLEMELNLRINDNE 332
Query 163 -----------QTSRKPDEIYRMIERMRPGTLKIELFGRQHNIRNNWITLG 202
+ + KP EI+ +I+R G K+ELFG +IRN W+T+G
Sbjct 333 DIQMQMDRYLTKKNTKPQEIFDIIDRFCLGRRKLELFGSDDSIRNGWVTVG 383
> dre:404603 mettl14, MGC77296, zgc:77296; methyltransferase like
14 (EC:2.1.1.-)
Length=455
Score = 100 bits (250), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 62/200 (31%), Positives = 103/200 (51%), Gaps = 19/200 (9%)
Query 24 PAQWIRCDIRTLDFSIFKPFVDVVMADPPWDIHMDLPYGTMTD------DEMRSLRVDLI 77
P +++ D T D K DV++ +PP + + G + + D++ L ++ I
Sbjct 165 PPMYLQADPDTFDLRELKCKFDVILIEPPLEEYY-RESGIIANERFWNWDDIMKLNIEEI 223
Query 78 QD-EGLLFLWV-TGRAMELARECLQLWGYRRVEEILWVKTNQLQ-RIIRTGRTGHWLNHS 134
+FLW +G ++L R CL+ WG+RR E+I W+KTN+ +T +
Sbjct 224 SSIRSFVFLWCGSGEGLDLGRMCLRKWGFRRCEDICWIKTNKNNPGKTKTLDPKAVFQRT 283
Query 135 KEHCLVGVKGNPK-------VNRNIDCDVIVSEVRQTSR--KPDEIYRMIERMRPGTLKI 185
KEHCL+G+KG + ++ N+D D+I++E + KP EI+ +IE G ++
Sbjct 284 KEHCLMGIKGTVRRSTDGDFIHANVDIDLIITEEPEMGNIEKPVEIFHIIEHFCLGRRRL 343
Query 186 ELFGRQHNIRNNWITLGNQL 205
LFGR IR W+T+G L
Sbjct 344 HLFGRDSTIRPGWLTVGPTL 363
> xla:414467 mettl14; methyltransferase like 14
Length=456
Score = 98.2 bits (243), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 60/200 (30%), Positives = 101/200 (50%), Gaps = 19/200 (9%)
Query 24 PAQWIRCDIRTLDFSIFKPFVDVVMADPPWDIHMDLPYGTMTDD------EMRSLRVDLI 77
P +++ D+ T D K DV++ +PP + + G ++ ++ L ++ I
Sbjct 166 PPMYLQADLETFDLRELKSEFDVILLEPPLEEYF-RETGIAANEKWWTWEDIMKLDIEGI 224
Query 78 Q-DEGLLFLWV-TGRAMELARECLQLWGYRRVEEILWVKTNQLQ-RIIRTGRTGHWLNHS 134
+FLW +G ++ R CL+ WG+RR E+I W+KTN+ +T +
Sbjct 225 AGSRAFVFLWCGSGEGLDFGRMCLRKWGFRRSEDICWIKTNKDNPGKTKTLDPKAIFQRT 284
Query 135 KEHCLVGVKGNPK-------VNRNIDCDVIVSEVRQTSR--KPDEIYRMIERMRPGTLKI 185
KEHCL+G+KG ++ N+D D+I++E + KP EI+ +IE G ++
Sbjct 285 KEHCLMGIKGTVHRSTDGDFIHANVDIDLIITEEPEIGNIEKPVEIFHIIEHFCLGRRRL 344
Query 186 ELFGRQHNIRNNWITLGNQL 205
LFGR IR W+T+G L
Sbjct 345 HLFGRDSTIRPGWLTVGPTL 364
> ath:AT4G09980 EMB1691 (EMBRYO DEFECTIVE 1691); S-adenosylmethionine-dependent
methyltransferase/ methyltransferase/ nucleic
acid binding
Length=775
Score = 89.4 bits (220), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 54/157 (34%), Positives = 87/157 (55%), Gaps = 28/157 (17%)
Query 27 WIRCDIRTLDFS--IFKPFVDVVMADPPWDIHMDLPYGTM------TDDEMRSLRVDLIQ 78
+++ D+ ++ S +F DV++ DPPW+ ++ G T +++ +L+++ I
Sbjct 619 YLKGDLHEVELSPELFGTKFDVILVDPPWEEYVHRAPGVSDSMEYWTFEDIINLKIEAIA 678
Query 79 DE-GLLFLWVT-GRAMELARECLQLWGYRRVEEILWVKTNQ------LQRIIRTGRTGHW 130
D LFLWV G +E R+CL+ WG+RR E+I WVKTN+ L+ RT
Sbjct 679 DTPSFLFLWVGDGVGLEQGRQCLKKWGFRRCEDICWVKTNKSNAAPTLRHDSRT-----V 733
Query 131 LNHSKEHCLVGVKGNPK-------VNRNIDCDVIVSE 160
SKEHCL+G+KG + ++ NID DVI++E
Sbjct 734 FQRSKEHCLMGIKGTVRRSTDGHIIHANIDTDVIIAE 770
> mmu:76781 Mettl4, 2410198H06Rik, A730091E08Rik, AV296509, HsT661;
methyltransferase like 4
Length=471
Score = 41.6 bits (96), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 47/195 (24%), Positives = 78/195 (40%), Gaps = 30/195 (15%)
Query 36 DFSIFKPFV------DVVMADPPWD---IHMDLPYGTMTDDEMRSLRV-DLIQDEGLLFL 85
D S +P + D ++ DPPW+ + Y +++ +++ + + L + L+
Sbjct 266 DISCMQPLLNCGKTFDAIVIDPPWENKSVKRSNRYSSLSPQQIKRMPIPKLAAADCLIVT 325
Query 86 WVTGRAMELA---RECLQLWGYRRVEEILWVKTNQLQRIIRTGRTGHWLNHSKEHCLVGV 142
WVT R L E W V E WVK + + H CLV
Sbjct 326 WVTNRQKHLCFVKEELYPSWSVEVVAEWYWVKITNSGEFVFPLDSPH---KKPYECLVLG 382
Query 143 K---GNPKVNRNIDCDV--------IVS-EVRQTSRKPDEIYRMIERMRPGTLKIELFGR 190
+ P RN D + IVS S KP + + ++PG +ELF R
Sbjct 383 RVKEKTPLALRNPDVRIPPVPDQKLIVSVPCVLHSHKPPLTEVLRDYIKPGGQCLELFAR 442
Query 191 QHNIRNNWITLGNQL 205
N++ W++ GN++
Sbjct 443 --NLQPGWMSWGNEV 455
Lambda K H
0.319 0.136 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9596524200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40