bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0781_orf1
Length=192
Score E
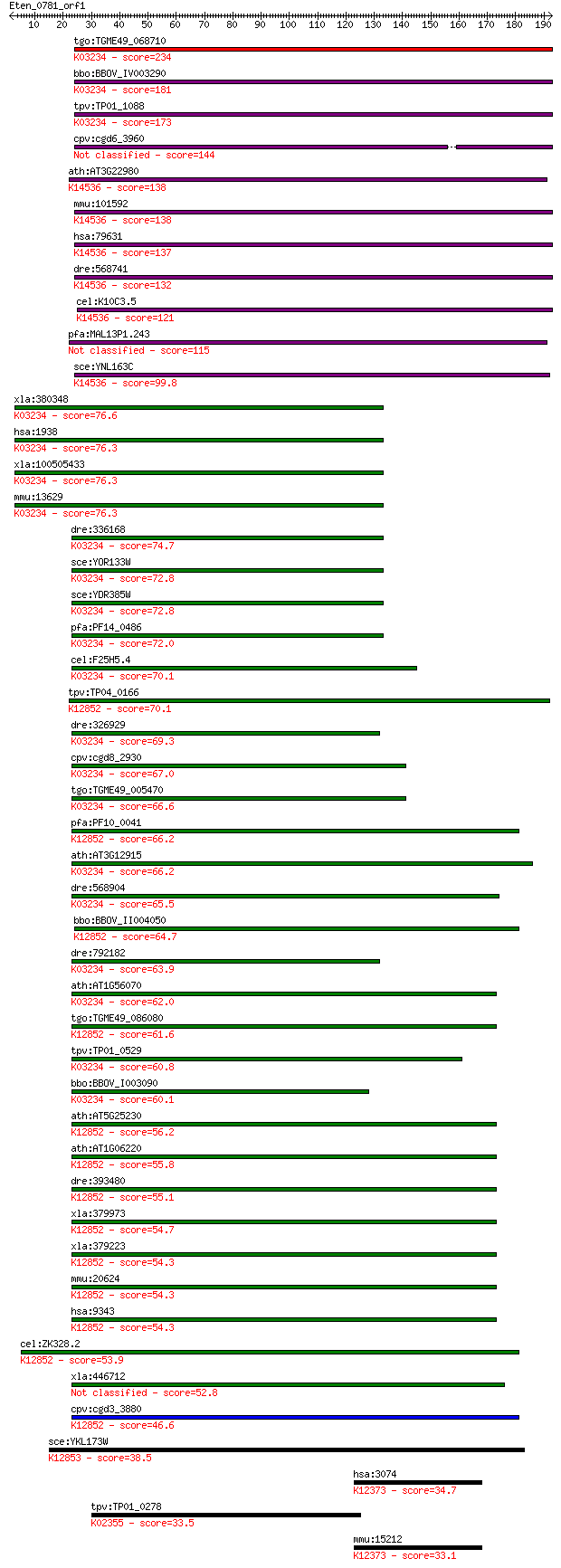
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_068710 elongation factor Tu GTP-binding domain-cont... 234 1e-61
bbo:BBOV_IV003290 21.m02927; Elongation factor Tu-like protein... 181 1e-45
tpv:TP01_1088 elongation factor Tu; K03234 elongation factor 2 173 3e-43
cpv:cgd6_3960 elongation factor-like protein 144 2e-34
ath:AT3G22980 elongation factor Tu family protein; K14536 ribo... 138 1e-32
mmu:101592 Eftud1, 4932434J20Rik, 6030468D11Rik, AI451340, AU0... 138 1e-32
hsa:79631 EFTUD1, FAM42A, FLJ13119, HsT19294, RIA1; elongation... 137 2e-32
dre:568741 Elongation FacTor family member (eft-2)-like; K1453... 132 1e-30
cel:K10C3.5 hypothetical protein; K14536 ribosome assembly pro... 121 1e-27
pfa:MAL13P1.243 elongation factor Tu, putative 115 8e-26
sce:YNL163C RIA1, EFL1; Cytoplasmic GTPase involved in biogene... 99.8 5e-21
xla:380348 eef2.1, MGC53560, eef-2, eef2, ef2; eukaryotic tran... 76.6 5e-14
hsa:1938 EEF2, EEF-2, EF2; eukaryotic translation elongation f... 76.3 6e-14
xla:100505433 hypothetical protein LOC100505433; K03234 elonga... 76.3 6e-14
mmu:13629 Eef2, Ef-2, MGC98463; eukaryotic translation elongat... 76.3 6e-14
dre:336168 eef2l2, fj53d02, si:ch211-113n10.4, wu:fj53d02; euk... 74.7 2e-13
sce:YOR133W EFT1; Eft1p; K03234 elongation factor 2 72.8
sce:YDR385W EFT2; Eft2p; K03234 elongation factor 2 72.8
pfa:PF14_0486 elongation factor 2; K03234 elongation factor 2 72.0
cel:F25H5.4 eft-2; Elongation FacTor family member (eft-2); K0... 70.1 4e-12
tpv:TP04_0166 U5 small nuclear ribonucleoprotein; K12852 116 k... 70.1 5e-12
dre:326929 eef2b, EEF2, MGC63584, eef2l, fe49h02, wu:fe49h02, ... 69.3 8e-12
cpv:cgd8_2930 Eft2p GTpase; translation elongation factor 2 (E... 67.0 4e-11
tgo:TGME49_005470 elongation factor 2, putative ; K03234 elong... 66.6 5e-11
pfa:PF10_0041 U5 small nuclear ribonuclear protein, putative; ... 66.2 6e-11
ath:AT3G12915 GTP binding / GTPase; K03234 elongation factor 2 66.2
dre:568904 eef2a.2, si:dkey-110c1.4; eukaryotic translation el... 65.5 1e-10
bbo:BBOV_II004050 18.m06335; u5 small nuclear ribonuclear prot... 64.7 2e-10
dre:792182 eef2a.1, MGC113191, si:dkey-110c1.3, zgc:113191; eu... 63.9 3e-10
ath:AT1G56070 LOS1; LOS1; copper ion binding / translation elo... 62.0 1e-09
tgo:TGME49_086080 U5 small nuclear ribonucleoprotein, putative... 61.6 1e-09
tpv:TP01_0529 elongation factor 2; K03234 elongation factor 2 60.8
bbo:BBOV_I003090 19.m02240; elongation factor 2, EF-2; K03234 ... 60.1 4e-09
ath:AT5G25230 elongation factor Tu family protein; K12852 116 ... 56.2 7e-08
ath:AT1G06220 MEE5; MEE5 (MATERNAL EFFECT EMBRYO ARREST 5); GT... 55.8 8e-08
dre:393480 eftud2, MGC66214, wu:fi20f05, zgc:66214; elongation... 55.1 2e-07
xla:379973 snrp116-pending, MGC52678; U5 snRNP-specific protei... 54.7 2e-07
xla:379223 eftud2, MGC53479, snrp116, snu114; elongation facto... 54.3 2e-07
mmu:20624 Eftud2, 116kDa, Snrp116, U5-116kD; elongation factor... 54.3 2e-07
hsa:9343 EFTUD2, DKFZp686E24196, FLJ44695, KIAA0031, Snrp116, ... 54.3 3e-07
cel:ZK328.2 eft-1; Elongation FacTor family member (eft-1); K1... 53.9 4e-07
xla:446712 eef2.2, MGC84492, eef-2, ef2, eft-2; eukaryotic tra... 52.8 7e-07
cpv:cgd3_3880 Snu114p GTpase, U5 snRNP-specific protein, 116 k... 46.6 5e-05
sce:YKL173W SNU114, GIN10; GTPase component of U5 snRNP involv... 38.5 0.016
hsa:3074 HEXB, ENC-1AS; hexosaminidase B (beta polypeptide) (E... 34.7 0.21
tpv:TP01_0278 translation elongation factor G 2; K02355 elonga... 33.5 0.44
mmu:15212 Hexb; hexosaminidase B (EC:3.2.1.52); K12373 hexosam... 33.1 0.66
> tgo:TGME49_068710 elongation factor Tu GTP-binding domain-containing
protein (EC:2.7.7.4); K03234 elongation factor 2
Length=1697
Score = 234 bits (598), Expect = 1e-61, Method: Compositional matrix adjust.
Identities = 121/170 (71%), Positives = 147/170 (86%), Gaps = 1/170 (0%)
Query 24 SGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEVP 83
SGQV+S MKEACR A+LQRG CRI EAM+RFT++C+QRV+GK Y VL+RRR KI KE +
Sbjct 1528 SGQVMSMMKEACRRALLQRGRCRIYEAMIRFTVTCEQRVLGKVYGVLSRRRSKIYKEGLL 1587
Query 84 EGQTS-FVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
+GQTS FVV G LPT+EA GI++E+RSKASGH SLQ+QFSHWEVL ++PFPEACMTE+EL
Sbjct 1588 DGQTSLFVVDGCLPTSEAVGIARELRSKASGHVSLQMQFSHWEVLDDDPFPEACMTEEEL 1647
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDAEKQRTLSRNK 192
E+EG A +++LA+Q AR IINSIRK+KGLPTEEKVV AEKQRTL++NK
Sbjct 1648 EEEGVAGIAALASQSCARKIINSIRKTKGLPTEEKVVAAAEKQRTLTKNK 1697
> bbo:BBOV_IV003290 21.m02927; Elongation factor Tu-like protein;
K03234 elongation factor 2
Length=1222
Score = 181 bits (460), Expect = 1e-45, Method: Composition-based stats.
Identities = 91/172 (52%), Positives = 118/172 (68%), Gaps = 7/172 (4%)
Query 24 SGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEVP 83
+G +IS M+ CR A++QRG RI E +LR + CDQ V+GK YSVL +RR +I+ E V
Sbjct 1055 TGNIISTMRSVCRKALMQRGRPRIYEVLLRLEIQCDQCVLGKIYSVLQKRRTQIVSENVR 1114
Query 84 EGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQELE 143
G +F++ G +P +E+ G++Q++RSKASG LQFSHWE+ ++PFPEA MT++E E
Sbjct 1115 NGTNTFMIEGLIPASESFGLAQDLRSKASGGVIFHLQFSHWEMNPDDPFPEASMTDEEFE 1174
Query 144 DEGEAALSSLATQIHA---RAIINSIRKSKGLPTEEKVVNDAEKQRTLSRNK 192
DEG +L I A R IIN IRK KGLPTEEKVV AEKQRTLS K
Sbjct 1175 DEG----FNLGAMIQANIPRKIINYIRKMKGLPTEEKVVVSAEKQRTLSTKK 1222
> tpv:TP01_1088 elongation factor Tu; K03234 elongation factor
2
Length=1210
Score = 173 bits (439), Expect = 3e-43, Method: Composition-based stats.
Identities = 86/169 (50%), Positives = 115/169 (68%), Gaps = 1/169 (0%)
Query 24 SGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEVP 83
S ++++ M+ CR A +QRG RI E +LR L C+Q V+GK Y+VL +RR +I+ E V
Sbjct 1043 SWRIMTNMRHLCRKAYMQRGRTRIYEVILRLDLQCEQNVLGKIYNVLQKRRTQILSENVK 1102
Query 84 EGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQELE 143
EG T+FV+ +P +E+ G++Q++RSKASG LQFSHWE+L ++PFPE MT+ ELE
Sbjct 1103 EGTTTFVIEATMPASESFGLAQDLRSKASGGVIFHLQFSHWEMLPDDPFPETTMTDSELE 1162
Query 144 DEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDAEKQRTLSRNK 192
D+G L + I R IIN +RK KGL TEEKVV EKQRTLS K
Sbjct 1163 DDGFNITLLLQSNI-PRKIINHLRKIKGLATEEKVVVAPEKQRTLSTKK 1210
> cpv:cgd6_3960 elongation factor-like protein
Length=1100
Score = 144 bits (363), Expect = 2e-34, Method: Composition-based stats.
Identities = 65/132 (49%), Positives = 90/132 (68%), Gaps = 0/132 (0%)
Query 24 SGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEVP 83
S Q+ + KE CR A LQRG RI E L + C+Q V+GK YSV+ +RRG + EE+
Sbjct 875 SNQLTTTTKELCRKAFLQRGNVRIYEIYLNLVIYCEQSVLGKVYSVINKRRGNVFNEELK 934
Query 84 EGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQELE 143
EG ++F + ++P E+ GISQE+RSKASG+ S L FSHWE+L E+PFPE+ MT +E E
Sbjct 935 EGTSTFKIEAYIPIIESLGISQELRSKASGNISFNLSFSHWELLDEDPFPESSMTMEEFE 994
Query 144 DEGEAALSSLAT 155
DEG + ++ L +
Sbjct 995 DEGFSKVNLLIS 1006
Score = 50.8 bits (120), Expect = 3e-06, Method: Composition-based stats.
Identities = 23/34 (67%), Positives = 28/34 (82%), Gaps = 0/34 (0%)
Query 159 ARAIINSIRKSKGLPTEEKVVNDAEKQRTLSRNK 192
AR IINSIR+ KGLPT+ K+V AEKQRTL++ K
Sbjct 1067 ARMIINSIRQRKGLPTQHKIVMAAEKQRTLNKKK 1100
> ath:AT3G22980 elongation factor Tu family protein; K14536 ribosome
assembly protein 1 [EC:3.6.5.-]
Length=1015
Score = 138 bits (348), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 79/171 (46%), Positives = 114/171 (66%), Gaps = 9/171 (5%)
Query 22 VGSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEE 81
+ +GQV++A+K+ACRAA+LQ RI EAM L+ +G Y+VL+RRR +I+KEE
Sbjct 850 IFTGQVMTAVKDACRAAVLQTN-PRIVEAMYFCELNTAPEYLGPMYAVLSRRRARILKEE 908
Query 82 VPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPF--PEACMTE 139
+ EG + F V ++P +E+ G + E+R SG AS + SHWE+L+E+PF P+ TE
Sbjct 909 MQEGSSLFTVHAYVPVSESFGFADELRKGTSGGASALMVLSHWEMLEEDPFFVPK---TE 965
Query 140 QELEDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDAEKQRTLSR 190
+E+E+ G+ A S L AR +IN++R+ KGL EEKVV A KQRTL+R
Sbjct 966 EEIEEFGDGA-SVLPNT--ARKLINAVRRRKGLHVEEKVVQYATKQRTLAR 1013
> mmu:101592 Eftud1, 4932434J20Rik, 6030468D11Rik, AI451340, AU019507,
AU022896, D7Ertd791e; elongation factor Tu GTP binding
domain containing 1; K14536 ribosome assembly protein 1
[EC:3.6.5.-]
Length=1127
Score = 138 bits (347), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 79/169 (46%), Positives = 107/169 (63%), Gaps = 6/169 (3%)
Query 24 SGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEVP 83
SGQ+I+ MKEACR A LQ R+ AM + V+G+ Y+VL++R G++++EE+
Sbjct 965 SGQLIATMKEACRYA-LQVKPQRLMAAMYTCDIMATSDVLGRVYAVLSKREGRVLQEEMK 1023
Query 84 EGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQELE 143
EG F++ LP AE+ G + EIR + SG AS QL FSHWEV+ +PF TE+E
Sbjct 1024 EGTDMFIIKAVLPVAESFGFADEIRKRTSGLASPQLVFSHWEVIPSDPF-WVPTTEEEYL 1082
Query 144 DEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDAEKQRTLSRNK 192
GE A S + AR +N++RK KGL EEK+V AEKQRTLS+NK
Sbjct 1083 HFGEKADS----ENQARKYMNAVRKRKGLYVEEKIVEHAEKQRTLSKNK 1127
> hsa:79631 EFTUD1, FAM42A, FLJ13119, HsT19294, RIA1; elongation
factor Tu GTP binding domain containing 1; K14536 ribosome
assembly protein 1 [EC:3.6.5.-]
Length=1069
Score = 137 bits (345), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 78/169 (46%), Positives = 107/169 (63%), Gaps = 6/169 (3%)
Query 24 SGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEVP 83
SGQ+I+ MKEACR A LQ R+ AM + V+G+ Y+VL++R G++++EE+
Sbjct 907 SGQLIATMKEACRYA-LQVKPQRLMAAMYTCDIMATGDVLGRVYAVLSKREGRVLQEEMK 965
Query 84 EGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQELE 143
EG F++ LP AE+ G + EIR + SG AS QL FSHWE++ +PF TE+E
Sbjct 966 EGTDMFIIKAVLPVAESFGFADEIRKRTSGLASPQLVFSHWEIIPSDPF-WVPTTEEEYL 1024
Query 144 DEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDAEKQRTLSRNK 192
GE A S + AR +N++RK KGL EEK+V AEKQRTLS+NK
Sbjct 1025 HFGEKADS----ENQARKYMNAVRKRKGLYVEEKIVEHAEKQRTLSKNK 1069
> dre:568741 Elongation FacTor family member (eft-2)-like; K14536
ribosome assembly protein 1 [EC:3.6.5.-]
Length=1115
Score = 132 bits (331), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 76/169 (44%), Positives = 105/169 (62%), Gaps = 6/169 (3%)
Query 24 SGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEVP 83
SGQ+I+A+KEACR A Q R+ AM + V+G+ Y+VL++R G++++EE+
Sbjct 953 SGQLIAAVKEACRYA-FQAKPQRLMAAMYTCEIMATAEVLGRVYAVLSKREGRVLQEEMK 1011
Query 84 EGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQELE 143
EG F++ LP AE+ G + EIR + SG AS QL FSHWEV+ +PF TE+E
Sbjct 1012 EGTDMFIIKAVLPVAESFGFADEIRKRTSGLASPQLIFSHWEVIGSDPF-WVPTTEEEYL 1070
Query 144 DEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDAEKQRTLSRNK 192
GE A S+ A +N++RK KGL EEK+V AEKQRTL +NK
Sbjct 1071 HFGEKADST----NQALKYMNAVRKRKGLYVEEKIVEHAEKQRTLGKNK 1115
> cel:K10C3.5 hypothetical protein; K14536 ribosome assembly protein
1 [EC:3.6.5.-]
Length=894
Score = 121 bits (304), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 64/168 (38%), Positives = 102/168 (60%), Gaps = 6/168 (3%)
Query 25 GQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEVPE 84
GQ+++A+K +C AA + L R+ AM R T++ + +GK ++VL++R+ K++ E++ E
Sbjct 733 GQMMTAIKASCSAAAKKLAL-RLVAAMYRCTVTTASQALGKVHAVLSQRKSKVLSEDINE 791
Query 85 GQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQELED 144
F V +P E+ ++R SG AS QLQFSHW+V+ E+P+ T LE+
Sbjct 792 ATNLFEVVSLMPVVESFSFCDQLRKFTSGMASAQLQFSHWQVIDEDPY----WTPSTLEE 847
Query 145 EGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDAEKQRTLSRNK 192
E L + HAR ++++R+ KGLPTE+ +V AEKQR L +NK
Sbjct 848 IEEFGLKGDSPN-HARGYMDAVRRRKGLPTEDLIVESAEKQRNLKKNK 894
> pfa:MAL13P1.243 elongation factor Tu, putative
Length=1394
Score = 115 bits (289), Expect = 8e-26, Method: Composition-based stats.
Identities = 63/169 (37%), Positives = 99/169 (58%), Gaps = 5/169 (2%)
Query 22 VGSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEE 81
+ +G +I+ MKEAC +M Q L RI E MLR L+C+ V+GK Y+VL +RR I+ EE
Sbjct 1231 INAGNIIALMKEACLNSMQQNKL-RIFEPMLRLNLTCESTVLGKVYNVLLKRRCSILSEE 1289
Query 82 VPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQE 141
+ +G + + +LP + +++E+RSK SG+ +QFSHW L E+ F +
Sbjct 1290 IKDGYFLYCIDAYLPLFNSFKLAEELRSKCSGNVIYDIQFSHWNKLNEDIFLLNDTSTFI 1349
Query 142 LEDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDAEKQRTLSR 190
+++ + L + A I+N IR++KGL T EK++ EKQ TL +
Sbjct 1350 YDEDFDVKL----MENTATDIVNYIRRAKGLETNEKIMQKPEKQCTLKK 1394
> sce:YNL163C RIA1, EFL1; Cytoplasmic GTPase involved in biogenesis
of the 60S ribosome; has similarity to translation elongation
factor 2 (Eft1p and Eft2p); K14536 ribosome assembly
protein 1 [EC:3.6.5.-]
Length=1110
Score = 99.8 bits (247), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 66/170 (38%), Positives = 96/170 (56%), Gaps = 10/170 (5%)
Query 24 SGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEVP 83
SG++I++ ++A A L RI A+ + V+GK Y+V+ +R GKII EE+
Sbjct 949 SGRLITSTRDAIHEAFLDWS-PRIMWAIYSCDIQTSVDVLGKVYAVILQRHGKIISEEMK 1007
Query 84 EGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPF--PEACMTEQE 141
EG F + +P EA G+S++IR + SG A QL FS +E + +PF P +E
Sbjct 1008 EGTPFFQIEAHVPVVEAFGLSEDIRKRTSGAAQPQLVFSGFECIDLDPFWVPTTEEELEE 1067
Query 142 LEDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDAEKQRTLSRN 191
L D + + AR +N+IR+ KGL EEKVV +AEKQRTL +N
Sbjct 1068 LGDTAD-------RENIARKHMNAIRRRKGLFIEEKVVENAEKQRTLKKN 1110
> xla:380348 eef2.1, MGC53560, eef-2, eef2, ef2; eukaryotic translation
elongation factor 2, gene 1; K03234 elongation factor
2
Length=858
Score = 76.6 bits (187), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 41/133 (30%), Positives = 67/133 (50%), Gaps = 4/133 (3%)
Query 3 ESKLTGKQTTTGPLSVHS---YVGSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCD 59
E L G + +++H+ + G GQ+I + A +L R+ E + + C
Sbjct 694 EENLRGVRFDVHDVTLHADAIHRGGGQIIPTARRVLYACVLT-AQPRLMEPIYLVEIQCP 752
Query 60 QRVIGKAYSVLARRRGKIIKEEVPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQL 119
++V+G Y VL R+RG + +E G FVV +LP E+ G + ++RS G A Q
Sbjct 753 EQVVGGIYGVLNRKRGHVFEESQVAGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQC 812
Query 120 QFSHWEVLQEEPF 132
F HW++L +PF
Sbjct 813 VFDHWQILPGDPF 825
> hsa:1938 EEF2, EEF-2, EF2; eukaryotic translation elongation
factor 2; K03234 elongation factor 2
Length=858
Score = 76.3 bits (186), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 40/133 (30%), Positives = 68/133 (51%), Gaps = 4/133 (3%)
Query 3 ESKLTGKQTTTGPLSVHS---YVGSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCD 59
E + G + +++H+ + G GQ+I + A++L R+ E + + C
Sbjct 694 EENMRGVRFDVHDVTLHADAIHRGGGQIIPTARRCLYASVLT-AQPRLMEPIYLVEIQCP 752
Query 60 QRVIGKAYSVLARRRGKIIKEEVPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQL 119
++V+G Y VL R+RG + +E G FVV +LP E+ G + ++RS G A Q
Sbjct 753 EQVVGGIYGVLNRKRGHVFEESQVAGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQC 812
Query 120 QFSHWEVLQEEPF 132
F HW++L +PF
Sbjct 813 VFDHWQILPGDPF 825
> xla:100505433 hypothetical protein LOC100505433; K03234 elongation
factor 2
Length=858
Score = 76.3 bits (186), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 40/133 (30%), Positives = 68/133 (51%), Gaps = 4/133 (3%)
Query 3 ESKLTGKQTTTGPLSVHS---YVGSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCD 59
E + G + +++H+ + G GQ+I + A++L R+ E + + C
Sbjct 694 EENMRGVRFDVHDVTLHADAIHRGGGQIIPTARRCLYASVLT-AQPRLMEPIYLVEIQCP 752
Query 60 QRVIGKAYSVLARRRGKIIKEEVPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQL 119
++V+G Y VL R+RG + +E G FVV +LP E+ G + ++RS G A Q
Sbjct 753 EQVVGGIYGVLNRKRGHVFEESQVAGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQC 812
Query 120 QFSHWEVLQEEPF 132
F HW++L +PF
Sbjct 813 VFDHWQILPGDPF 825
> mmu:13629 Eef2, Ef-2, MGC98463; eukaryotic translation elongation
factor 2; K03234 elongation factor 2
Length=858
Score = 76.3 bits (186), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 40/133 (30%), Positives = 68/133 (51%), Gaps = 4/133 (3%)
Query 3 ESKLTGKQTTTGPLSVHS---YVGSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCD 59
E + G + +++H+ + G GQ+I + A++L R+ E + + C
Sbjct 694 EENMRGVRFDVHDVTLHADAIHRGGGQIIPTARRCLYASVLT-AQPRLMEPIYLVEIQCP 752
Query 60 QRVIGKAYSVLARRRGKIIKEEVPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQL 119
++V+G Y VL R+RG + +E G FVV +LP E+ G + ++RS G A Q
Sbjct 753 EQVVGGIYGVLNRKRGHVFEESQVAGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQC 812
Query 120 QFSHWEVLQEEPF 132
F HW++L +PF
Sbjct 813 VFDHWQILPGDPF 825
> dre:336168 eef2l2, fj53d02, si:ch211-113n10.4, wu:fj53d02; eukaryotic
translation elongation factor 2, like 2; K03234 elongation
factor 2
Length=861
Score = 74.7 bits (182), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 36/110 (32%), Positives = 58/110 (52%), Gaps = 1/110 (0%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I + A++L R+ E + + C ++V+G Y VL R+RG + +E
Sbjct 720 GGGQIIPTARRVLYASVLT-AQPRLMEPIYLVEIQCPEQVVGGIYGVLNRKRGHVFEESQ 778
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPF 132
G FVV +LP E+ G + ++RS G A Q F HW++L +P+
Sbjct 779 VAGTPIFVVKAYLPVNESFGFTADLRSNTGGQAFPQCVFDHWQILPGDPY 828
> sce:YOR133W EFT1; Eft1p; K03234 elongation factor 2
Length=842
Score = 72.8 bits (177), Expect = 6e-13, Method: Composition-based stats.
Identities = 37/110 (33%), Positives = 56/110 (50%), Gaps = 1/110 (0%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I M+ A A L +I E + + C ++ +G YSVL ++RG+++ EE
Sbjct 701 GGGQIIPTMRRATYAGFLLAD-PKIQEPVFLVEIQCPEQAVGGIYSVLNKKRGQVVSEEQ 759
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPF 132
G F V +LP E+ G + E+R G A Q+ F HW L +P
Sbjct 760 RPGTPLFTVKAYLPVNESFGFTGELRQATGGQAFPQMVFDHWSTLGSDPL 809
> sce:YDR385W EFT2; Eft2p; K03234 elongation factor 2
Length=842
Score = 72.8 bits (177), Expect = 6e-13, Method: Composition-based stats.
Identities = 37/110 (33%), Positives = 56/110 (50%), Gaps = 1/110 (0%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I M+ A A L +I E + + C ++ +G YSVL ++RG+++ EE
Sbjct 701 GGGQIIPTMRRATYAGFLLAD-PKIQEPVFLVEIQCPEQAVGGIYSVLNKKRGQVVSEEQ 759
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPF 132
G F V +LP E+ G + E+R G A Q+ F HW L +P
Sbjct 760 RPGTPLFTVKAYLPVNESFGFTGELRQATGGQAFPQMVFDHWSTLGSDPL 809
> pfa:PF14_0486 elongation factor 2; K03234 elongation factor
2
Length=832
Score = 72.0 bits (175), Expect = 1e-12, Method: Composition-based stats.
Identities = 39/110 (35%), Positives = 57/110 (51%), Gaps = 1/110 (0%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G+GQ++ A K+ A L R+ E + +SC Q V+ Y VL +RRG +I EE
Sbjct 691 GAGQIMPACKKCIYACELT-AFPRLVEPIYLVDISCPQDVVSGVYGVLNKRRGIVISEEQ 749
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPF 132
G + LP +E+ G + +R+ SG A Q F HW VL ++PF
Sbjct 750 KLGTPLLKIQSHLPVSESFGFTSALRAATSGQAFPQCVFDHWSVLYDDPF 799
> cel:F25H5.4 eft-2; Elongation FacTor family member (eft-2);
K03234 elongation factor 2
Length=852
Score = 70.1 bits (170), Expect = 4e-12, Method: Composition-based stats.
Identities = 39/122 (31%), Positives = 58/122 (47%), Gaps = 1/122 (0%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I + A++L R+ E + + C + +G Y VL RRRG + +E
Sbjct 711 GGGQIIPTARRVFYASVLT-AEPRLLEPVYLVEIQCPEAAVGGIYGVLNRRRGHVFEESQ 769
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G FVV +LP E+ G + ++RS G A Q F HW+VL +P Q +
Sbjct 770 VTGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQCVFDHWQVLPGDPLEAGTKPNQIV 829
Query 143 ED 144
D
Sbjct 830 LD 831
> tpv:TP04_0166 U5 small nuclear ribonucleoprotein; K12852 116
kDa U5 small nuclear ribonucleoprotein component
Length=1028
Score = 70.1 bits (170), Expect = 5e-12, Method: Composition-based stats.
Identities = 48/173 (27%), Positives = 82/173 (47%), Gaps = 14/173 (8%)
Query 22 VGSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEE 81
+ GQ+I A + C ++ L R+ E +L + C + +AY +L++RRG ++K+
Sbjct 862 ITPGQIIPATRRLCYSSFLL-STPRLMEPVLFSEIHCPADCVSEAYKILSKRRGHVLKDM 920
Query 82 VPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQE 141
G +VV +LP E+ G ++R SG A F HW ++ +P ++ +
Sbjct 921 PKPGTPFYVVHAYLPAIESFGFETDLRVDTSGQAFCLSMFDHWNIVPGDPLDKSIV---- 976
Query 142 LEDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVN---DAEKQRTLSRN 191
L A + L AR + R+ KGL TE+ +N D E +L+ N
Sbjct 977 LRTLEPAPVPHL-----AREFLVKTRRRKGL-TEDVSINSFFDEEMLSSLAEN 1023
> dre:326929 eef2b, EEF2, MGC63584, eef2l, fe49h02, wu:fe49h02,
zgc:63584; eukaryotic translation elongation factor 2b; K03234
elongation factor 2
Length=858
Score = 69.3 bits (168), Expect = 8e-12, Method: Composition-based stats.
Identities = 36/109 (33%), Positives = 55/109 (50%), Gaps = 1/109 (0%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I + A L R+ E + + C ++V+G Y VL R+RG + +E
Sbjct 717 GGGQIIPTARRVLYACQLT-AEPRLMEPIYLVEIQCPEQVVGGIYGVLNRKRGHVFEESQ 775
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEP 131
G FVV +LP E+ G + ++RS G A Q F HW++L +P
Sbjct 776 VMGTPMFVVKAYLPVNESFGFTADLRSNTGGQAFPQCVFDHWQILPGDP 824
> cpv:cgd8_2930 Eft2p GTpase; translation elongation factor 2
(EF-2) ; K03234 elongation factor 2
Length=836
Score = 67.0 bits (162), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 37/121 (30%), Positives = 57/121 (47%), Gaps = 7/121 (5%)
Query 23 GSGQVISAMKEACRAAMLQRGLC---RICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIK 79
G+GQ+ CR M L R+ E M +S Q V+G Y+ L +RRG +
Sbjct 695 GAGQITPT----CRRVMYAAALTASPRLLEPMFLVEISAPQEVVGGIYATLNQRRGHVFH 750
Query 80 EEVPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTE 139
EE G + +LP A++ + +R+ SG A Q F HWE++ +P + TE
Sbjct 751 EEPKSGTPQVEIKAYLPVADSFKFTTVLRAATSGKAFPQCVFDHWELINGDPLEKGSKTE 810
Query 140 Q 140
+
Sbjct 811 E 811
> tgo:TGME49_005470 elongation factor 2, putative ; K03234 elongation
factor 2
Length=832
Score = 66.6 bits (161), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 36/118 (30%), Positives = 54/118 (45%), Gaps = 1/118 (0%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G+GQ++ + A L R+ E M ++C Q +G YS L RRG + EE
Sbjct 691 GAGQIMPTCRRVLYACQLASA-PRLQEPMFLVDITCPQDAVGGIYSTLNTRRGHVFHEEQ 749
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQ 140
G + +LP AE+ G + +R+ SG A Q F HW L +P + E+
Sbjct 750 RSGTPLVEIKAYLPVAESFGFTTALRAATSGQAFPQCVFDHWSTLNGDPLEKGSKMEE 807
> pfa:PF10_0041 U5 small nuclear ribonuclear protein, putative;
K12852 116 kDa U5 small nuclear ribonucleoprotein component
Length=1235
Score = 66.2 bits (160), Expect = 6e-11, Method: Composition-based stats.
Identities = 41/158 (25%), Positives = 75/158 (47%), Gaps = 11/158 (6%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G+GQ+I + A +++L R+ E +L + C + Y+VL+RRRG ++K+
Sbjct 1070 GAGQIIPTARRAIYSSVLL-ATPRLLEPILLTEIICSGDSVSAVYNVLSRRRGHVLKDFP 1128
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G ++V ++P E+ G ++R+ SG A F HW ++ +P ++ +
Sbjct 1129 KVGTPLYMVHAYIPAIESFGFETDLRTHTSGQAFCISMFDHWHIVPGDPLDKSVILR--- 1185
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVN 180
L Q AR + R+ KGL +E+ +N
Sbjct 1186 ------PLEPAPIQHLAREFLLKTRRRKGL-SEDVTIN 1216
> ath:AT3G12915 GTP binding / GTPase; K03234 elongation factor
2
Length=820
Score = 66.2 bits (160), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 44/163 (26%), Positives = 73/163 (44%), Gaps = 22/163 (13%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+IS + A A+ L R+ E + + + +G YSVL ++RG + +E
Sbjct 679 GCGQMISTARRAIYASQLT-AKPRLLEPVYMVEIQAPEGALGGIYSVLNQKRGHVFEEMQ 737
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G + + +LP E+ G S ++R+ SG A Q F HW+++ +P
Sbjct 738 RPGTPLYNIKAYLPVVESFGFSGQLRAATSGQAFPQCVFDHWDMMSSDP----------- 786
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDAEKQ 185
L T A ++ IRK KGL + ++D E +
Sbjct 787 ----------LETGSQAATLVADIRKRKGLKLQMTPLSDYEDK 819
> dre:568904 eef2a.2, si:dkey-110c1.4; eukaryotic translation
elongation factor 2a, tandem duplicate 2; K03234 elongation
factor 2
Length=853
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 42/151 (27%), Positives = 70/151 (46%), Gaps = 22/151 (14%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQVI+A + L R+ E + + C + VIG ++VL +RRG + E
Sbjct 712 GPGQVITATRRVLYGCQLT-AEPRLSEPVYLVEMQCPESVIGIIHAVLVKRRGVVFLESQ 770
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G +++ +LP +E+ G + ++ + SG A Q F HW+++ +P
Sbjct 771 VTGTPIYLLKAYLPVSESFGFTADLCANTSGQAFSQCVFDHWQIMPGDP----------- 819
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGLP 173
L++ + H I+ IRK KGLP
Sbjct 820 -------LNTTSKTAH---IMADIRKRKGLP 840
> bbo:BBOV_II004050 18.m06335; u5 small nuclear ribonuclear protein;
K12852 116 kDa U5 small nuclear ribonucleoprotein component
Length=999
Score = 64.7 bits (156), Expect = 2e-10, Method: Composition-based stats.
Identities = 44/157 (28%), Positives = 74/157 (47%), Gaps = 11/157 (7%)
Query 24 SGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEVP 83
+GQ+I A + A L R+ E ++ ++C + AYS+L+RRRG ++K+
Sbjct 835 AGQIIPAARRGVYGAFLL-STPRLMEPVVYSEITCAADCVSAAYSILSRRRGHVLKDLPK 893
Query 84 EGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQELE 143
G + V +LP E+ G ++R G A F HW ++ +P ++ + L+
Sbjct 894 PGTPFYEVHAYLPAIESFGFETDLRVHTHGQAFCITFFDHWNIVPGDPLDKSII----LK 949
Query 144 DEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVN 180
A + L AR + RK KGL TE+ +N
Sbjct 950 TLEPAPIPHL-----AREFMVKTRKRKGL-TEDITIN 980
> dre:792182 eef2a.1, MGC113191, si:dkey-110c1.3, zgc:113191;
eukaryotic translation elongation factor 2a, tandem duplicate
1; K03234 elongation factor 2
Length=854
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 33/109 (30%), Positives = 53/109 (48%), Gaps = 1/109 (0%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I A + L R+ E + + C + VIG Y L RRRG + E
Sbjct 715 GPGQIIVATRRVLYGCQLT-AEPRLSEPIYLVEMQCPESVIGNVYGELVRRRGVVFSESQ 773
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEP 131
G +++ +LP +E+ G + ++ + SG A Q F HW++L +P
Sbjct 774 VMGTPVYLLKAYLPVSESFGFTADLCANTSGQAFSQCVFDHWQILPGDP 822
> ath:AT1G56070 LOS1; LOS1; copper ion binding / translation elongation
factor/ translation factor, nucleic acid binding;
K03234 elongation factor 2
Length=843
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 40/150 (26%), Positives = 64/150 (42%), Gaps = 22/150 (14%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQVI + A+ + R+ E + + + +G YSVL ++RG + +E
Sbjct 702 GGGQVIPTARRVIYASQIT-AKPRLLEPVYMVEIQAPEGALGGIYSVLNQKRGHVFEEMQ 760
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G + + +LP E+ G S ++R+ SG A Q F HWE++ +P
Sbjct 761 RPGTPLYNIKAYLPVVESFGFSSQLRAATSGQAFPQCVFDHWEMMSSDP----------- 809
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGL 172
L A ++ IRK KGL
Sbjct 810 ----------LEPGTQASVLVADIRKRKGL 829
> tgo:TGME49_086080 U5 small nuclear ribonucleoprotein, putative
(EC:2.7.7.4); K12852 116 kDa U5 small nuclear ribonucleoprotein
component
Length=1008
Score = 61.6 bits (148), Expect = 1e-09, Method: Composition-based stats.
Identities = 41/150 (27%), Positives = 67/150 (44%), Gaps = 10/150 (6%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQVI + +A+L R+ E + + C + Y+VLARRRG + ++
Sbjct 843 GGGQVIPTARRVAYSALLL-ATPRLMEPVYFTEIQCPADCVSAIYTVLARRRGNVSRDMP 901
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G ++V +LP E+ G ++R+ G A F HW ++ +P +A + + L
Sbjct 902 KPGTPLYIVHAYLPAIESFGFETDLRTHTCGQAFCLSMFDHWAIVPGDPLDKAILL-RPL 960
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGL 172
E L AR + R+ KGL
Sbjct 961 EPAPAPHL--------AREFLLKTRRRKGL 982
> tpv:TP01_0529 elongation factor 2; K03234 elongation factor
2
Length=825
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 37/138 (26%), Positives = 62/138 (44%), Gaps = 13/138 (9%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
GSGQ++ + A L ++ E + ++C Q +G YS L +RRG + EE
Sbjct 684 GSGQILPTCRRCLYACQLT-AQPKLQEPIFLVDINCPQDAVGGVYSTLNQRRGHVFHEEN 742
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G + +LP +E+ G + +R+ SG A Q F HW+++ +
Sbjct 743 RSGTPLVEIKAYLPVSESFGFTTALRASTSGQAFPQCVFDHWQLVSGDAL---------- 792
Query 143 EDEGEAALSSLATQIHAR 160
E + L+ + TQI R
Sbjct 793 --EKGSKLNEIITQIRVR 808
> bbo:BBOV_I003090 19.m02240; elongation factor 2, EF-2; K03234
elongation factor 2
Length=833
Score = 60.1 bits (144), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 31/105 (29%), Positives = 52/105 (49%), Gaps = 1/105 (0%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G+GQ++ + A L ++ E + ++C Q +G YS L +RRG + EE
Sbjct 692 GAGQIMPTCRRCLYACELT-AQPKLQEPIFLVDINCPQDAVGGVYSTLNQRRGHVFHEEN 750
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVL 127
G + +LP AE+ G + +R+ SG A Q F HW+++
Sbjct 751 RAGTPLIEIKAYLPVAESFGFTTALRASTSGQAFPQCVFDHWQLM 795
> ath:AT5G25230 elongation factor Tu family protein; K12852 116
kDa U5 small nuclear ribonucleoprotein component
Length=973
Score = 56.2 bits (134), Expect = 7e-08, Method: Composition-based stats.
Identities = 38/150 (25%), Positives = 62/150 (41%), Gaps = 10/150 (6%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
GSGQ+I + +A L R+ E + + + Y+VL+RRRG + +
Sbjct 804 GSGQMIPTARRVAYSAFLM-ATPRLMEPVYYVEIQTPIDCVTAIYTVLSRRRGYVTSDVP 862
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G +++V FLP E+ G ++R G A F HW ++ +P +A
Sbjct 863 QPGTPAYIVKAFLPVIESFGFETDLRYHTQGQAFCLSVFDHWAIVPGDPLDKAIQLR--- 919
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGL 172
L Q AR + R+ KG+
Sbjct 920 ------PLEPAPIQHLAREFMVKTRRRKGM 943
> ath:AT1G06220 MEE5; MEE5 (MATERNAL EFFECT EMBRYO ARREST 5);
GTP binding / GTPase/ translation elongation factor/ translation
factor, nucleic acid binding; K12852 116 kDa U5 small nuclear
ribonucleoprotein component
Length=987
Score = 55.8 bits (133), Expect = 8e-08, Method: Composition-based stats.
Identities = 38/150 (25%), Positives = 62/150 (41%), Gaps = 10/150 (6%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
GSGQ+I + +A L R+ E + + + Y+VL+RRRG + +
Sbjct 818 GSGQMIPTARRVAYSAFLM-ATPRLMEPVYYVEIQTPIDCVTAIYTVLSRRRGHVTSDVP 876
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G +++V FLP E+ G ++R G A F HW ++ +P +A
Sbjct 877 QPGTPAYIVKAFLPVIESFGFETDLRYHTQGQAFCLSVFDHWAIVPGDPLDKAIQLR--- 933
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGL 172
L Q AR + R+ KG+
Sbjct 934 ------PLEPAPIQHLAREFMVKTRRRKGM 957
> dre:393480 eftud2, MGC66214, wu:fi20f05, zgc:66214; elongation
factor Tu GTP binding domain containing 2; K12852 116 kDa
U5 small nuclear ribonucleoprotein component
Length=973
Score = 55.1 bits (131), Expect = 2e-07, Method: Composition-based stats.
Identities = 37/150 (24%), Positives = 65/150 (43%), Gaps = 10/150 (6%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQVI + +A L R+ E + + Y+VLARRRG + ++
Sbjct 805 GGGQVIPTARRVVYSAFLM-ATPRLMEPYYFVEVQAPADCVSAVYTVLARRRGHVTQDAP 863
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G + + F+P ++ G ++R+ G A F HW+++ +P ++ + + L
Sbjct 864 IPGSPLYTIKAFIPAIDSFGFETDLRTHTQGQAFALSVFHHWQIVPGDPLDKSIVI-RPL 922
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGL 172
E + L AR + R+ KGL
Sbjct 923 EPQPAPHL--------AREFMIKTRRRKGL 944
> xla:379973 snrp116-pending, MGC52678; U5 snRNP-specific protein,
116 kD; K12852 116 kDa U5 small nuclear ribonucleoprotein
component
Length=974
Score = 54.7 bits (130), Expect = 2e-07, Method: Composition-based stats.
Identities = 36/150 (24%), Positives = 65/150 (43%), Gaps = 10/150 (6%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I + +A L R+ E + + Y+VLARRRG + ++
Sbjct 806 GGGQIIPTARRVVYSAFLM-ATPRLMEPYYFVEVQAPADCVSAVYTVLARRRGHVTQDAP 864
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G + + F+P ++ G ++R+ G A F HW+++ +P ++ + + L
Sbjct 865 IPGSPLYTIKAFIPAVDSFGFETDLRTHTQGQAFSLSVFHHWQIVPGDPLDKSIII-RPL 923
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGL 172
E + L AR + R+ KGL
Sbjct 924 EPQPAPHL--------AREFMIKTRRRKGL 945
> xla:379223 eftud2, MGC53479, snrp116, snu114; elongation factor
Tu GTP binding domain containing 2; K12852 116 kDa U5 small
nuclear ribonucleoprotein component
Length=974
Score = 54.3 bits (129), Expect = 2e-07, Method: Composition-based stats.
Identities = 36/150 (24%), Positives = 65/150 (43%), Gaps = 10/150 (6%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I + +A L R+ E + + Y+VLARRRG + ++
Sbjct 806 GGGQIIPTARRVVYSAFLM-ATPRLMEPYYFVEVQAPADCVSAVYTVLARRRGHVTQDAP 864
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G + + F+P ++ G ++R+ G A F HW+++ +P ++ + + L
Sbjct 865 IPGSPLYTIKAFIPAIDSFGFETDLRTHTQGQAFSLSVFHHWQIVPGDPLDKSIII-RPL 923
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGL 172
E + L AR + R+ KGL
Sbjct 924 EPQPAPHL--------AREFMIKTRRRKGL 945
> mmu:20624 Eftud2, 116kDa, Snrp116, U5-116kD; elongation factor
Tu GTP binding domain containing 2; K12852 116 kDa U5 small
nuclear ribonucleoprotein component
Length=972
Score = 54.3 bits (129), Expect = 2e-07, Method: Composition-based stats.
Identities = 36/150 (24%), Positives = 65/150 (43%), Gaps = 10/150 (6%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I + +A L R+ E + + Y+VLARRRG + ++
Sbjct 804 GGGQIIPTARRVVYSAFLM-ATPRLMEPYYFVEVQAPADCVSAVYTVLARRRGHVTQDAP 862
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G + + F+P ++ G ++R+ G A F HW+++ +P ++ + + L
Sbjct 863 IPGSPLYTIKAFIPAIDSFGFETDLRTHTQGQAFSLSVFHHWQIVPGDPLDKSIVI-RPL 921
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGL 172
E + L AR + R+ KGL
Sbjct 922 EPQPAPHL--------AREFMIKTRRRKGL 943
> hsa:9343 EFTUD2, DKFZp686E24196, FLJ44695, KIAA0031, Snrp116,
Snu114, U5-116KD; elongation factor Tu GTP binding domain
containing 2; K12852 116 kDa U5 small nuclear ribonucleoprotein
component
Length=937
Score = 54.3 bits (129), Expect = 3e-07, Method: Composition-based stats.
Identities = 36/150 (24%), Positives = 65/150 (43%), Gaps = 10/150 (6%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I + +A L R+ E + + Y+VLARRRG + ++
Sbjct 769 GGGQIIPTARRVVYSAFLM-ATPRLMEPYYFVEVQAPADCVSAVYTVLARRRGHVTQDAP 827
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G + + F+P ++ G ++R+ G A F HW+++ +P ++ + + L
Sbjct 828 IPGSPLYTIKAFIPAIDSFGFETDLRTHTQGQAFSLSVFHHWQIVPGDPLDKSIVI-RPL 886
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGL 172
E + L AR + R+ KGL
Sbjct 887 EPQPAPHL--------AREFMIKTRRRKGL 908
> cel:ZK328.2 eft-1; Elongation FacTor family member (eft-1);
K12852 116 kDa U5 small nuclear ribonucleoprotein component
Length=974
Score = 53.9 bits (128), Expect = 4e-07, Method: Composition-based stats.
Identities = 42/176 (23%), Positives = 72/176 (40%), Gaps = 15/176 (8%)
Query 5 KLTGKQTTTGPLSVHSYVGSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIG 64
KL T PL Y G GQ+I + +A L R+ E + +
Sbjct 794 KLLDAAIATEPL----YRGGGQMIPTARRCAYSAFLM-ATPRLMEPYYTVEVVAPADCVA 848
Query 65 KAYSVLARRRGKIIKEEVPEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHW 124
Y+VLA+RRG + + G + + ++P ++ G ++R G A F HW
Sbjct 849 AVYTVLAKRRGHVTTDAPMPGSPMYTISAYIPVMDSFGFETDLRIHTQGQAFCMSAFHHW 908
Query 125 EVLQEEPFPEACMTEQELEDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVN 180
+++ +P ++ + + L T AR + R+ KGL +E+ VN
Sbjct 909 QLVPGDPLDKSIVIK---------TLDVQPTPHLAREFMIKTRRRKGL-SEDVSVN 954
> xla:446712 eef2.2, MGC84492, eef-2, ef2, eft-2; eukaryotic translation
elongation factor 2, gene 2
Length=850
Score = 52.8 bits (125), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 42/153 (27%), Positives = 60/153 (39%), Gaps = 22/153 (14%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G GQ+I + C A + I E + + ++G YS L ++RG I EE
Sbjct 709 GGGQIIGTARR-CFYACVLTAQPAILEPVYLVEIQGPDTILGGIYSTLNKKRGVIQSEER 767
Query 83 PEGQTSFVVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQEL 142
G V FLP E+ G + ++R+ G A Q F HW+ P +
Sbjct 768 VAGMPVCCVKAFLPVNESFGFTTDLRANTGGQAFPQCVFDHWQQYPGNPLDPS------- 820
Query 143 EDEGEAALSSLATQIHARAIINSIRKSKGLPTE 175
G+A L +IRK KGL E
Sbjct 821 SKPGQAVL--------------AIRKRKGLSDE 839
> cpv:cgd3_3880 Snu114p GTpase, U5 snRNP-specific protein, 116
kDa ; K12852 116 kDa U5 small nuclear ribonucleoprotein component
Length=1035
Score = 46.6 bits (109), Expect = 5e-05, Method: Composition-based stats.
Identities = 40/159 (25%), Positives = 72/159 (45%), Gaps = 12/159 (7%)
Query 23 GSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEV 82
G+GQ++ A + AC +M +I E + + C + +++++RRG KE +
Sbjct 875 GTGQIVPASRRACYTSMFLAS-PKILEPISLVEIICPSGLDEFINNIVSKRRGHAGKE-I 932
Query 83 PEGQTSFV-VFGFLPTAEAPGISQEIRSKASGHASLQLQFSHWEVLQEEPFPEACMTEQE 141
P + V + F+P E G ++R SG A F HW ++ P + ++ +
Sbjct 933 PIPASPLVTILAFVPAIETFGFETDLRIHTSGQAFCTSCFDHWAIVPGNPL-DRNISLRL 991
Query 142 LEDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVN 180
LE +A + L AR + R+ KGL + + N
Sbjct 992 LE---KAPIPHL-----ARDFLLKTRRRKGLSDDVNIQN 1022
> sce:YKL173W SNU114, GIN10; GTPase component of U5 snRNP involved
in mRNA splicing via spliceosome; binds directly to U5
snRNA; proposed to be involved in conformational changes of
the spliceosome; similarity to ribosomal translocation factor
EF-2; K12853 114 kDa U5 small nuclear ribonucleoprotein component
Length=1008
Score = 38.5 bits (88), Expect = 0.016, Method: Composition-based stats.
Identities = 41/176 (23%), Positives = 74/176 (42%), Gaps = 23/176 (13%)
Query 15 PLSVHSYVGSGQVISAMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRR 74
P V+ V Q+I MK+AC +L + + E + ++ ++ ++ +RR
Sbjct 826 PSDVNIDVMKSQIIPLMKKACYVGLLT-AIPILLEPIYEVDITVHAPLLPIVEELMKKRR 884
Query 75 GKIIKEEVPEGQTSFV-VFGFLPTAEAPGISQEIRSKASGHASLQLQFSH--W-----EV 126
G I + + T + V G +P E+ G ++R +G QL F H W +V
Sbjct 885 GSRIYKTIKVAGTPLLEVRGQVPVIESAGFETDLRLSTNGLGMCQLYFWHKIWRKVPGDV 944
Query 127 LQEEPFPEACMTEQELEDEGEAALSSLATQIHARAIINSIRKSKGLPTEEKVVNDA 182
L ++ F + A ++SL +R + R+ KG+ T + ND
Sbjct 945 LDKDAF---------IPKLKPAPINSL-----SRDFVMKTRRRKGISTGGFMSNDG 986
> hsa:3074 HEXB, ENC-1AS; hexosaminidase B (beta polypeptide)
(EC:3.2.1.52); K12373 hexosaminidase [EC:3.2.1.52]
Length=556
Score = 34.7 bits (78), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 123 HWEVLQEEPFPEACMTEQELEDEGEAALSSLATQIHARAIINSIR 167
HW ++ ++ FP +T EL ++G +LS + T R +I R
Sbjct 235 HWHIVDDQSFPYQSITFPELSNKGSYSLSHVYTPNDVRMVIEYAR 279
> tpv:TP01_0278 translation elongation factor G 2; K02355 elongation
factor G
Length=803
Score = 33.5 bits (75), Expect = 0.44, Method: Composition-based stats.
Identities = 22/95 (23%), Positives = 44/95 (46%), Gaps = 6/95 (6%)
Query 30 AMKEACRAAMLQRGLCRICEAMLRFTLSCDQRVIGKAYSVLARRRGKIIKEEVPEGQTSF 89
+KEA R + + ++ E +++ +++C G+ L+RRRG++ + G T
Sbjct 697 GIKEAARLSGM-----KLLEPIMKVSITCPTDNFGEVVCDLSRRRGRVTNTKQGYG-TVK 750
Query 90 VVFGFLPTAEAPGISQEIRSKASGHASLQLQFSHW 124
+ G P E G +R + G ++ SH+
Sbjct 751 EIEGEAPLREMTGYMTTLRKISQGRGFYTMEMSHY 785
> mmu:15212 Hexb; hexosaminidase B (EC:3.2.1.52); K12373 hexosaminidase
[EC:3.2.1.52]
Length=536
Score = 33.1 bits (74), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 23/45 (51%), Gaps = 0/45 (0%)
Query 123 HWEVLQEEPFPEACMTEQELEDEGEAALSSLATQIHARAIINSIR 167
HW ++ ++ FP T EL ++G +LS + T R ++ R
Sbjct 214 HWHIVDDQSFPYQSTTFPELSNKGSYSLSHVYTPNDVRMVLEYAR 258
Lambda K H
0.315 0.128 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5558593832
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40