bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0791_orf1
Length=237
Score E
Sequences producing significant alignments: (Bits) Value
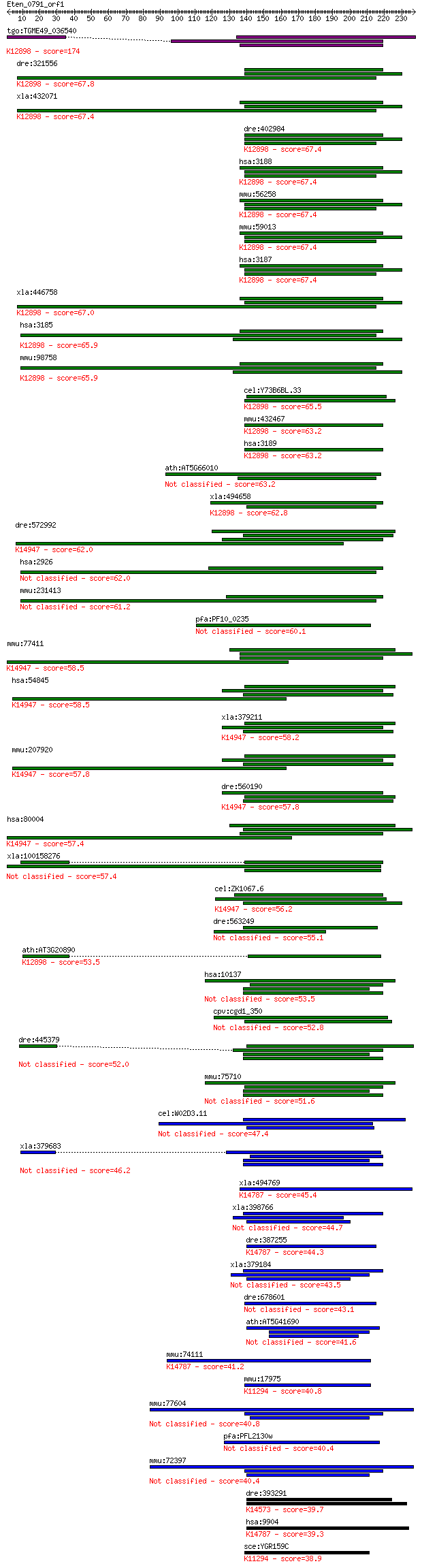
tgo:TGME49_036540 RRM domain-containing protein ; K12898 heter... 174 3e-43
dre:321556 hnrnph1l, fb19a06, wu:fb19a06, zgc:55399, zgc:85960... 67.8 3e-11
xla:432071 hnrnph1-a, MGC78776, hnrnph, hnrnph2, hnrph, hnrph1... 67.4 4e-11
dre:402984 hnrnph1, MGC77712, zgc:77712; heterogeneous nuclear... 67.4 4e-11
hsa:3188 HNRNPH2, FTP3, HNRPH', HNRPH2, hnRNPH'; heterogeneous... 67.4 4e-11
mmu:56258 Hnrnph2, DXHXS1271E, Ftp-3, Ftp3, H', HNRNP, Hnrph2;... 67.4 4e-11
mmu:59013 Hnrnph1, AI642080, E430005G16Rik, Hnrnph, Hnrph1; he... 67.4 4e-11
hsa:3187 HNRNPH1, DKFZp686A15170, HNRPH, HNRPH1, hnRNPH; heter... 67.4 5e-11
xla:446758 hnrnph1-b, MGC130700, MGC80081, hnrnph, hnrnph1, hn... 67.0 5e-11
hsa:3185 HNRNPF, HNRPF, MGC110997, OK/SW-cl.23, mcs94-1; heter... 65.9 1e-10
mmu:98758 Hnrnpf, 4833420I20Rik, AA407306, Hnrpf, MGC101981, M... 65.9 1e-10
cel:Y73B6BL.33 hypothetical protein; K12898 heterogeneous nucl... 65.5 2e-10
mmu:432467 Hnrnph3, AA693301, AI666703, Hnrph3; heterogeneous ... 63.2 8e-10
hsa:3189 HNRNPH3, 2H9, FLJ34092, HNRPH3; heterogeneous nuclear... 63.2 8e-10
ath:AT5G66010 RNA binding / nucleic acid binding / nucleotide ... 63.2 9e-10
xla:494658 hnrnph3, hnrph3; heterogeneous nuclear ribonucleopr... 62.8 9e-10
dre:572992 esrp2, FLJ20171, MGC55721, MGC77254, cb404, fa07a06... 62.0 2e-09
hsa:2926 GRSF1, FLJ13125; G-rich RNA sequence binding factor 1 62.0
mmu:231413 Grsf1, B130010H02, BB232551, C80280, D5Wsu31e; G-ri... 61.2 3e-09
pfa:PF10_0235 RNA binding protein, putative 60.1 7e-09
mmu:77411 Esrp2, 9530027K23Rik, Rbm35b; epithelial splicing re... 58.5 2e-08
hsa:54845 ESRP1, FLJ20171, RBM35A, RMB35A; epithelial splicing... 58.5 2e-08
xla:379211 esrp1, MGC53361, rbm35a; epithelial splicing regula... 58.2 3e-08
mmu:207920 Esrp1, 2210008M09Rik, A630065D16, BC031468, MGC2580... 57.8 3e-08
dre:560190 esrp1, rbm35a, wu:fi28a07, zgc:154050; epithelial s... 57.8 3e-08
hsa:80004 ESRP2, FLJ21918, FLJ22248, RBM35B; epithelial splici... 57.4 4e-08
xla:100158276 grsf1; G-rich RNA sequence binding factor 1 57.4
cel:ZK1067.6 sym-2; SYnthetic lethal with Mec family member (s... 56.2 1e-07
dre:563249 grsf1, MGC153305, wu:fb62c04, zgc:153305; G-rich RN... 55.1 2e-07
ath:AT3G20890 RNA binding / nucleic acid binding / nucleotide ... 53.5 6e-07
hsa:10137 RBM12, HRIHFB2091, KIAA0765, SWAN; RNA binding motif... 53.5 7e-07
cpv:cgd1_350 CG8205/fusilli (animal)- like, 2x RRM domains, in... 52.8 1e-06
dre:445379 rbm12, zgc:193560, zgc:193570; RNA binding motif pr... 52.0 2e-06
mmu:75710 Rbm12, 5730420G12Rik, 9430070C08Rik, AI852903, MGC30... 51.6 3e-06
cel:W02D3.11 hrpf-1; HnRNP F homolog family member (hrpf-1) 47.4 4e-05
xla:379683 rbm12, MGC68861, SWAN; RNA binding motif protein 12 46.2
xla:494769 rbm19; RNA binding motif protein 19; K14787 multipl... 45.4 2e-04
xla:398766 rbm12b, MGC68792; RNA binding motif protein 12B 44.7 3e-04
dre:387255 rbm19, npo; RNA binding motif protein 19; K14787 mu... 44.3 4e-04
xla:379184 MGC132368; hypothetical protein MGC53694 43.5 7e-04
dre:678601 MGC136953; zgc:136953 43.1 8e-04
ath:AT5G41690 RNA binding / nucleic acid binding / nucleotide ... 41.6 0.003
mmu:74111 Rbm19, 1200009A02Rik, AV302714, KIAA0682, NPO; RNA b... 41.2 0.004
mmu:17975 Ncl, B530004O11Rik, C23, D0Nds28, D1Nds28, Nucl; nuc... 40.8 0.004
mmu:77604 C430048L16Rik, AV299215, MGC91091, Rbm12bb; RIKEN cD... 40.8 0.005
pfa:PFL2130w conserved Plasmodium protein 40.4 0.006
mmu:72397 Rbm12b, 3000004N20Rik, Rbm12ba; RNA binding motif pr... 40.4 0.006
dre:393291 rbm28, MGC56258, zgc:56258; RNA binding motif prote... 39.7 0.009
hsa:9904 RBM19, DKFZp586F1023, KIAA0682; RNA binding motif pro... 39.3 0.014
sce:YGR159C NSR1, SHE5; Nsr1p; K11294 nucleolin 38.9 0.018
> tgo:TGME49_036540 RRM domain-containing protein ; K12898 heterogeneous
nuclear ribonucleoprotein F/H
Length=513
Score = 174 bits (441), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 80/104 (76%), Positives = 89/104 (85%), Gaps = 0/104 (0%)
Query 134 GWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWR 193
W+ VLRLRG+P++ANEQHIVQFF GFHM AILPSTIPIDGRPSGEAYVQF D +E R
Sbjct 410 NWSAQVLRLRGLPYSANEQHIVQFFHGFHMAAILPSTIPIDGRPSGEAYVQFVDAAEALR 469
Query 194 ALQAKNGARMDRRYIELFPASKQEMTFAAQGGDPRAFRERDRTF 237
A QAKNG RMD+R IELFP+SKQEM FAAQGGDPR FRER+R +
Sbjct 470 AFQAKNGGRMDKRMIELFPSSKQEMEFAAQGGDPRVFRERNRGY 513
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 24/34 (70%), Positives = 31/34 (91%), Gaps = 0/34 (0%)
Query 1 ADAAKEKLHRKYLGRRFVEVYPSTREEMHRAKRP 34
A+ A+E LH+KY+GRRF+EVYP+TRE+M RAKRP
Sbjct 221 AEKAREVLHKKYMGRRFIEVYPATREDMQRAKRP 254
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 38/130 (29%), Positives = 67/130 (51%), Gaps = 10/130 (7%)
Query 96 PSDIYGGYGGATQQAAYTQPPAQVVQSPPVQDVAPIPVGWNYT----VLRLRGMPFNANE 151
P ++ +Q A ++ PA+V SP ++ P + V+RLRG+P++ E
Sbjct 8 PVEVVADLAPEEEQPAPSEVPAEVQASP--EEEKPSNGDERLSKPSFVVRLRGLPWDVQE 65
Query 152 QHIVQFFQ---GFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAKNGARMDRRYI 208
++++ FF+ +L I D R +GEAYVQ D +A++ +G + R+I
Sbjct 66 ENVIAFFKPVVALENDNVL-ICIGFDKRTTGEAYVQLPDPGLREQAIKDLHGRLLGTRWI 124
Query 209 ELFPASKQEM 218
E+F AS++E
Sbjct 125 EVFRASEEEF 134
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 33/89 (37%), Positives = 56/89 (62%), Gaps = 8/89 (8%)
Query 136 NYTVLRLRGMPFNANEQHIVQFFQ---GFHM-TAILPSTIPIDGRPSGEAYVQFS--DVS 189
N V++LRG+P++ +E IV+FF+ GF + + + + DGR SG A+V+ DV+
Sbjct 162 NLNVVKLRGLPWSCSENEIVRFFKAEGGFDIHSDDVVLGVTGDGRLSGIAFVELPSPDVA 221
Query 190 ETWRALQAKNGARMDRRYIELFPASKQEM 218
E R + K M RR+IE++PA++++M
Sbjct 222 EKAREVLHKK--YMGRRFIEVYPATREDM 248
> dre:321556 hnrnph1l, fb19a06, wu:fb19a06, zgc:55399, zgc:85960;
heterogeneous nuclear ribonucleoprotein H1, like; K12898
heterogeneous nuclear ribonucleoprotein F/H
Length=407
Score = 67.8 bits (164), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 36/84 (42%), Positives = 57/84 (67%), Gaps = 8/84 (9%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPS--TIPID--GRPSGEAYVQFSDVSETWRA 194
++RLRG+PF +++ IVQFF G I+P+ T+P+D GR +GEA+VQF+ +A
Sbjct 108 LVRLRGLPFGCSKEEIVQFFSGLE---IVPNGITLPVDFQGRSTGEAFVQFASQDIAEKA 164
Query 195 LQAKNGARMDRRYIELFPASKQEM 218
L+ K+ R+ RYIE+F +S+ E+
Sbjct 165 LK-KHKERIGHRYIEIFKSSRAEV 187
Score = 53.5 bits (127), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 53/94 (56%), Gaps = 4/94 (4%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMT---AILPSTIPIDGRPSGEAYVQFSDVSETWRAL 195
V+R+RG+P++ + + + +FF G ++ + + T +GRPSGEA+V+ + A+
Sbjct 8 VVRVRGLPWSCSAEEVSRFFSGCKISSNGSAIHFTYTREGRPSGEAFVELESEDDLKIAV 67
Query 196 QAKNGARMDRRYIELFPASKQEMTFAAQGGDPRA 229
+ K+ M RY+E+F ++ EM + + P
Sbjct 68 K-KDRESMGHRYVEVFKSNNVEMDWVLKHTGPNC 100
Score = 47.8 bits (112), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 54/208 (25%), Positives = 80/208 (38%), Gaps = 45/208 (21%)
Query 7 KLHRKYLGRRFVEVYPSTREEMHRAKRPGRIIPPDARMMGATAQGPYSGGYAPAPQPYGT 66
K H++ +G R++E++ S+R E+ P R + MG GPY P+
Sbjct 166 KKHKERIGHRYIEIFKSSRAEVRTHYEPPR------KGMGMQRPGPYD---RPS------ 210
Query 67 MATGMGSAQGYGGMDAMGAPQGGYGGYGAPSDIYGGYGGATQQAAYTQPPAQVVQSPPVQ 126
G +GY GM G+ G G YG + T
Sbjct 211 -----GGGRGYNGMSRGGSFDRMRRGGYGGGVSDGRYGDGGNFQSTTG------------ 253
Query 127 DVAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFS 186
+ +RG+P+ A E I FF + + P DGR +GEA V+F+
Sbjct 254 -----------HCVHMRGLPYRATEPDIYNFFSPLNPVRVHIEIGP-DGRVTGEADVEFA 301
Query 187 DVSETWRALQAKNGARMDRRYIELFPAS 214
E A + + A M RY+ELF S
Sbjct 302 -THEDAVAAMSNDKANMQHRYVELFLNS 328
> xla:432071 hnrnph1-a, MGC78776, hnrnph, hnrnph2, hnrph, hnrph1;
heterogeneous nuclear ribonucleoprotein H1 (H); K12898 heterogeneous
nuclear ribonucleoprotein F/H
Length=441
Score = 67.4 bits (163), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 37/87 (42%), Positives = 57/87 (65%), Gaps = 8/87 (9%)
Query 136 NYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPS--TIPID--GRPSGEAYVQFSDVSET 191
N +RLRG+PF +++ IVQFF G I+P+ T+P+D GR +GEA+VQF+
Sbjct 108 NDGFVRLRGLPFGCSKEEIVQFFSGLE---IVPNGITLPVDFQGRSTGEAFVQFASQEIA 164
Query 192 WRALQAKNGARMDRRYIELFPASKQEM 218
+AL+ K+ R+ RYIE+F +S+ E+
Sbjct 165 EKALK-KHKERIGHRYIEIFKSSRAEV 190
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 51/94 (54%), Gaps = 4/94 (4%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI---DGRPSGEAYVQFSDVSETWRAL 195
V+++RG+P++ + I FF + L I +GRPSGEA+V+F + A+
Sbjct 11 VVKVRGLPWSCSHDEIENFFSESKIANGLSGVHFIYTREGRPSGEAFVEFETEDDLQLAV 70
Query 196 QAKNGARMDRRYIELFPASKQEMTFAAQGGDPRA 229
+ K+ A M RY+E+F ++ EM + + P +
Sbjct 71 K-KDRATMAHRYVEVFKSNSVEMDWVLKHTGPNS 103
Score = 48.5 bits (114), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 55/216 (25%), Positives = 85/216 (39%), Gaps = 41/216 (18%)
Query 7 KLHRKYLGRRFVEVYPSTREEMHRAKRPGRIIPPDARMMGATAQGPYS-----GGYAPAP 61
K H++ +G R++E++ S+R E+ P R ++ G GPY GY
Sbjct 169 KKHKERIGHRYIEIFKSSRAEVRTNYDPPR------KLFGMQRPGPYDRPGAGRGYNNLG 222
Query 62 QPYGTMATGMGSAQGYGGMDAMGAPQGGYGG---YGAPSDIYGGYGGATQQAAYTQPPAQ 118
+ + M G G D G + +G +G SD G G Q
Sbjct 223 RGFDRMRRGAYGGGYSGYEDYNGYNEYAFGADQRFGRVSDNRYGDGSTFQSTT------- 275
Query 119 VVQSPPVQDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPS 178
+ +RG+P+ A E I FF + + I DGR +
Sbjct 276 ------------------GHCVHMRGLPYRATETDIYTFFSPLNPVRVHIE-IGADGRVT 316
Query 179 GEAYVQFSDVSETWRALQAKNGARMDRRYIELFPAS 214
GEA V+F+ + A+ +K+ A M RY+ELF S
Sbjct 317 GEADVEFASHEDAVAAM-SKDKANMQHRYVELFLNS 351
> dre:402984 hnrnph1, MGC77712, zgc:77712; heterogeneous nuclear
ribonucleoprotein H1; K12898 heterogeneous nuclear ribonucleoprotein
F/H
Length=403
Score = 67.4 bits (163), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 36/84 (42%), Positives = 57/84 (67%), Gaps = 8/84 (9%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPS--TIPID--GRPSGEAYVQFSDVSETWRA 194
++RLRG+PF +++ IVQFF G I+P+ T+P+D GR +GEA+VQF+ +A
Sbjct 108 LVRLRGLPFGCSKEEIVQFFAGLE---IVPNGITLPVDFQGRSTGEAFVQFASQDIAEKA 164
Query 195 LQAKNGARMDRRYIELFPASKQEM 218
L+ K+ R+ RYIE+F +S+ E+
Sbjct 165 LK-KHKERIGHRYIEIFKSSRAEV 187
Score = 50.4 bits (119), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 50/94 (53%), Gaps = 4/94 (4%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPS---TIPIDGRPSGEAYVQFSDVSETWRAL 195
V+R+RG+P++ + + +FF + + S T +GRPSGEA+V+F + E +
Sbjct 8 VVRVRGLPWSCSVDEVQRFFSECKIASNGTSIHFTYTREGRPSGEAFVEF-ESEEDLKIA 66
Query 196 QAKNGARMDRRYIELFPASKQEMTFAAQGGDPRA 229
K+ M RY+E+F ++ EM + + P
Sbjct 67 VKKDRETMGHRYVEVFKSNSVEMDWVLKHTGPNC 100
Score = 46.6 bits (109), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 28/76 (36%), Positives = 40/76 (52%), Gaps = 2/76 (2%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAK 198
+ +RG+P+ A E I FF + + P DGR +GEA V+F+ E A +K
Sbjct 252 CVHMRGLPYRATETDIYNFFSPLNPVRVHLEIGP-DGRVTGEADVEFA-THEDAVAAMSK 309
Query 199 NGARMDRRYIELFPAS 214
+ A M RY+ELF S
Sbjct 310 DKANMQHRYVELFLNS 325
> hsa:3188 HNRNPH2, FTP3, HNRPH', HNRPH2, hnRNPH'; heterogeneous
nuclear ribonucleoprotein H2 (H'); K12898 heterogeneous nuclear
ribonucleoprotein F/H
Length=449
Score = 67.4 bits (163), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 37/87 (42%), Positives = 57/87 (65%), Gaps = 8/87 (9%)
Query 136 NYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPS--TIPID--GRPSGEAYVQFSDVSET 191
N +RLRG+PF +++ IVQFF G I+P+ T+P+D GR +GEA+VQF+
Sbjct 109 NDGFVRLRGLPFGCSKEEIVQFFSGLE---IVPNGMTLPVDFQGRSTGEAFVQFASQEIA 165
Query 192 WRALQAKNGARMDRRYIELFPASKQEM 218
+AL+ K+ R+ RYIE+F +S+ E+
Sbjct 166 EKALK-KHKERIGHRYIEIFKSSRAEV 191
Score = 50.8 bits (120), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 52/94 (55%), Gaps = 4/94 (4%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHM---TAILPSTIPIDGRPSGEAYVQFSDVSETWRAL 195
V+++RG+P++ + +++FF + T+ + +GRPSGEA+V+ E AL
Sbjct 12 VVKVRGLPWSCSADEVMRFFSDCKIQNGTSGIRFIYTREGRPSGEAFVELESEEEVKLAL 71
Query 196 QAKNGARMDRRYIELFPASKQEMTFAAQGGDPRA 229
+ K+ M RY+E+F ++ EM + + P +
Sbjct 72 K-KDRETMGHRYVEVFKSNSVEMDWVLKHTGPNS 104
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 29/76 (38%), Positives = 40/76 (52%), Gaps = 2/76 (2%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAK 198
+ +RG+P+ A E I FF + + P DGR +GEA V+F+ E A AK
Sbjct 290 CVHMRGLPYRATENDIYNFFSPLNPMRVHIEIGP-DGRVTGEADVEFA-THEDAVAAMAK 347
Query 199 NGARMDRRYIELFPAS 214
+ A M RY+ELF S
Sbjct 348 DKANMQHRYVELFLNS 363
> mmu:56258 Hnrnph2, DXHXS1271E, Ftp-3, Ftp3, H', HNRNP, Hnrph2;
heterogeneous nuclear ribonucleoprotein H2; K12898 heterogeneous
nuclear ribonucleoprotein F/H
Length=449
Score = 67.4 bits (163), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 37/87 (42%), Positives = 57/87 (65%), Gaps = 8/87 (9%)
Query 136 NYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPS--TIPID--GRPSGEAYVQFSDVSET 191
N +RLRG+PF +++ IVQFF G I+P+ T+P+D GR +GEA+VQF+
Sbjct 109 NDGFVRLRGLPFGCSKEEIVQFFSGLE---IVPNGMTLPVDFQGRSTGEAFVQFASQEIA 165
Query 192 WRALQAKNGARMDRRYIELFPASKQEM 218
+AL+ K+ R+ RYIE+F +S+ E+
Sbjct 166 EKALK-KHKERIGHRYIEIFKSSRAEV 191
Score = 51.2 bits (121), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 53/94 (56%), Gaps = 4/94 (4%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHM---TAILPSTIPIDGRPSGEAYVQFSDVSETWRAL 195
V+++RG+P++ + + +++FF + T+ + +GRPSGEA+V+ E AL
Sbjct 12 VVKVRGLPWSCSAEEVMRFFSDCKIQNGTSGVRFIYTREGRPSGEAFVELESEDEVKLAL 71
Query 196 QAKNGARMDRRYIELFPASKQEMTFAAQGGDPRA 229
+ K+ M RY+E+F ++ EM + + P +
Sbjct 72 K-KDRETMGHRYVEVFKSNSVEMDWVLKHTGPNS 104
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 29/76 (38%), Positives = 40/76 (52%), Gaps = 2/76 (2%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAK 198
+ +RG+P+ A E I FF + + P DGR +GEA V+F+ E A AK
Sbjct 290 CVHMRGLPYRATENDIYNFFSPLNPMRVHIEIGP-DGRVTGEADVEFA-THEDAVAAMAK 347
Query 199 NGARMDRRYIELFPAS 214
+ A M RY+ELF S
Sbjct 348 DKANMQHRYVELFLNS 363
> mmu:59013 Hnrnph1, AI642080, E430005G16Rik, Hnrnph, Hnrph1;
heterogeneous nuclear ribonucleoprotein H1; K12898 heterogeneous
nuclear ribonucleoprotein F/H
Length=449
Score = 67.4 bits (163), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 37/87 (42%), Positives = 57/87 (65%), Gaps = 8/87 (9%)
Query 136 NYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPS--TIPID--GRPSGEAYVQFSDVSET 191
N +RLRG+PF +++ IVQFF G I+P+ T+P+D GR +GEA+VQF+
Sbjct 109 NDGFVRLRGLPFGCSKEEIVQFFSGLE---IVPNGITLPVDFQGRSTGEAFVQFASQEIA 165
Query 192 WRALQAKNGARMDRRYIELFPASKQEM 218
+AL+ K+ R+ RYIE+F +S+ E+
Sbjct 166 EKALK-KHKERIGHRYIEIFKSSRAEV 191
Score = 47.4 bits (111), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 49/94 (52%), Gaps = 4/94 (4%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI---DGRPSGEAYVQFSDVSETWRAL 195
V+++RG+P++ + + +FF + I +GRPSGEA+V+ E AL
Sbjct 12 VVKVRGLPWSCSADEVQRFFSDCKIQNGAQGIRFIYTREGRPSGEAFVELESEDEVKLAL 71
Query 196 QAKNGARMDRRYIELFPASKQEMTFAAQGGDPRA 229
+ K+ M RY+E+F ++ EM + + P +
Sbjct 72 K-KDRETMGHRYVEVFKSNNVEMDWVLKHTGPNS 104
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 28/76 (36%), Positives = 40/76 (52%), Gaps = 2/76 (2%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAK 198
+ +RG+P+ A E I FF + + P DGR +GEA V+F+ E A +K
Sbjct 290 CVHMRGLPYRATENDIYNFFSPLNPVRVHIEIGP-DGRVTGEADVEFA-THEDAVAAMSK 347
Query 199 NGARMDRRYIELFPAS 214
+ A M RY+ELF S
Sbjct 348 DKANMQHRYVELFLNS 363
> hsa:3187 HNRNPH1, DKFZp686A15170, HNRPH, HNRPH1, hnRNPH; heterogeneous
nuclear ribonucleoprotein H1 (H); K12898 heterogeneous
nuclear ribonucleoprotein F/H
Length=449
Score = 67.4 bits (163), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 37/87 (42%), Positives = 57/87 (65%), Gaps = 8/87 (9%)
Query 136 NYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPS--TIPID--GRPSGEAYVQFSDVSET 191
N +RLRG+PF +++ IVQFF G I+P+ T+P+D GR +GEA+VQF+
Sbjct 109 NDGFVRLRGLPFGCSKEEIVQFFSGLE---IVPNGITLPVDFQGRSTGEAFVQFASQEIA 165
Query 192 WRALQAKNGARMDRRYIELFPASKQEM 218
+AL+ K+ R+ RYIE+F +S+ E+
Sbjct 166 EKALK-KHKERIGHRYIEIFKSSRAEV 191
Score = 47.4 bits (111), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 49/94 (52%), Gaps = 4/94 (4%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI---DGRPSGEAYVQFSDVSETWRAL 195
V+++RG+P++ + + +FF + I +GRPSGEA+V+ E AL
Sbjct 12 VVKVRGLPWSCSADEVQRFFSDCKIQNGAQGIRFIYTREGRPSGEAFVELESEDEVKLAL 71
Query 196 QAKNGARMDRRYIELFPASKQEMTFAAQGGDPRA 229
+ K+ M RY+E+F ++ EM + + P +
Sbjct 72 K-KDRETMGHRYVEVFKSNNVEMDWVLKHTGPNS 104
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 28/76 (36%), Positives = 40/76 (52%), Gaps = 2/76 (2%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAK 198
+ +RG+P+ A E I FF + + P DGR +GEA V+F+ E A +K
Sbjct 290 CVHMRGLPYRATENDIYNFFSPLNPVRVHIEIGP-DGRVTGEADVEFA-THEDAVAAMSK 347
Query 199 NGARMDRRYIELFPAS 214
+ A M RY+ELF S
Sbjct 348 DKANMQHRYVELFLNS 363
> xla:446758 hnrnph1-b, MGC130700, MGC80081, hnrnph, hnrnph1,
hnrph, hnrph1, hnrph2; heterogeneous nuclear ribonucleoprotein
H1 (H); K12898 heterogeneous nuclear ribonucleoprotein F/H
Length=456
Score = 67.0 bits (162), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 37/87 (42%), Positives = 57/87 (65%), Gaps = 8/87 (9%)
Query 136 NYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPS--TIPID--GRPSGEAYVQFSDVSET 191
N +RLRG+PF +++ IVQFF G I+P+ T+P+D GR +GEA+VQF+
Sbjct 108 NDGFVRLRGLPFGCSKEEIVQFFSGLE---IVPNGITLPVDFQGRSTGEAFVQFASQEIA 164
Query 192 WRALQAKNGARMDRRYIELFPASKQEM 218
+AL+ K+ R+ RYIE+F +S+ E+
Sbjct 165 EKALK-KHKERIGHRYIEIFKSSRAEV 190
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 50/94 (53%), Gaps = 4/94 (4%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI---DGRPSGEAYVQFSDVSETWRAL 195
V+++RG+P++ + I FF + L I +GRPSGEA+V+F + E +
Sbjct 11 VVKVRGLPWSCSHDEIENFFSECKIANGLSGIHFIYTREGRPSGEAFVEF-ETEEDLKLG 69
Query 196 QAKNGARMDRRYIELFPASKQEMTFAAQGGDPRA 229
K+ A M RY+E+F ++ EM + + P +
Sbjct 70 LKKDRATMGHRYVEVFKSNNVEMDWVLKHTGPNS 103
Score = 48.5 bits (114), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 57/216 (26%), Positives = 86/216 (39%), Gaps = 41/216 (18%)
Query 7 KLHRKYLGRRFVEVYPSTREEMHRAKRPGRIIPPDARMMGATAQGPYS-----GGYAPAP 61
K H++ +G R++E++ S+R E+ P R ++ G GPY GY
Sbjct 169 KKHKERIGHRYIEIFKSSRAEVRTHYDPPR------KLFGMQRPGPYDRPGAGRGYNNLG 222
Query 62 QPYGTMATGMGSAQGYGGMDAMGAPQGGYGG---YGAPSDIYGGYGGATQQAAYTQPPAQ 118
+ + M G G D G + +G +G SD YG T + T
Sbjct 223 RGFDRMRRGAYGGGYSGYEDYNGYNEYAFGTDQRFGRVSD--SRYGDGTSFQSTTGH--- 277
Query 119 VVQSPPVQDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPS 178
+ +RG+P+ A E I FF + + I DGR +
Sbjct 278 --------------------CVHMRGLPYRATETDIYTFFSPLNPVRVHIE-IGADGRVT 316
Query 179 GEAYVQFSDVSETWRALQAKNGARMDRRYIELFPAS 214
GEA V+F+ E A +K+ A M RY+ELF S
Sbjct 317 GEADVEFA-THEDAVAAMSKDKANMQHRYVELFLNS 351
> hsa:3185 HNRNPF, HNRPF, MGC110997, OK/SW-cl.23, mcs94-1; heterogeneous
nuclear ribonucleoprotein F; K12898 heterogeneous
nuclear ribonucleoprotein F/H
Length=415
Score = 65.9 bits (159), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 36/87 (41%), Positives = 56/87 (64%), Gaps = 8/87 (9%)
Query 136 NYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPS--TIPID--GRPSGEAYVQFSDVSET 191
N +RLRG+PF ++ IVQFF G I+P+ T+P+D G+ +GEA+VQF+
Sbjct 109 NDGFVRLRGLPFGCTKEEIVQFFSGLE---IVPNGITLPVDPEGKITGEAFVQFASQELA 165
Query 192 WRALQAKNGARMDRRYIELFPASKQEM 218
+AL K+ R+ RYIE+F +S++E+
Sbjct 166 EKAL-GKHKERIGHRYIEVFKSSQEEV 191
Score = 61.2 bits (147), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 62/212 (29%), Positives = 94/212 (44%), Gaps = 26/212 (12%)
Query 9 HRKYLGRRFVEVYPSTREEMHRAKRPGRIIPPDARMMGATAQGPYSGGYAPAPQPYGTMA 68
H++ +G R++EV+ S++EE+ P + M GPY +P GT
Sbjct 172 HKERIGHRYIEVFKSSQEEVRSYS------DPPLKFMSVQRPGPYD-------RP-GTAR 217
Query 69 TGMGSAQGYGGMDAM--GAPQGGYGGYGAPSDIYGGYGGAT----QQAAYTQPPAQVVQS 122
+G + G++ M GA GYGGY S + GYG T + +Y +
Sbjct 218 RYIGIVK-QAGLERMRPGAYSTGYGGYEEYSGLSDGYGFTTDLFGRDLSYCLSGMYDHRY 276
Query 123 PPVQDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAY 182
+ G + +RG+P+ A E I FF + + P DGR +GEA
Sbjct 277 GDSEFTVQSTTGH---CVHMRGLPYKATENDIYNFFSPLNPVRVHIEIGP-DGRVTGEAD 332
Query 183 VQFSDVSETWRALQAKNGARMDRRYIELFPAS 214
V+F+ E A+ +K+ A M RYIELF S
Sbjct 333 VEFATHEEAVAAM-SKDRANMQHRYIELFLNS 363
Score = 45.4 bits (106), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 50/101 (49%), Gaps = 4/101 (3%)
Query 132 PVGWNYTVLRLRGMPFNANEQHIVQFFQGFHM---TAILPSTIPIDGRPSGEAYVQFSDV 188
P G V++LRG+P++ + + + F + A + +GR SGEA+V+
Sbjct 5 PEGGEGFVVKLRGLPWSCSVEDVQNFLSDCTIHDGAAGVHFIYTREGRQSGEAFVELGSE 64
Query 189 SETWRALQAKNGARMDRRYIELFPASKQEMTFAAQGGDPRA 229
+ AL+ K+ M RYIE+F + + EM + + P +
Sbjct 65 DDVKMALK-KDRESMGHRYIEVFKSHRTEMDWVLKHSGPNS 104
> mmu:98758 Hnrnpf, 4833420I20Rik, AA407306, Hnrpf, MGC101981,
MGC36971; heterogeneous nuclear ribonucleoprotein F; K12898
heterogeneous nuclear ribonucleoprotein F/H
Length=415
Score = 65.9 bits (159), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 36/87 (41%), Positives = 56/87 (64%), Gaps = 8/87 (9%)
Query 136 NYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPS--TIPID--GRPSGEAYVQFSDVSET 191
N +RLRG+PF ++ IVQFF G I+P+ T+P+D G+ +GEA+VQF+
Sbjct 109 NDGFVRLRGLPFGCTKEEIVQFFSGLE---IVPNGITLPVDPEGKITGEAFVQFASQELA 165
Query 192 WRALQAKNGARMDRRYIELFPASKQEM 218
+AL K+ R+ RYIE+F +S++E+
Sbjct 166 EKAL-GKHKERIGHRYIEVFKSSQEEV 191
Score = 63.9 bits (154), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 63/212 (29%), Positives = 94/212 (44%), Gaps = 26/212 (12%)
Query 9 HRKYLGRRFVEVYPSTREEMHRAKRPGRIIPPDARMMGATAQGPYSGGYAPAPQPYGTMA 68
H++ +G R++EV+ S++EE+ P + M GPY +P GT
Sbjct 172 HKERIGHRYIEVFKSSQEEVRSYS------DPPLKFMSVQRPGPYD-------RP-GTAR 217
Query 69 TGMGSAQGYGGMDAM--GAPQGGYGGYGAPSDIYGGYGGAT----QQAAYTQPPAQVVQS 122
+G + G+D M GA GYGGY S + GYG T + +Y +
Sbjct 218 RYIGIVK-QAGLDRMRSGAYSAGYGGYEEYSGLSDGYGFTTDLFGRDLSYCLSGMYDHRY 276
Query 123 PPVQDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAY 182
+ G + +RG+P+ A E I FF + + P DGR +GEA
Sbjct 277 GDSEFTVQSTTGH---CVHMRGLPYKATENDIYNFFSPLNPVRVHIEIGP-DGRVTGEAD 332
Query 183 VQFSDVSETWRALQAKNGARMDRRYIELFPAS 214
V+F+ E A+ +K+ A M RYIELF S
Sbjct 333 VEFATHEEAVAAM-SKDRANMQHRYIELFLNS 363
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 50/101 (49%), Gaps = 4/101 (3%)
Query 132 PVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI---DGRPSGEAYVQFSDV 188
P G V++LRG+P++ + + + F + + I +GR SGEA+V+
Sbjct 5 PEGGEGYVVKLRGLPWSCSIEDVQNFLSDCTIHDGVAGVHFIYTREGRQSGEAFVELESE 64
Query 189 SETWRALQAKNGARMDRRYIELFPASKQEMTFAAQGGDPRA 229
+ AL+ K+ M RYIE+F + + EM + + P +
Sbjct 65 DDVKLALK-KDRESMGHRYIEVFKSHRTEMDWVLKHSGPNS 104
> cel:Y73B6BL.33 hypothetical protein; K12898 heterogeneous nuclear
ribonucleoprotein F/H
Length=610
Score = 65.5 bits (158), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 31/81 (38%), Positives = 47/81 (58%), Gaps = 3/81 (3%)
Query 140 LRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAKN 199
+RLRG+PFNA E+ I +FF G + + + GRP+GEAYV+F + E +A++ +
Sbjct 66 IRLRGLPFNATEKDIHEFFAGLTIERV--KFVCTTGRPNGEAYVEFKNTEEAGKAME-ND 122
Query 200 GARMDRRYIELFPASKQEMTF 220
+ RYIE+F E F
Sbjct 123 RKEISNRYIEVFTVEADEAEF 143
Score = 54.7 bits (130), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 34/93 (36%), Positives = 50/93 (53%), Gaps = 10/93 (10%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDG------RPSGEAYVQFSDVSETW 192
V+RLRG+P++ E + +FF+G P+ I I G RPSGEA+V+F+
Sbjct 158 VIRLRGVPWSCKEDDVRKFFEGLEPP---PAEIVIGGTGGPRSRPSGEAFVRFTTQDAAE 214
Query 193 RALQAKNGARMDRRYIELFPASKQEMTFAAQGG 225
+A+ N M RY+E+F +S E A GG
Sbjct 215 KAMDYNN-RHMGSRYVEVFMSSMVEFNRAKGGG 246
> mmu:432467 Hnrnph3, AA693301, AI666703, Hnrph3; heterogeneous
nuclear ribonucleoprotein H3; K12898 heterogeneous nuclear
ribonucleoprotein F/H
Length=346
Score = 63.2 bits (152), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 35/84 (41%), Positives = 52/84 (61%), Gaps = 8/84 (9%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI----DGRPSGEAYVQFSDVSETWRA 194
+RLRG+PF +++ IVQFFQG I+P+ I + GR +GEA+VQF+ A
Sbjct 17 TVRLRGLPFGCSKEEIVQFFQGL---EIVPNGITLTMDYQGRSTGEAFVQFASKEIAENA 73
Query 195 LQAKNGARMDRRYIELFPASKQEM 218
L K+ R+ RYIE+F +S+ E+
Sbjct 74 L-GKHKERIGHRYIEIFRSSRSEI 96
> hsa:3189 HNRNPH3, 2H9, FLJ34092, HNRPH3; heterogeneous nuclear
ribonucleoprotein H3 (2H9); K12898 heterogeneous nuclear
ribonucleoprotein F/H
Length=346
Score = 63.2 bits (152), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 35/84 (41%), Positives = 52/84 (61%), Gaps = 8/84 (9%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI----DGRPSGEAYVQFSDVSETWRA 194
+RLRG+PF +++ IVQFFQG I+P+ I + GR +GEA+VQF+ A
Sbjct 17 TVRLRGLPFGCSKEEIVQFFQGL---EIVPNGITLTMDYQGRSTGEAFVQFASKEIAENA 73
Query 195 LQAKNGARMDRRYIELFPASKQEM 218
L K+ R+ RYIE+F +S+ E+
Sbjct 74 L-GKHKERIGHRYIEIFRSSRSEI 96
> ath:AT5G66010 RNA binding / nucleic acid binding / nucleotide
binding
Length=255
Score = 63.2 bits (152), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 42/128 (32%), Positives = 67/128 (52%), Gaps = 7/128 (5%)
Query 93 YGAPSDIYGGYG---GATQQAAYTQPPAQVVQSPPVQDVAPIPVGWNYTVLRLRGMPFNA 149
YG+ ++G G G+ +Q P V + P P + V+RLRG+PFN
Sbjct 2 YGSRGAMFGSGGYEVGSKRQRMMQSNPYLAVGTGPT-SFPPFGYAGGFPVVRLRGLPFNC 60
Query 150 NEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAKNGARMDRRYIE 209
+ I +FF G ++ +L + +G+ SGEA+V F+ + ALQ ++ M RRY+E
Sbjct 61 ADIDIFEFFAGLNIVDVL--LVSKNGKFSGEAFVVFAGPMQVEIALQ-RDRHNMGRRYVE 117
Query 210 LFPASKQE 217
+F SKQ+
Sbjct 118 VFRCSKQD 125
Score = 58.2 bits (139), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 30/82 (36%), Positives = 50/82 (60%), Gaps = 3/82 (3%)
Query 135 WNYT-VLRLRGMPFNANEQHIVQFFQGFHMT-AILPSTIPIDGRPSGEAYVQFSDVSETW 192
YT VL++RG+P++ N+ I++FF G+ + + DG+ +GEA+V+F E
Sbjct 162 LEYTEVLKMRGLPYSVNKPQIIEFFSGYKVIQGRVQVVCRPDGKATGEAFVEFETGEEAR 221
Query 193 RALQAKNGARMDRRYIELFPAS 214
RA+ AK+ + RY+ELFP +
Sbjct 222 RAM-AKDKMSIGSRYVELFPTT 242
> xla:494658 hnrnph3, hnrph3; heterogeneous nuclear ribonucleoprotein
H3 (2H9); K12898 heterogeneous nuclear ribonucleoprotein
F/H
Length=342
Score = 62.8 bits (151), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 39/104 (37%), Positives = 59/104 (56%), Gaps = 12/104 (11%)
Query 119 VVQSPPVQDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPS--TIPID-- 174
V++ DV G +RLRG+PF +++ IVQFF G I+P+ T+ +D
Sbjct 19 VLKHNSTDDVETDSDG----TVRLRGLPFGCSKEEIVQFFSGLR---IVPNGITLTVDYQ 71
Query 175 GRPSGEAYVQFSDVSETWRALQAKNGARMDRRYIELFPASKQEM 218
GR +GEA+VQF+ AL K+ R+ RYIE+F +S+ E+
Sbjct 72 GRSTGEAFVQFASKEIAENAL-GKHKERIGHRYIEIFKSSRGEI 114
Score = 47.8 bits (112), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 42/79 (53%), Gaps = 10/79 (12%)
Query 140 LRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI----DGRPSGEAYVQFSDVSETWRAL 195
+ +RG+PF A+E I FF + + P + I DGR +GEA V+F+ E A
Sbjct 213 VHMRGLPFRASESDIANFF-----SPLTPIRVHIDVGADGRATGEADVEFA-THEDAVAA 266
Query 196 QAKNGARMDRRYIELFPAS 214
+K+ M RYIELF S
Sbjct 267 MSKDKNNMQHRYIELFLNS 285
> dre:572992 esrp2, FLJ20171, MGC55721, MGC77254, cb404, fa07a06,
rbm35b, sb:cb404, zgc:77254; epithelial splicing regulatory
protein 2; K14947 epithelial splicing regulatory protein
1/2
Length=736
Score = 62.0 bits (149), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/110 (31%), Positives = 55/110 (50%), Gaps = 10/110 (9%)
Query 120 VQSPPVQDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI----DG 175
+ +PP+ I G +RLRG+P+ A + I++F G H I P + + G
Sbjct 437 LATPPL-----ITTGNTRDCIRLRGLPYTAAIEDILEFM-GEHTIDIKPHGVHMVLNQQG 490
Query 176 RPSGEAYVQFSDVSETWRALQAKNGARMDRRYIELFPASKQEMTFAAQGG 225
RPSG+A++Q + Q + M RY+E+F S +EM+F GG
Sbjct 491 RPSGDAFIQMKSADRAFMVAQKCHKKMMKDRYVEVFQCSTEEMSFVLMGG 540
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 29/88 (32%), Positives = 50/88 (56%), Gaps = 2/88 (2%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMTA-ILPSTIPIDGRPSGEAYVQFSDVSETWRALQ 196
TV+R RG+P+ +++Q I +FF+G ++ + + GR +GEA V+F + AL
Sbjct 224 TVIRARGLPWQSSDQDIARFFKGLNIAKGGVALCLNAQGRRNGEALVRFINSEHRDMALD 283
Query 197 AKNGARMDRRYIELFPASKQEMTFAAQG 224
++ M RYIE++ A+ +E A G
Sbjct 284 -RHKHHMGSRYIEVYKATGEEFLKIAGG 310
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/100 (32%), Positives = 51/100 (51%), Gaps = 11/100 (11%)
Query 126 QDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFF-------QGFHMTAILPSTIPIDGRPS 178
+VA N ++R+RG+PF A Q ++ F G +L P DGRP+
Sbjct 313 NEVAQFLSKENQMIIRMRGLPFTATPQDVLGFLGPECPVTDG--TEGLLFVKYP-DGRPT 369
Query 179 GEAYVQFSDVSETWRALQAKNGARMDRRYIELFPASKQEM 218
G+A+V F+ AL+ K+ + +RYIELF ++ E+
Sbjct 370 GDAFVLFACEEYAQNALK-KHKQILGKRYIELFRSTAAEV 408
Score = 38.9 bits (89), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 54/231 (23%), Positives = 88/231 (38%), Gaps = 65/231 (28%)
Query 6 EKLHRKYLGRRFVEVYPSTREEMHRAKRPGRI-----IPPDARM---------------- 44
+K H+K + R+VEV+ + EEM G + PP ++
Sbjct 511 QKCHKKMMKDRYVEVFQCSTEEMSFVLMGGTLNRSGLSPPPCKLPCLSPPTYAAFPAAPA 570
Query 45 ------------MGATAQGPYSGGYAPAPQPYGTMATGMGSAQGYGGMDAMGAPQGGYGG 92
+ AT + P + ++PAP A S Q Y M+ Y
Sbjct 571 MLPTEAGLYQPPLLATPRTPQAPTHSPAP------AFAYYSPQLYMNMNM------SYTT 618
Query 93 Y-----GAPSDIYGGYGGATQQAAYTQPPAQVVQSPPVQDVAPIPVGWNYTVLRLRGMPF 147
Y +PS + + + PP V + Q I ++R++G+P+
Sbjct 619 YYPSPPVSPSTV----------SYFAAPPGSVAAAVAAQPTPAILPPQPGALVRMQGLPY 668
Query 148 NANEQHIVQFFQGFHMTAILPSTIPI---DGRPSGEAYVQFSDVSETWRAL 195
NA + I+ FFQG+ + A S + + G+ SGEA V F RA+
Sbjct 669 NAGVKDILSFFQGYQLQA--DSVLVLYNWSGQRSGEALVTFPSEKAARRAV 717
> hsa:2926 GRSF1, FLJ13125; G-rich RNA sequence binding factor
1
Length=318
Score = 62.0 bits (149), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 40/104 (38%), Positives = 57/104 (54%), Gaps = 17/104 (16%)
Query 118 QVVQSPPVQDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPID--- 174
QV SP V D V+RLRG+P++ NE+ IV FF G ++ I T +D
Sbjct 78 QVKSSPVVND----------GVVRLRGLPYSCNEKDIVDFFAGLNIVDI---TFVMDYRG 124
Query 175 GRPSGEAYVQFSDVSETWRALQAKNGARMDRRYIELFPASKQEM 218
R +GEAYVQF + +AL K+ + RYIE+FP+ + E+
Sbjct 125 RRKTGEAYVQFEEPEMANQAL-LKHREEIGNRYIEIFPSRRNEV 167
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 51/210 (24%), Positives = 82/210 (39%), Gaps = 48/210 (22%)
Query 9 HRKYLGRRFVEVYPSTREEMHRAKRPGRIIPPDARMMGATAQGPYSGGYAPAPQPYGTMA 68
HR+ +G R++E++PS R E+ T G Y G A P
Sbjct 148 HREEIGNRYIEIFPSRRNEVR------------------THVGSYKGK-KIASFPTAKYI 188
Query 69 TGMGSAQGYGGMDAMGAPQGGYGGYGAPSDIYGGYGGATQQAAYTQPPAQVVQSPPVQDV 128
T P+ + + DI +++ + P +V + P +
Sbjct 189 T---------------EPEMVFEEHEVNEDIQPMTAFESEKE--IELPKEVPEKLP--EA 229
Query 129 APIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPID----GRPSGEAYVQ 184
A + + +RG+PF AN Q I+ FF + P I ++ G+ +GEA V
Sbjct 230 ADFGTTSSLHFVHMRGLPFQANAQDIINFF-----APLKPVRITMEYSSSGKATGEADVH 284
Query 185 FSDVSETWRALQAKNGARMDRRYIELFPAS 214
F + E A K+ + + RYIELF S
Sbjct 285 F-ETHEDAVAAMLKDRSHVHHRYIELFLNS 313
> mmu:231413 Grsf1, B130010H02, BB232551, C80280, D5Wsu31e; G-rich
RNA sequence binding factor 1
Length=362
Score = 61.2 bits (147), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 38/94 (40%), Positives = 56/94 (59%), Gaps = 8/94 (8%)
Query 128 VAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPID---GRPSGEAYVQ 184
V P PV + V+RLRG+P++ NE+ IV FF G ++ I T +D R +GEAYVQ
Sbjct 123 VKPSPV-LSDGVVRLRGLPYSCNEKDIVDFFAGLNIVDI---TFVMDYRGRRKTGEAYVQ 178
Query 185 FSDVSETWRALQAKNGARMDRRYIELFPASKQEM 218
F + +AL K+ + RYIE+FP+ + E+
Sbjct 179 FEEPEMANQAL-LKHREEIGNRYIEIFPSRRNEV 211
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 49/210 (23%), Positives = 79/210 (37%), Gaps = 48/210 (22%)
Query 9 HRKYLGRRFVEVYPSTREEMHRAKRPGRIIPPDARMMGATAQGPYSGGYAPAPQPYGTMA 68
HR+ +G R++E++PS R E+ T G + G + P +
Sbjct 192 HREEIGNRYIEIFPSRRNEVR------------------THVGSHKGKKMTSSPPTKYIT 233
Query 69 TGMGSAQGYGGMDAMGAPQGGYGGYGAPSDIYGGYGGATQQAAYTQPPAQVVQSPPVQDV 128
P+ + + DI + + P + P D
Sbjct 234 ----------------EPEVVFEEHEVNEDIRPMTAFESDKEI-ELPKEMSEKLPEAVDF 276
Query 129 APIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPID----GRPSGEAYVQ 184
+P + + +RG+PF AN Q I+ FF + P I ++ G+ +GEA V
Sbjct 277 GTLP---SLHFVHMRGLPFQANAQDIINFF-----APLKPVRITMEYSSSGKATGEADVH 328
Query 185 FSDVSETWRALQAKNGARMDRRYIELFPAS 214
F D E A K+ + + RYIELF S
Sbjct 329 F-DTHEDAVAAMLKDRSHVQHRYIELFLNS 357
> pfa:PF10_0235 RNA binding protein, putative
Length=160
Score = 60.1 bits (144), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 33/104 (31%), Positives = 54/104 (51%), Gaps = 3/104 (2%)
Query 111 AYTQPPAQVVQSPPVQDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTA-ILPS 169
+++ P +++ + I N L+LRG+PF+A+E+ I FF+ F +T P
Sbjct 28 SFSSSPQKIMSVRNFSEYINIEEKINLPRLKLRGLPFDASEEEIKNFFRDFQLTKQAYPI 87
Query 170 TIP--IDGRPSGEAYVQFSDVSETWRALQAKNGARMDRRYIELF 211
I I +P+G AYV F D E A QA N + R++E++
Sbjct 88 HIIKGIKNKPTGHAYVYFDDEEEARNACQAMNRKYIRDRFVEIY 131
> mmu:77411 Esrp2, 9530027K23Rik, Rbm35b; epithelial splicing
regulatory protein 2; K14947 epithelial splicing regulatory
protein 1/2
Length=717
Score = 58.5 bits (140), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 48/105 (45%), Gaps = 15/105 (14%)
Query 130 PIPVGWNYTVLRLRGMPFNANEQHIVQFF---------QGFHMTAILPSTIPIDGRPSGE 180
P+ G +RLRG+P+ A + I+ F G HM + GRPSG+
Sbjct 457 PLAGGTGRDCVRLRGLPYTATIEDILSFLGEAAADIRPHGVHMV------LNQQGRPSGD 510
Query 181 AYVQFSDVSETWRALQAKNGARMDRRYIELFPASKQEMTFAAQGG 225
A++Q V A Q + M RY+E+ P S +EM+ GG
Sbjct 511 AFIQMMSVERALAAAQRCHKKMMKERYVEVVPCSTEEMSRVLMGG 555
Score = 53.1 bits (126), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 32/101 (31%), Positives = 55/101 (54%), Gaps = 2/101 (1%)
Query 136 NYTVLRLRGMPFNANEQHIVQFFQGFHMT-AILPSTIPIDGRPSGEAYVQFSDVSETWRA 194
N TV+R RG+P+ +++Q + +FF+G ++ + + GR +GEA ++F D + A
Sbjct 245 NETVVRARGLPWQSSDQDVARFFKGLNIARGGVALCLNAQGRRNGEALIRFVDSEQRDLA 304
Query 195 LQAKNGARMDRRYIELFPASKQEMTFAAQGGDPRAFRERDR 235
LQ ++ M RYIE++ A+ +E A G R R
Sbjct 305 LQ-RHKHHMGVRYIEVYKATGEEFVKIAGGTSLEVARFLSR 344
Score = 40.8 bits (94), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 46/96 (47%), Gaps = 23/96 (23%)
Query 136 NYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI-------------DGRPSGEAY 182
+ +LRLRG+PF+A ++ F L P+ DGRP+G+A+
Sbjct 346 DQVILRLRGLPFSAGPTDVLGF---------LGPECPVTGGADGLLFVRHPDGRPTGDAF 396
Query 183 VQFSDVSETWRALQAKNGARMDRRYIELFPASKQEM 218
F+ E +A ++ + +RYIELF ++ E+
Sbjct 397 ALFA-CEELAQAALRRHKGMLGKRYIELFRSTAAEV 431
Score = 38.5 bits (88), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 42/177 (23%), Positives = 69/177 (38%), Gaps = 22/177 (12%)
Query 1 ADAAKEKLHRKYLGRRFVEVYPSTREEMHRAKRPGR-----IIPPDARMMGATAQGPYSG 55
A AA ++ H+K + R+VEV P + EEM R G + PP ++ + +
Sbjct 521 ALAAAQRCHKKMMKERYVEVVPCSTEEMSRVLMGGSLSRSGLSPPPCKLPCLSPPTYATF 580
Query 56 GYAPAPQPYGTMATGMGSAQGYGG-MDAMGAPQGGYGGYGAPSDIYGGYGG--------A 106
PA P T A SA + A P Y G + +Y Y
Sbjct 581 QATPALIPTETTALYPSSALLPAARVPAAATPLAYYPGPA--TQLYMNYTAYYPSPPVSP 638
Query 107 TQQAAYTQPPAQVVQSPPVQDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHM 163
T T PP + +P P ++R++G+P+ A + ++ FQ + +
Sbjct 639 TTVGYLTTPPTALASTPTTMLSQP------GALVRMQGVPYTAGMKDLLSVFQAYQL 689
> hsa:54845 ESRP1, FLJ20171, RBM35A, RMB35A; epithelial splicing
regulatory protein 1; K14947 epithelial splicing regulatory
protein 1/2
Length=677
Score = 58.5 bits (140), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 31/96 (32%), Positives = 45/96 (46%), Gaps = 15/96 (15%)
Query 139 VLRLRGMPFNANEQHIVQFF---------QGFHMTAILPSTIPIDGRPSGEAYVQFSDVS 189
+RLRG+P+ A + I+ F G HM + GRPSG+A++Q
Sbjct 446 CIRLRGLPYAATIEDILDFLGEFATDIRTHGVHMV------LNHQGRPSGDAFIQMKSAD 499
Query 190 ETWRALQAKNGARMDRRYIELFPASKQEMTFAAQGG 225
+ A Q + M RY+E+F S +EM F GG
Sbjct 500 RAFMAAQKCHKKNMKDRYVEVFQCSAEEMNFVLMGG 535
Score = 54.7 bits (130), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 36/99 (36%), Positives = 54/99 (54%), Gaps = 9/99 (9%)
Query 126 QDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHM------TAILPSTIPIDGRPSG 179
+VA N ++R+RG+PF A + +V FF G H IL T P DGRP+G
Sbjct 314 NEVAQFLSKENQVIVRMRGLPFTATAEEVVAFF-GQHCPITGGKEGILFVTYP-DGRPTG 371
Query 180 EAYVQFSDVSETWRALQAKNGARMDRRYIELFPASKQEM 218
+A+V F+ AL+ K+ + +RYIELF ++ E+
Sbjct 372 DAFVLFACEEYAQNALR-KHKDLLGKRYIELFRSTAAEV 409
Score = 47.4 bits (111), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 33/91 (36%), Positives = 53/91 (58%), Gaps = 8/91 (8%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMT---AILPSTIPIDGRPSGEAYVQFSDVSETWRA 194
TV+R RG+P+ +++Q I +FF+G ++ A L + GR +GEA V+F VSE R
Sbjct 225 TVVRARGLPWQSSDQDIARFFKGLNIAKGGAAL--CLNAQGRRNGEALVRF--VSEEHRD 280
Query 195 LQA-KNGARMDRRYIELFPASKQEMTFAAQG 224
L ++ M RYIE++ A+ ++ A G
Sbjct 281 LALQRHKHHMGTRYIEVYKATGEDFLKIAGG 311
Score = 33.1 bits (74), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 40/167 (23%), Positives = 65/167 (38%), Gaps = 25/167 (14%)
Query 4 AKEKLHRKYLGRRFVEVYPSTREEMHRAKRPGRIIPPDARMMGATAQGPYSGGYAPAPQP 63
A +K H+K + R+VEV+ + EEM+ G + + Y+ P P
Sbjct 504 AAQKCHKKNMKDRYVEVFQCSAEEMNFVLMGGTLNRNGLSPPPCLSPPSYT-----FPAP 558
Query 64 YGTMATGMGSAQGYGGMDAMGAPQGGYGGYGAPSDIYGGYGGATQQAAYTQPPAQVVQSP 123
+ T Q ++ A Q Y A + ++ Y A Y PP SP
Sbjct 559 AAVIPTEAAIYQPSVILNPR-ALQPSTAYYPAGTQLFMNY-----TAYYPSPPG----SP 608
Query 124 PVQDVAPIPVGWNY--------TVLRLRGMPFNANEQHIVQFFQGFH 162
+ P N TV+R++G+ +N + I+ FFQG+
Sbjct 609 --NSLGYFPTAANLSGVPPQPGTVVRMQGLAYNTGVKEILNFFQGYQ 653
> xla:379211 esrp1, MGC53361, rbm35a; epithelial splicing regulatory
protein 1; K14947 epithelial splicing regulatory protein
1/2
Length=688
Score = 58.2 bits (139), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 46/96 (47%), Gaps = 15/96 (15%)
Query 139 VLRLRGMPFNANEQHIVQFF---------QGFHMTAILPSTIPIDGRPSGEAYVQFSDVS 189
+RLRG+P+ A + I++F G HM + GRPSG++++Q
Sbjct 447 CIRLRGLPYAATIEDILEFLGEFSADIRTHGVHMV------LNHQGRPSGDSFIQMKSAD 500
Query 190 ETWRALQAKNGARMDRRYIELFPASKQEMTFAAQGG 225
+ A Q + M RY+E+F S +EM F GG
Sbjct 501 RAYLAAQKCHKKTMKDRYVEVFQCSAEEMNFVLMGG 536
Score = 51.2 bits (121), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 34/98 (34%), Positives = 54/98 (55%), Gaps = 7/98 (7%)
Query 126 QDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFF-QGFHMT----AILPSTIPIDGRPSGE 180
+VA N ++R+RG+PF A + ++ FF Q +T IL T P D RP+G+
Sbjct 315 NEVAQFLSKENQVIVRMRGLPFTATAEEVLAFFGQQCPVTGGKEGILFVTYP-DNRPTGD 373
Query 181 AYVQFSDVSETWRALQAKNGARMDRRYIELFPASKQEM 218
A+V F+ AL+ K+ + +RYIELF ++ E+
Sbjct 374 AFVLFACEEYAQNALK-KHKELLGKRYIELFRSTAAEV 410
Score = 47.4 bits (111), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 32/91 (35%), Positives = 53/91 (58%), Gaps = 8/91 (8%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMT---AILPSTIPIDGRPSGEAYVQFSDVSETWRA 194
T++R RG+P+ +++Q I +FF+G ++ A L + GR +GEA V+F VSE R
Sbjct 226 TIIRARGLPWQSSDQDIARFFKGLNIAKGGAAL--CLNAQGRRNGEALVRF--VSEEHRD 281
Query 195 LQA-KNGARMDRRYIELFPASKQEMTFAAQG 224
L ++ M RYIE++ A+ ++ A G
Sbjct 282 LALQRHKHHMGNRYIEVYKATGEDFLKIAGG 312
> mmu:207920 Esrp1, 2210008M09Rik, A630065D16, BC031468, MGC25805,
Rbm35a; epithelial splicing regulatory protein 1; K14947
epithelial splicing regulatory protein 1/2
Length=681
Score = 57.8 bits (138), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 31/96 (32%), Positives = 45/96 (46%), Gaps = 15/96 (15%)
Query 139 VLRLRGMPFNANEQHIVQFF---------QGFHMTAILPSTIPIDGRPSGEAYVQFSDVS 189
+RLRG+P+ A + I+ F G HM + GRPSG+A++Q
Sbjct 446 CIRLRGLPYAATIEDILDFLGEFSTDIRTHGVHMV------LNHQGRPSGDAFIQMKSTD 499
Query 190 ETWRALQAKNGARMDRRYIELFPASKQEMTFAAQGG 225
+ A Q + M RY+E+F S +EM F GG
Sbjct 500 RAFMAAQKYHKKTMKDRYVEVFQCSAEEMNFVLMGG 535
Score = 54.7 bits (130), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 36/99 (36%), Positives = 54/99 (54%), Gaps = 9/99 (9%)
Query 126 QDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHM------TAILPSTIPIDGRPSG 179
+VA N ++R+RG+PF A + +V FF G H IL T P DGRP+G
Sbjct 314 NEVAQFLSKENQVIVRMRGLPFTATAEEVVAFF-GQHCPITGGKEGILFVTYP-DGRPTG 371
Query 180 EAYVQFSDVSETWRALQAKNGARMDRRYIELFPASKQEM 218
+A+V F+ AL+ K+ + +RYIELF ++ E+
Sbjct 372 DAFVLFACEEYAQNALR-KHKELLGKRYIELFRSTAAEV 409
Score = 47.4 bits (111), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 33/91 (36%), Positives = 53/91 (58%), Gaps = 8/91 (8%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMT---AILPSTIPIDGRPSGEAYVQFSDVSETWRA 194
TV+R RG+P+ +++Q I +FF+G ++ A L + GR +GEA V+F VSE R
Sbjct 225 TVVRARGLPWQSSDQDIARFFKGLNIAKGGAAL--CLNAQGRRNGEALVRF--VSEEHRD 280
Query 195 LQA-KNGARMDRRYIELFPASKQEMTFAAQG 224
L ++ M RYIE++ A+ ++ A G
Sbjct 281 LALQRHKHHMGTRYIEVYKATGEDFLKIAGG 311
Score = 33.1 bits (74), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 43/172 (25%), Positives = 69/172 (40%), Gaps = 31/172 (18%)
Query 4 AKEKLHRKYLGRRFVEVYPSTREEMHRAKRPGRI-----IPPDARMMGATAQGPYSGGYA 58
A +K H+K + R+VEV+ + EEM+ G + PP ++ P S +
Sbjct 504 AAQKYHKKTMKDRYVEVFQCSAEEMNFVLMGGTLNRNGLSPPPCKL---PCLSPPSYTF- 559
Query 59 PAPQPYGTMATGMGSAQGYGGMDAMGAPQGGYGGYGAPSDIYGGYGGATQQAAYTQPPAQ 118
P P + T Q ++ A Q Y A + ++ Y A Y PP
Sbjct 560 --PAPTAVIPTEAAIYQPSLLLNPR-ALQPSTAYYPAGTQLFMNY-----TAYYPSPPG- 610
Query 119 VVQSPPVQDVAPIPVGWNY--------TVLRLRGMPFNANEQHIVQFFQGFH 162
SP + P N TV+R++G+ +N + I+ FFQG+
Sbjct 611 ---SP--NSLGYFPTAANLSSVPPQPGTVVRMQGLAYNTGVKEILNFFQGYQ 657
> dre:560190 esrp1, rbm35a, wu:fi28a07, zgc:154050; epithelial
splicing regulatory protein 1; K14947 epithelial splicing regulatory
protein 1/2
Length=714
Score = 57.8 bits (138), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 46/96 (47%), Gaps = 15/96 (15%)
Query 139 VLRLRGMPFNANEQHIVQFF---------QGFHMTAILPSTIPIDGRPSGEAYVQFSDVS 189
+RLRG+P++A+ Q I+ F G HM + GRPSGEA++Q
Sbjct 451 CVRLRGLPYDASIQDILVFLGEYGADIKTHGVHMV------LNHQGRPSGEAFIQMRSAE 504
Query 190 ETWRALQAKNGARMDRRYIELFPASKQEMTFAAQGG 225
+ A Q + M RY+E+F S QE+ GG
Sbjct 505 RAFLAAQRCHKRSMKERYVEVFACSAQEVNIVLMGG 540
Score = 57.8 bits (138), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 36/98 (36%), Positives = 55/98 (56%), Gaps = 7/98 (7%)
Query 126 QDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQ-GFHMT----AILPSTIPIDGRPSGE 180
+VA N ++R+RG+PFNA + ++QFF +T IL P DGRP+G+
Sbjct 314 NEVASFLSRENQIIVRMRGLPFNATAEQVLQFFSPACPVTDGSEGILFVRFP-DGRPTGD 372
Query 181 AYVQFSDVSETWRALQAKNGARMDRRYIELFPASKQEM 218
A+V FS AL+ K+ + +RYIELF ++ E+
Sbjct 373 AFVLFSCEEHAQNALK-KHKDMLGKRYIELFKSTAAEV 409
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 51/90 (56%), Gaps = 6/90 (6%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMT---AILPSTIPIDGRPSGEAYVQFSDVSETWRA 194
TV+R RG+P+ +++Q I +FF+G ++ A L + GR +GEA V+F A
Sbjct 225 TVIRARGLPWQSSDQDIARFFRGLNIAKGGAAL--CLNAQGRRNGEALVRFESEEHRDLA 282
Query 195 LQAKNGARMDRRYIELFPASKQEMTFAAQG 224
LQ ++ M RYIE++ A+ ++ A G
Sbjct 283 LQ-RHKHHMGGRYIEVYKATGEDFLKIAGG 311
> hsa:80004 ESRP2, FLJ21918, FLJ22248, RBM35B; epithelial splicing
regulatory protein 2; K14947 epithelial splicing regulatory
protein 1/2
Length=717
Score = 57.4 bits (137), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 32/105 (30%), Positives = 48/105 (45%), Gaps = 15/105 (14%)
Query 130 PIPVGWNYTVLRLRGMPFNANEQHIVQFF---------QGFHMTAILPSTIPIDGRPSGE 180
P+ G +RLRG+P+ A + I+ F G HM + GRPSG+
Sbjct 457 PLAPGTGRDCVRLRGLPYTATIEDILSFLGEAAADIRPHGVHMV------LNQQGRPSGD 510
Query 181 AYVQFSDVSETWRALQAKNGARMDRRYIELFPASKQEMTFAAQGG 225
A++Q + A Q + M RY+E+ P S +EM+ GG
Sbjct 511 AFIQMTSAERALAAAQRCHKKVMKERYVEVVPCSTEEMSRVLMGG 555
Score = 50.8 bits (120), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 31/99 (31%), Positives = 54/99 (54%), Gaps = 2/99 (2%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMT-AILPSTIPIDGRPSGEAYVQFSDVSETWRALQ 196
TV+R RG+P+ +++Q + +FF+G ++ + + GR +GEA ++F D + ALQ
Sbjct 247 TVVRARGLPWQSSDQDVARFFKGLNVARGGVALCLNAQGRRNGEALIRFVDSEQRDLALQ 306
Query 197 AKNGARMDRRYIELFPASKQEMTFAAQGGDPRAFRERDR 235
++ M RYIE++ A+ +E A G R R
Sbjct 307 -RHKHHMGVRYIEVYKATGEEFVKIAGGTSLEVARFLSR 344
Score = 40.8 bits (94), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 47/87 (54%), Gaps = 5/87 (5%)
Query 136 NYTVLRLRGMPFNANEQHIVQFF-QGFHMTAILPSTIPI---DGRPSGEAYVQFSDVSET 191
+ +LRLRG+PF+A ++ F +T + + DGRP+G+A+ F+ E
Sbjct 346 DQVILRLRGLPFSAGPTDVLGFLGPECPVTGGTEGLLFVRHPDGRPTGDAFALFA-CEEL 404
Query 192 WRALQAKNGARMDRRYIELFPASKQEM 218
+A ++ + +RYIELF ++ E+
Sbjct 405 AQAALRRHKGMLGKRYIELFRSTAAEV 431
Score = 35.8 bits (81), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 45/182 (24%), Positives = 74/182 (40%), Gaps = 28/182 (15%)
Query 1 ADAAKEKLHRKYLGRRFVEVYPSTREEMHRAKRPGRI-----IPPDARMMGATAQGPYSG 55
A AA ++ H+K + R+VEV P + EEM R G + PP ++ + +
Sbjct 521 ALAAAQRCHKKVMKERYVEVVPCSTEEMSRVLMGGTLGRSGMSPPPCKLPCLSPPTYTTF 580
Query 56 GYAPAPQPYGTMATGMGSAQGYGGMDAMGAPQGGYGGYGAPSDIYGGYGGATQ-----QA 110
P P T A SA + A P AP+ + G ATQ A
Sbjct 581 QATPTLIPTETAALYPSSAL----LPAARVP-------AAPTPVAYYPGPATQLYLNYTA 629
Query 111 AYTQPPAQVVQ----SPPVQDVAPIP---VGWNYTVLRLRGMPFNANEQHIVQFFQGFHM 163
Y PP + P +A P + + ++R++G+P+ A + ++ FQ + +
Sbjct 630 YYPSPPVSPTTVGYLTTPTAALASAPTSVLSQSGALVRMQGVPYTAGMKDLLSVFQAYQL 689
Query 164 TA 165
A
Sbjct 690 PA 691
> xla:100158276 grsf1; G-rich RNA sequence binding factor 1
Length=348
Score = 57.4 bits (137), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 32/82 (39%), Positives = 50/82 (60%), Gaps = 4/82 (4%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPID--GRPSGEAYVQFSDVSETWRALQ 196
++RLRG+P++ +EQ I+ FF G + A T +D GR SGEA+VQF +AL
Sbjct 119 IVRLRGLPYSCSEQDIIHFFSGLDI-ADEGITFVLDQRGRKSGEAFVQFLSQEHADQAL- 176
Query 197 AKNGARMDRRYIELFPASKQEM 218
K+ + RYIE+FP+ + ++
Sbjct 177 LKHKQEIGSRYIEIFPSRRNDV 198
Score = 45.4 bits (106), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 54/222 (24%), Positives = 84/222 (37%), Gaps = 52/222 (23%)
Query 1 ADAAKEKLHRKYLGRRFVEVYPSTREEMHRAKRPGRIIPPDARMMGATAQGPYSGGYAPA 60
AD A K H++ +G R++E++PS R ++ A+ P R R G T Y P
Sbjct 172 ADQALLK-HKQEIGSRYIEIFPSRRNDVQTARFPFR------RRKGVTFAPTIKDLYDPD 224
Query 61 PQPYGTMATGMGSAQGYGGMDAMGAPQGGYGGYGAPSDIYGGYGGATQQAAYTQPPAQVV 120
T + P+ G+ Y + +A +
Sbjct 225 NCINNTSKDLLSDV-----------PENGH---------INDY--VKEMSAKSVDVHDFT 262
Query 121 QSPPVQDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPID-----G 175
PV D+ +RG+PF+A+ Q I FF I+P I I+ G
Sbjct 263 VMSPVHDI------------HIRGLPFHASGQDIANFFH-----PIMPLKISIEYSADAG 305
Query 176 RPSGEAYVQFSDVSETWRALQAKNGARMDRRYIELFPASKQE 217
+GEA V+F + A+ AKN Y+EL+ S +
Sbjct 306 GATGEAVVRFLTHDDAVAAM-AKNRCHSQHGYLELYLNSSTD 346
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/82 (24%), Positives = 47/82 (57%), Gaps = 4/82 (4%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHM---TAILPSTIPIDGRPSGEAYVQFSDVSETWRAL 195
++R+RG+P++ ++ FF ++ T + DG+P G+A ++F + +A+
Sbjct 3 IVRVRGLPWSCTADDVLNFFDDSNVRNGTDGVHFIFNRDGKPRGDAVIEFESAEDVQKAV 62
Query 196 QAKNGARMDRRYIELFPASKQE 217
+ ++ M +RY+E+F +++E
Sbjct 63 E-QHKKYMGQRYVEVFEMNQKE 83
Score = 30.4 bits (67), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 21/35 (60%), Gaps = 7/35 (20%)
Query 9 HRKYLGRRFVEVYPSTREE-------MHRAKRPGR 36
H+KY+G+R+VEV+ ++E MH A P R
Sbjct 65 HKKYMGQRYVEVFEMNQKEAESLLNRMHSALSPTR 99
> cel:ZK1067.6 sym-2; SYnthetic lethal with Mec family member
(sym-2); K14947 epithelial splicing regulatory protein 1/2
Length=618
Score = 56.2 bits (134), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 28/86 (32%), Positives = 49/86 (56%), Gaps = 1/86 (1%)
Query 133 VGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETW 192
V N ++R+RG+P++ + I FF+ +T + DGRP+G+A+VQF +
Sbjct 276 VSANAIIVRMRGLPYDCTDAQIRTFFEPLKLTDKILFITRTDGRPTGDAFVQFETEEDAQ 335
Query 193 RALQAKNGARMDRRYIELFPASKQEM 218
+ L K+ + +RYIELF ++ E+
Sbjct 336 QGL-LKHRQVIGQRYIELFKSTAAEV 360
Score = 50.8 bits (120), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 36/112 (32%), Positives = 48/112 (42%), Gaps = 19/112 (16%)
Query 122 SPPVQDVAPIPVGWNYTVLRLRGMPFNANEQHIVQF---------FQGFHMTAILPSTIP 172
SP V + P +RLRG+P+ A QHIV F FQG HM
Sbjct 373 SPAVANAVEAPEEKKKDCVRLRGLPYEATVQHIVTFLGDFATMVKFQGVHM------VYN 426
Query 173 IDGRPSGEAYVQFSDVSETWRALQAKNGARMD----RRYIELFPASKQEMTF 220
G PSGEA++Q + + M +RYIE+F AS +E+
Sbjct 427 NQGHPSGEAFIQMINEQAASACAAGVHNNFMSVGKKKRYIEVFQASAEELNL 478
Score = 49.7 bits (117), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 35/97 (36%), Positives = 48/97 (49%), Gaps = 10/97 (10%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI----DGRPSGEAYVQFSDVSETWR 193
V R RG+P+ A++ H+ QFF G I+P I + +GR +GE VQFS S+ R
Sbjct 181 VVCRARGLPWQASDHHVAQFFAGL---DIVPGGIALCLSSEGRRNGEVLVQFS--SQESR 235
Query 194 ALQAKNGAR-MDRRYIELFPASKQEMTFAAQGGDPRA 229
L K + RYIE++ A E A G A
Sbjct 236 DLALKRHRNFLLSRYIEVYKAGLDEFMHVATGSSTEA 272
> dre:563249 grsf1, MGC153305, wu:fb62c04, zgc:153305; G-rich
RNA sequence binding factor 1
Length=301
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 32/83 (38%), Positives = 50/83 (60%), Gaps = 10/83 (12%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFH-----MTAILPSTIPIDGRPSGEAYVQFSDVSETW 192
TV+RLRG+P++ E I++FF G +T IL G+ SG+A+V+F+ +
Sbjct 134 TVVRLRGLPYSCTEGDIIRFFSGLDVVEDGVTIILNRR----GKSSGDAFVEFATKAMAE 189
Query 193 RALQAKNGARMDRRYIELFPASK 215
+AL+ K+ + RYIE+FPA K
Sbjct 190 KALK-KDREILGNRYIEIFPAMK 211
Score = 38.5 bits (88), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 36/65 (55%), Gaps = 6/65 (9%)
Query 121 QSPPVQDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGE 180
+ PP+++ A V+ +RG+PF+A + IV+FF + ++ P +G+P+GE
Sbjct 232 EDPPLRNTA-----VTKNVIHMRGLPFDAKAEDIVKFFAPVRLMKVVVEFGP-EGKPTGE 285
Query 181 AYVQF 185
A F
Sbjct 286 AEAYF 290
> ath:AT3G20890 RNA binding / nucleic acid binding / nucleotide
binding; K12898 heterogeneous nuclear ribonucleoprotein F/H
Length=278
Score = 53.5 bits (127), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 29/77 (37%), Positives = 45/77 (58%), Gaps = 3/77 (3%)
Query 141 RLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAKNG 200
RLRG+PF+ E +V+FF G + +L + + + +GEA+ + ALQ KN
Sbjct 46 RLRGLPFDCAELDVVEFFHGLDVVDVL--FVHRNNKVTGEAFCVLGYPLQVDFALQ-KNR 102
Query 201 ARMDRRYIELFPASKQE 217
M RRY+E+F ++KQE
Sbjct 103 QNMGRRYVEVFRSTKQE 119
Score = 30.8 bits (68), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 10 RKYLGRRFVEVYPSTREEMHRAKRPGR 36
RK LG R++E++PS+ EE+ A GR
Sbjct 252 RKTLGSRYIELFPSSVEELEEALSRGR 278
> hsa:10137 RBM12, HRIHFB2091, KIAA0765, SWAN; RNA binding motif
protein 12
Length=932
Score = 53.5 bits (127), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 34/112 (30%), Positives = 59/112 (52%), Gaps = 8/112 (7%)
Query 116 PAQVVQSPPVQDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAI--LPSTIPI 173
P +Q V P+P+ + + + GMPF+A E + FF G + A+ L +
Sbjct 282 PVNPIQMNSQSSVKPLPINPDDLYVSVHGMPFSAMENDVRDFFHGLRVDAVHLLKDHV-- 339
Query 174 DGRPSGEAYVQFSDVSETWRALQAKNGARMDRRYIELFPASKQEMTFAAQGG 225
GR +G V+F +T+ AL+ +N M +RY+E+ PA++++ + A GG
Sbjct 340 -GRNNGNGLVKFLSPQDTFEALK-RNRMLMIQRYVEVSPATERQ--WVAAGG 387
Score = 46.6 bits (109), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 48/81 (59%), Gaps = 8/81 (9%)
Query 142 LRGMPFNANEQHIVQFFQGFHMTAILPSTIPI----DGRPSGEAYVQFSDVSETWRALQA 197
L+G+PF A +H++ FF+ I+ +I I +G+ +GE +V+F + ++ ++A
Sbjct 434 LKGLPFEAENKHVIDFFKKLD---IVEDSIYIAYGPNGKATGEGFVEFRNEAD-YKAALC 489
Query 198 KNGARMDRRYIELFPASKQEM 218
++ M R+I++ P +K+ M
Sbjct 490 RHKQYMGNRFIQVHPITKKGM 510
Score = 39.7 bits (91), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 22/77 (28%), Positives = 39/77 (50%), Gaps = 7/77 (9%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI----DGRPSGEAYVQFSDVSETWR 193
TV++++ MPF + I+ FF G+ ++P ++ + G P+GEA V F E
Sbjct 856 TVIKVQNMPFTVSIDEILDFFYGYQ---VIPGSVCLKYNEKGMPTGEAMVAFESRDEATA 912
Query 194 ALQAKNGARMDRRYIEL 210
A+ N + R ++L
Sbjct 913 AVIDLNDRPIGSRKVKL 929
Score = 35.8 bits (81), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 36/81 (44%), Gaps = 4/81 (4%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQA 197
V+RL+G+P A I FF G I + I G GEA++ F+ E R
Sbjct 3 VVIRLQGLPIVAGTMDIRHFFSGL---TIPDGGVHIVGGELGEAFIVFA-TDEDARLGMM 58
Query 198 KNGARMDRRYIELFPASKQEM 218
+ G + + L +SK EM
Sbjct 59 RTGGTIKGSKVTLLLSSKTEM 79
> cpv:cgd1_350 CG8205/fusilli (animal)- like, 2x RRM domains,
involved in RNA metabolism
Length=569
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 37/107 (34%), Positives = 58/107 (54%), Gaps = 7/107 (6%)
Query 121 QSPPV-QDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFF-----QGFHMTAILPSTIPID 174
Q+P + D I +N +VLRLRG+P++ E IVQFF G + + + I +
Sbjct 412 QNPSIFNDNKIIDRYYNRSVLRLRGLPWSTTEIEIVQFFISGGIYGLNASDVFLG-ITEN 470
Query 175 GRPSGEAYVQFSDVSETWRALQAKNGARMDRRYIELFPASKQEMTFA 221
R SGEA++ + + A + N + +RYIE+F +S QE+T A
Sbjct 471 QRASGEAWIILPHKCDAFDAQRILNRRVIGKRYIEVFISSFQELTTA 517
Score = 50.8 bits (120), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 31/89 (34%), Positives = 46/89 (51%), Gaps = 6/89 (6%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI----DGRPSGEAYVQFSDVSETWRA 194
V+RLRG+P+ A I+ FF + I I I DG+ +GEAYV + +
Sbjct 317 VVRLRGLPWKAAVLDIIAFFNP--ICRISSYDIAISYNKDGKMTGEAYVLLPSIKAYELS 374
Query 195 LQAKNGARMDRRYIELFPASKQEMTFAAQ 223
L +G RM +R+IE+ P+S +E Q
Sbjct 375 LTLLHGKRMGKRWIEVLPSSTKEFLICLQ 403
> dre:445379 rbm12, zgc:193560, zgc:193570; RNA binding motif
protein 12
Length=876
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 34/103 (33%), Positives = 54/103 (52%), Gaps = 10/103 (9%)
Query 140 LRLRGMPFNANEQHIVQFFQGFHMTAI--LPSTIPIDGRPSGEAYVQFSDVSETWRALQA 197
+ L G+PF+ E I FF G + +I L + GR SG A V+F E++ AL+
Sbjct 304 VHLHGLPFSVLEHEIRDFFHGLGIESIRLLKDNL---GRNSGRALVKFYSPHESFEALK- 359
Query 198 KNGARMDRRYIELFPAS----KQEMTFAAQGGDPRAFRERDRT 236
+N + +RYIE+ PA+ ++ + + GGD R R R+
Sbjct 360 RNAGMIGQRYIEVSPATERQWRESVGHSKAGGDSEHNRHRRRS 402
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 49/91 (53%), Gaps = 9/91 (9%)
Query 132 PVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI----DGRPSGEAYVQFSD 187
P +Y V L+G+P+ A + I +FF+ I+ +I I +GR +GE +V+F +
Sbjct 419 PHKLDYCVY-LKGLPYEAENKQIFEFFKNLD---IVEDSIYIAYGPNGRATGEGFVEFRN 474
Query 188 VSETWRALQAKNGARMDRRYIELFPASKQEM 218
+ ++A + M R+I++ P +K+ M
Sbjct 475 EMD-YKAALGCHMQYMGSRFIQVHPITKKNM 504
Score = 40.0 bits (92), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 38/77 (49%), Gaps = 7/77 (9%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI----DGRPSGEAYVQFSDVSETWR 193
T+++++ MPF I+ FF G+ +LP ++ + G P+GEA V F E
Sbjct 800 TIVKIQNMPFTVTVDEIIDFFYGYQ---VLPGSVCLQFSDKGLPTGEAMVAFDSHDEAMA 856
Query 194 ALQAKNGARMDRRYIEL 210
A+ N + R +++
Sbjct 857 AVMDLNDRPIGARKVKI 873
Score = 36.2 bits (82), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 36/81 (44%), Gaps = 4/81 (4%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQA 197
V+RL+G+P A I FF G I + I G GEA++ F+ E R
Sbjct 3 VVIRLQGLPIVAGTMDIRHFFSGL---TIPDGGVHIVGGEHGEAFIVFA-TDEDARLGMM 58
Query 198 KNGARMDRRYIELFPASKQEM 218
+ G + + L +SK EM
Sbjct 59 RTGGSIKGSKVSLLLSSKTEM 79
Score = 29.6 bits (65), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 9/22 (40%), Positives = 17/22 (77%), Gaps = 0/22 (0%)
Query 8 LHRKYLGRRFVEVYPSTREEMH 29
H +Y+G RF++V+P T++ M+
Sbjct 484 CHMQYMGSRFIQVHPITKKNMY 505
> mmu:75710 Rbm12, 5730420G12Rik, 9430070C08Rik, AI852903, MGC30712,
MGC38279, SWAN, mKIAA0765; RNA binding motif protein
12
Length=992
Score = 51.6 bits (122), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 33/112 (29%), Positives = 59/112 (52%), Gaps = 8/112 (7%)
Query 116 PAQVVQSPPVQDVAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAI--LPSTIPI 173
P +Q V +P+ + + + GMPF+A E + +FF G + A+ L +
Sbjct 282 PVNSIQMNSQSSVKSLPINPDDLYVSVHGMPFSAMENDVREFFHGLRVDAVHLLKDHV-- 339
Query 174 DGRPSGEAYVQFSDVSETWRALQAKNGARMDRRYIELFPASKQEMTFAAQGG 225
GR +G V+F +T+ AL+ +N M +RY+E+ PA++++ + A GG
Sbjct 340 -GRNNGNGLVKFLSPQDTFEALK-RNRMLMIQRYVEVSPATERQ--WVAAGG 387
Score = 46.2 bits (108), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 23/84 (27%), Positives = 49/84 (58%), Gaps = 8/84 (9%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI----DGRPSGEAYVQFSDVSETWRA 194
+ L+G+PF A +H++ FF+ I+ +I I +G+ +GE +V+F + ++ ++A
Sbjct 431 CVYLKGLPFEAENKHVIDFFKKLD---IVEDSIYIAYGPNGKATGEGFVEFRNDAD-YKA 486
Query 195 LQAKNGARMDRRYIELFPASKQEM 218
++ M R+I++ P +K+ M
Sbjct 487 ALCRHKQYMGNRFIQVHPITKKGM 510
Score = 39.7 bits (91), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 39/77 (50%), Gaps = 7/77 (9%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI----DGRPSGEAYVQFSDVSETWR 193
T+++++ MPF + I+ FF G+ ++P ++ + G P+GEA V F E
Sbjct 916 TIIKVQNMPFTVSIDEILDFFYGYQ---VIPGSVCLKYNEKGMPTGEAMVAFESRDEATA 972
Query 194 ALQAKNGARMDRRYIEL 210
A+ N + R ++L
Sbjct 973 AVIDLNDRPIGSRKVKL 989
Score = 36.2 bits (82), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 36/81 (44%), Gaps = 4/81 (4%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQA 197
V+RL+G+P A I FF G I + I G GEA++ F+ E R
Sbjct 3 VVIRLQGLPIVAGTMDIRHFFSGL---TIPDGGVHIVGGELGEAFIVFA-TDEDARLGMM 58
Query 198 KNGARMDRRYIELFPASKQEM 218
+ G + + L +SK EM
Sbjct 59 RTGGTIKGSKVTLLLSSKTEM 79
> cel:W02D3.11 hrpf-1; HnRNP F homolog family member (hrpf-1)
Length=549
Score = 47.4 bits (111), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 51/97 (52%), Gaps = 6/97 (6%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMT---AILPSTIPIDGRPSGEAYVQFSDVSETWRA 194
+++RLRG+PF+ + I F + +LP RP GEAYV F + E+ +
Sbjct 123 SIVRLRGLPFSVTSRDISDFLAPLPIVRDGILLPDQ--QRARPGGEAYVCF-ETMESVQI 179
Query 195 LQAKNGARMDRRYIELFPASKQEMTFAAQGGDPRAFR 231
+ ++ + RYIE+F A+ ++++ A+ D R R
Sbjct 180 AKQRHMKNIGHRYIEVFEATHRDLSRFAEENDLRVPR 216
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 39/135 (28%), Positives = 59/135 (43%), Gaps = 20/135 (14%)
Query 89 GYGGYGAPSDIYGGYGGATQQAAYTQPPAQVVQSPPVQDVAPIPVGWN-----------Y 137
G GGY + GGA +++ Y +P + + P P G++ +
Sbjct 419 GSGGYASEPYAQREAGGAMRRSEYGRPDDRYSK--------PEPYGYSRDRDYGAPQNQH 470
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQA 197
VLR+RG+PF A+E + FF + RPSG+A V F + + AL
Sbjct 471 FVLRMRGVPFRASEADVYDFFHPIRPNQVELLRDHQFQRPSGDARVIFYNRKDYDDALM- 529
Query 198 KNGARMDRRYIELFP 212
K+ M RYIE+ P
Sbjct 530 KDKQYMGERYIEMIP 544
Score = 44.3 bits (103), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 44/75 (58%), Gaps = 5/75 (6%)
Query 140 LRLRGMPFNANEQHIVQFFQGFHMTAILPSTIP-IDGRPSGEAYVQFSDVSETWRALQAK 198
++ RG+P+ A EQ + FF + ++ IP +GR SG+A V F++ + AL+ K
Sbjct 7 VQCRGLPWEATEQELRDFFGNNGIESL---EIPRRNGRTSGDAKVVFTNEEDYNNALK-K 62
Query 199 NGARMDRRYIELFPA 213
+ + RYIE+FPA
Sbjct 63 DREHLGSRYIEVFPA 77
> xla:379683 rbm12, MGC68861, SWAN; RNA binding motif protein
12
Length=877
Score = 46.2 bits (108), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 27/90 (30%), Positives = 50/90 (55%), Gaps = 2/90 (2%)
Query 128 VAPIPVGWNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSD 187
+ P P+ + + L G+P +E I + F G + I+ T P+ GR +G A V+
Sbjct 294 MKPPPLNHDDPYVCLHGLPLPVSEADIKELFHGLRIDGIVILTDPM-GRHNGSALVKLIT 352
Query 188 VSETWRALQAKNGARMDRRYIELFPASKQE 217
+T+ AL+ +N M +R+IE+ PA++++
Sbjct 353 PHDTFEALK-RNRMLMGQRFIEVSPATERQ 381
Score = 44.7 bits (104), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 20/81 (24%), Positives = 48/81 (59%), Gaps = 8/81 (9%)
Query 142 LRGMPFNANEQHIVQFFQGFHMTAILPSTIPI----DGRPSGEAYVQFSDVSETWRALQA 197
L+G+P+ A +H++ FF+ + I+ +I I +G+ +GE +++F + E +++
Sbjct 434 LKGLPYEAENKHVIDFFKKLN---IVEDSIYIAYGSNGKATGEGFLEFRN-EEDYKSALC 489
Query 198 KNGARMDRRYIELFPASKQEM 218
++ M R++++ P +K+ M
Sbjct 490 RHKQYMGNRFVQVHPITKKAM 510
Score = 40.0 bits (92), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 38/77 (49%), Gaps = 7/77 (9%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI----DGRPSGEAYVQFSDVSETWR 193
TV+R++ MPF I+ FF G+ + +P ++ + G P+GEA V F E
Sbjct 801 TVIRVQNMPFTVTVDEILDFFYGYQL---IPGSVCLKFSDKGMPTGEAMVAFESRDEAMA 857
Query 194 ALQAKNGARMDRRYIEL 210
A+ N + R ++L
Sbjct 858 AVVDLNERPIGSRKVKL 874
Score = 35.0 bits (79), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 36/81 (44%), Gaps = 4/81 (4%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQA 197
V+RL+G+P A I FF G I + I G GEA++ F+ E R
Sbjct 3 VVIRLQGLPIVAGTMDIRHFFSGL---TIPDGGVHIVGGELGEAFIVFA-TDEDARLGMM 58
Query 198 KNGARMDRRYIELFPASKQEM 218
+ G + + L +SK EM
Sbjct 59 RTGGTIKGSKVSLLLSSKTEM 79
Score = 30.8 bits (68), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 10/20 (50%), Positives = 17/20 (85%), Gaps = 0/20 (0%)
Query 9 HRKYLGRRFVEVYPSTREEM 28
H++Y+G RFV+V+P T++ M
Sbjct 491 HKQYMGNRFVQVHPITKKAM 510
> xla:494769 rbm19; RNA binding motif protein 19; K14787 multiple
RNA-binding domain-containing protein 1
Length=920
Score = 45.4 bits (106), Expect = 2e-04, Method: Composition-based stats.
Identities = 32/104 (30%), Positives = 51/104 (49%), Gaps = 11/104 (10%)
Query 136 NYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPID----GRPSGEAYVQFSDVSET 191
+YTV +LRG PFN EQ++ +F + + P I I G +G +V S E
Sbjct 278 SYTV-KLRGAPFNVTEQNVKEF-----LVPLKPVAIRIARNTYGNKTGYVFVDLSSEEEV 331
Query 192 WRALQAKNGARMDRRYIELFPASKQEMTFAAQGGDPRAFRERDR 235
+AL+ +N M RYIE+F + + + R + +RD+
Sbjct 332 QKALK-RNKDYMGGRYIEVFRDNYTKSPSVQSKAESRPWEQRDK 374
> xla:398766 rbm12b, MGC68792; RNA binding motif protein 12B
Length=654
Score = 44.7 bits (104), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 42/81 (51%), Gaps = 4/81 (4%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQA 197
V+RL+G+P A I FF G H I + I G EA++ F+ + RA++
Sbjct 3 VVIRLQGLPVTAGSNDIRHFFSGLH---IPDGGVHITGGKYAEAFIIFATDEDARRAMRC 59
Query 198 KNGARMDRRYIELFPASKQEM 218
+G + + IELF +SK EM
Sbjct 60 -SGGFIKKSQIELFLSSKAEM 79
Score = 37.4 bits (85), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 36/65 (55%), Gaps = 2/65 (3%)
Query 132 PVGWN-YTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSE 190
P+ N Y + L G+P++ +E + FF GFH+ + S + +G G AYV+F+ V +
Sbjct 150 PIKDNSYGYVFLCGLPYSTSELDVKDFFHGFHVVDVHFS-VRSNGARDGNAYVKFASVQD 208
Query 191 TWRAL 195
+L
Sbjct 209 AKASL 213
Score = 32.7 bits (73), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 31/60 (51%), Gaps = 3/60 (5%)
Query 140 LRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAKN 199
+RL+ +P+ A ++I+ FF G++M +P ++ +D G A V + A+ N
Sbjct 582 IRLKNVPYTATIENILDFFYGYNM---VPDSVEMDYISKGTAIVHMENYDVAVAAVNELN 638
> dre:387255 rbm19, npo; RNA binding motif protein 19; K14787
multiple RNA-binding domain-containing protein 1
Length=926
Score = 44.3 bits (103), Expect = 4e-04, Method: Composition-based stats.
Identities = 29/75 (38%), Positives = 39/75 (52%), Gaps = 2/75 (2%)
Query 140 LRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAKN 199
++LRG PFN EQ + +F AI + DGR SG YV +E RAL+ +
Sbjct 291 VKLRGAPFNVKEQQVKEFMMPLKPVAIRFAKNS-DGRNSGYVYVDLRSEAEVERALRL-D 348
Query 200 GARMDRRYIELFPAS 214
M RYIE+F A+
Sbjct 349 KDYMGGRYIEVFRAN 363
> xla:379184 MGC132368; hypothetical protein MGC53694
Length=658
Score = 43.5 bits (101), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 41/81 (50%), Gaps = 4/81 (4%)
Query 138 TVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQA 197
V+RL+G+P A I FF G H I + I G GEA++ F + RA+
Sbjct 3 VVIRLQGLPLVAGSTDIRHFFSGLH---IPEGGVHITGGKHGEAFIIFPTDEDARRAMSC 59
Query 198 KNGARMDRRYIELFPASKQEM 218
+G + + I+LF +SK EM
Sbjct 60 -SGGFIKKSQIDLFLSSKAEM 79
Score = 38.1 bits (87), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 44/81 (54%), Gaps = 3/81 (3%)
Query 131 IPVGWN-YTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVS 189
+P+ N Y + L G+P+ A+E + +FF GF + I + +G G+AYV+F+
Sbjct 144 MPLKENGYGYVFLNGLPYTADEHDVKEFFHGFDVEDI-NFCVRQNGDKDGKAYVKFATFQ 202
Query 190 ETWRALQAKNGARMDRRYIEL 210
+ +L +++ + RYI L
Sbjct 203 DAKASL-SRHKEYIGHRYIFL 222
Score = 34.7 bits (78), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 30/60 (50%), Gaps = 3/60 (5%)
Query 140 LRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAKN 199
+RL +P+ A + I+ FF G++ ++P ++ +D G A V + E A+ N
Sbjct 586 IRLENVPYTATIEEILDFFHGYN---VVPDSVKMDYNSKGTAIVHMENYYEAAAAINELN 642
> dre:678601 MGC136953; zgc:136953
Length=209
Score = 43.1 bits (100), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 40/80 (50%), Gaps = 10/80 (12%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPID----GRPSGEAYVQFSDVSETWRA 194
+ +RG+PF A E + FF + P + ID G+ +GEA V+F + A
Sbjct 104 FVHMRGLPFRATESDVAHFF-----GPLTPVRVHIDMGPNGKSTGEADVEFRSHEDAVSA 158
Query 195 LQAKNGARMDRRYIELFPAS 214
+ +K+ M RYIELF S
Sbjct 159 M-SKDKNHMQHRYIELFLNS 177
> ath:AT5G41690 RNA binding / nucleic acid binding / nucleotide
binding
Length=567
Score = 41.6 bits (96), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/77 (28%), Positives = 39/77 (50%), Gaps = 0/77 (0%)
Query 140 LRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAKN 199
L + +P+ +I+ FF+ + + + G+ G +V+F+ V+E ALQ KN
Sbjct 70 LFVANLPYETKIPNIIDFFKKVGEVVRVQLIVNLKGKLVGCGFVEFASVNEAEEALQKKN 129
Query 200 GARMDRRYIELFPASKQ 216
G +D I L A+K+
Sbjct 130 GECLDNNKIFLDVANKK 146
Score = 34.7 bits (78), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 29/58 (50%), Gaps = 0/58 (0%)
Query 153 HIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAKNGARMDRRYIEL 210
HI++FF+ + + G G +V+F+ V+E +ALQ NG + R I L
Sbjct 451 HIIKFFKDVAEVVRVRLIVNHRGEHVGCGFVEFASVNEAQKALQKMNGENLRSREIFL 508
Score = 34.3 bits (77), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 29/56 (51%), Gaps = 8/56 (14%)
Query 153 HIVQFFQGF----HMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAKNGARMD 204
HI+ FF+ H+ IL T G+ G A+V+F +E AL+ KNG ++
Sbjct 323 HIINFFKDVGEVVHVRLILNHT----GKHVGCAFVEFGSANEAKMALETKNGEYLN 374
> mmu:74111 Rbm19, 1200009A02Rik, AV302714, KIAA0682, NPO; RNA
binding motif protein 19; K14787 multiple RNA-binding domain-containing
protein 1
Length=952
Score = 41.2 bits (95), Expect = 0.004, Method: Composition-based stats.
Identities = 38/122 (31%), Positives = 54/122 (44%), Gaps = 15/122 (12%)
Query 94 GAPSDIYGGYGGATQQAAYTQPPAQVVQSPPVQDVAPIPVGWNYTVLRLRGMPFNANEQH 153
+P+ G GA Q A V+ P Q P YTV +LRG PFN E++
Sbjct 254 ASPAKQGGVSRGAVPGVLRPQEAAGKVEKPVSQKEPTTP----YTV-KLRGAPFNVTEKN 308
Query 154 IVQFFQGFHMTAILPSTIPI----DGRPSGEAYVQFSDVSETWRALQAKNGARMDRRYIE 209
+++F + + P I I G +G +V S E +AL+ N M RYIE
Sbjct 309 VIEF-----LAPLKPVAIRIVRNAHGNKTGYVFVDLSSEEEVKKALKC-NRDYMGGRYIE 362
Query 210 LF 211
+F
Sbjct 363 VF 364
> mmu:17975 Ncl, B530004O11Rik, C23, D0Nds28, D1Nds28, Nucl; nucleolin;
K11294 nucleolin
Length=707
Score = 40.8 bits (94), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 36/73 (49%), Gaps = 4/73 (5%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAK 198
L + + FN E + + F+ A+ + DG+ G AY++F ++ + L+ K
Sbjct 396 TLLAKNLSFNITEDELKEVFED----AMEIRLVSQDGKSKGIAYIEFKSEADAEKNLEEK 451
Query 199 NGARMDRRYIELF 211
GA +D R + L+
Sbjct 452 QGAEIDGRSVSLY 464
> mmu:77604 C430048L16Rik, AV299215, MGC91091, Rbm12bb; RIKEN
cDNA C430048L16 gene
Length=834
Score = 40.8 bits (94), Expect = 0.005, Method: Composition-based stats.
Identities = 42/166 (25%), Positives = 68/166 (40%), Gaps = 18/166 (10%)
Query 84 GAPQGGYGGYG---------APSDIYGGYGGATQQAAYTQPPAQVVQSPPVQDVAPIPVG 134
G P G G G + Y GYG + + A + P + P+
Sbjct 94 GRPGSGASGVGNVYHFSDALKEEESYSGYGSSVNRDAGFHTNGTGLDLRP-RKTRPLKAE 152
Query 135 WNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRA 194
Y L LRG+P+ NE + FF G + ++ DGR +G+A V+F+ +
Sbjct 153 NPY--LFLRGLPYLVNEDDVRVFFSGLCVDGVILLK-HHDGRNNGDAIVKFASCVDASGG 209
Query 195 LQAKNGARMDRRYIELFPASKQEMT----FAAQGGDPRAFRERDRT 236
L+ + M R+IE+ S+Q+ A +GGD R + +
Sbjct 210 LKCHR-SFMGSRFIEVMQGSEQQWIEFGGTATEGGDTPRMRSEEHS 254
Score = 40.4 bits (93), Expect = 0.005, Method: Composition-based stats.
Identities = 28/80 (35%), Positives = 43/80 (53%), Gaps = 4/80 (5%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAK 198
V+RL G+PF A I FF+G I + I G GEA++ F+ + RA+ ++
Sbjct 4 VIRLLGLPFIAGPVDIRHFFKGL---TIPDGGVHIIGGKVGEAFIIFATDEDARRAI-SR 59
Query 199 NGARMDRRYIELFPASKQEM 218
+G + +ELF +SK EM
Sbjct 60 SGGFIKDSSVELFLSSKVEM 79
Score = 37.7 bits (86), Expect = 0.034, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 39/73 (53%), Gaps = 7/73 (9%)
Query 142 LRGMPFNANEQHIVQFFQGFHMTAILPSTIPI----DGRPSGEAYVQFSDVSETWRALQA 197
+ +PF AN I+ FF G+ ++P ++ I +G P GEA V ++ +E A++
Sbjct 762 ISNLPFKANANEILDFFHGY---KVIPDSVSIQYNEEGLPLGEAIVSMTNYNEALSAVKD 818
Query 198 KNGARMDRRYIEL 210
+G + R ++L
Sbjct 819 LSGRPVGPRKVKL 831
> pfa:PFL2130w conserved Plasmodium protein
Length=1335
Score = 40.4 bits (93), Expect = 0.006, Method: Composition-based stats.
Identities = 25/96 (26%), Positives = 51/96 (53%), Gaps = 7/96 (7%)
Query 127 DVAPIPV-----GWNYTVLRLRGMPFNANEQHIVQFFQGFH-MTAILPSTIPIDGRPSGE 180
++ P+PV G+N +L +G+PF+ + I++FF+ + M + G G+
Sbjct 170 NILPLPVEKNMEGYNCLIL-CKGLPFHVDNSQIIEFFKPYKIMEKYIIFMRDKKGHFFGD 228
Query 181 AYVQFSDVSETWRALQAKNGARMDRRYIELFPASKQ 216
V+F + + + AL+ KN + RYI+L+ +++
Sbjct 229 ILVRFINKEQKYLALKNKNYKFLLHRYIQLYNVNEE 264
> mmu:72397 Rbm12b, 3000004N20Rik, Rbm12ba; RNA binding motif
protein 12B
Length=836
Score = 40.4 bits (93), Expect = 0.006, Method: Composition-based stats.
Identities = 28/80 (35%), Positives = 43/80 (53%), Gaps = 4/80 (5%)
Query 139 VLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQAK 198
V+RL G+PF A I FF+G I + I G GEA++ F+ + RA+ ++
Sbjct 4 VIRLLGLPFIAGPVDIRHFFKGL---TIPDGGVHIIGGKVGEAFIIFATDEDARRAI-SR 59
Query 199 NGARMDRRYIELFPASKQEM 218
+G + +ELF +SK EM
Sbjct 60 SGGFIKDSSVELFLSSKVEM 79
Score = 40.4 bits (93), Expect = 0.006, Method: Composition-based stats.
Identities = 42/166 (25%), Positives = 68/166 (40%), Gaps = 18/166 (10%)
Query 84 GAPQGGYGGYG---------APSDIYGGYGGATQQAAYTQPPAQVVQSPPVQDVAPIPVG 134
G P G G G + Y GYG + + A + P + P+
Sbjct 94 GRPGSGASGVGNVYHFSDALKEEESYSGYGSSVNRDAGFHTNGTGLDLRP-RKTRPLKAE 152
Query 135 WNYTVLRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRA 194
Y L LRG+P+ NE + FF G + ++ DGR +G+A V+F+ +
Sbjct 153 NPY--LFLRGLPYLVNEDDVRVFFSGLCVDGVILLK-HHDGRNNGDAIVKFASCVDASGG 209
Query 195 LQAKNGARMDRRYIELFPASKQEMT----FAAQGGDPRAFRERDRT 236
L+ + M R+IE+ S+Q+ A +GGD R + +
Sbjct 210 LKCHR-SFMGSRFIEVMQGSEQQWIEFGGTATEGGDTPRMRSEEHS 254
Score = 36.6 bits (83), Expect = 0.087, Method: Composition-based stats.
Identities = 20/75 (26%), Positives = 40/75 (53%), Gaps = 7/75 (9%)
Query 140 LRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI----DGRPSGEAYVQFSDVSETWRAL 195
+++ +PF AN I+ FF G+ ++P ++ + G P GEA V ++ +E A+
Sbjct 762 VKISNLPFKANASEILDFFHGY---KVIPDSVSLQYNEQGLPIGEAIVAMTNYNEALAAV 818
Query 196 QAKNGARMDRRYIEL 210
+ +G + R ++L
Sbjct 819 KDLSGRPVGPRKVKL 833
> dre:393291 rbm28, MGC56258, zgc:56258; RNA binding motif protein
28; K14573 nucleolar protein 4
Length=864
Score = 39.7 bits (91), Expect = 0.009, Method: Composition-based stats.
Identities = 26/86 (30%), Positives = 42/86 (48%), Gaps = 4/86 (4%)
Query 140 LRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI--DGRPSGEAYVQFSDVSETWRALQA 197
L +R + F E + Q F F +L + IP+ DG+ G A+V F + E +AL A
Sbjct 238 LIIRNLSFKCEEDDLKQIFSEF--GTVLEAKIPLKPDGKKRGFAFVLFKRMPEAGKALTA 295
Query 198 KNGARMDRRYIELFPASKQEMTFAAQ 223
NG ++ R + + A ++ A Q
Sbjct 296 MNGKKIKDRQVAVDWAIAKDKYLATQ 321
Score = 38.9 bits (89), Expect = 0.015, Method: Composition-based stats.
Identities = 29/95 (30%), Positives = 42/95 (44%), Gaps = 9/95 (9%)
Query 140 LRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI--DGRPSGEAYVQFSDVSETWRALQA 197
L +R + F + + Q F F +L + IP+ DG+ G A+VQF VSE +A A
Sbjct 126 LIIRNLSFKCEDDELKQIFSKF--GTVLETRIPLKPDGKKKGFAFVQFKCVSEAEKARAA 183
Query 198 KNGARMDRRYIELFPASKQEMTFAAQGGDPRAFRE 232
M+R+ I P T + DP E
Sbjct 184 -----MNRKAIRDRPVEVDWTTSNTESADPEDTEE 213
> hsa:9904 RBM19, DKFZp586F1023, KIAA0682; RNA binding motif protein
19; K14787 multiple RNA-binding domain-containing protein
1
Length=960
Score = 39.3 bits (90), Expect = 0.014, Method: Composition-based stats.
Identities = 29/98 (29%), Positives = 48/98 (48%), Gaps = 10/98 (10%)
Query 140 LRLRGMPFNANEQHIVQFFQGFHMTAILPSTIPI----DGRPSGEAYVQFSDVSETWRAL 195
++LRG PFN E+++++F + + P I I G +G +V FS+ E +AL
Sbjct 296 VKLRGAPFNVTEKNVMEF-----LAPLKPVAIRIVRNAHGNKTGYIFVDFSNEEEVKQAL 350
Query 196 QAKNGARMDRRYIELFPASKQEMTFAAQGGDPRAFRER 233
+ N M RYIE+F T A ++++ R
Sbjct 351 KC-NREYMGGRYIEVFREKNVPTTKGAPKNTTKSWQGR 387
> sce:YGR159C NSR1, SHE5; Nsr1p; K11294 nucleolin
Length=414
Score = 38.9 bits (89), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 25/74 (33%), Positives = 38/74 (51%), Gaps = 3/74 (4%)
Query 139 VLRLRGMPFNANEQHIVQFF--QGFHMTAILPSTIPIDGRPSGEAYVQFSDVSETWRALQ 196
L L + FNA+ I + F G ++ +P T P +P G YVQFS++ + +AL
Sbjct 268 TLFLGNLSFNADRDAIFELFAKHGEVVSVRIP-THPETEQPKGFGYVQFSNMEDAKKALD 326
Query 197 AKNGARMDRRYIEL 210
A G +D R + L
Sbjct 327 ALQGEYIDNRPVRL 340
Lambda K H
0.318 0.136 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8210089472
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40