bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0818_orf3
Length=217
Score E
Sequences producing significant alignments: (Bits) Value
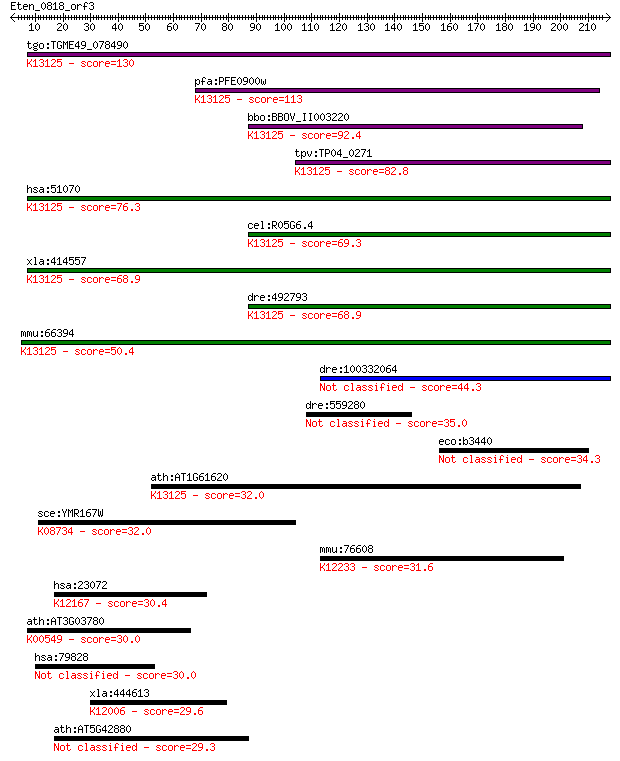
tgo:TGME49_078490 hypothetical protein ; K13125 nitric oxide s... 130 3e-30
pfa:PFE0900w conserved protein, unknown function; K13125 nitri... 113 4e-25
bbo:BBOV_II003220 18.m06270; hypothetical protein; K13125 nitr... 92.4 1e-18
tpv:TP04_0271 hypothetical protein; K13125 nitric oxide syntha... 82.8 9e-16
hsa:51070 NOSIP; nitric oxide synthase interacting protein; K1... 76.3 7e-14
cel:R05G6.4 hypothetical protein; K13125 nitric oxide synthase... 69.3 9e-12
xla:414557 nosip, MGC78783; nitric oxide synthase interacting ... 68.9 1e-11
dre:492793 nosip, wu:fb80a11, zgc:101669; nitric oxide synthas... 68.9 1e-11
mmu:66394 Nosip, 2310061K06Rik, CGI-25, MGC115777; nitric oxid... 50.4 4e-06
dre:100332064 nitric oxide synthase-interacting protein-like 44.3 4e-04
dre:559280 MGC171819; zgc:171819 35.0 0.20
eco:b3440 yhhX, ECK3425, JW3403; predicted oxidoreductase with... 34.3 0.31
ath:AT1G61620 phosphoinositide binding; K13125 nitric oxide sy... 32.0 1.8
sce:YMR167W MLH1, PMS2; Mlh1p; K08734 DNA mismatch repair prot... 32.0 1.9
mmu:76608 Hectd3, 1700064K09Rik, AI467540, AW743312; HECT doma... 31.6 2.0
hsa:23072 HECW1, KIAA0322, NEDL1; HECT, C2 and WW domain conta... 30.4 4.5
ath:AT3G03780 AtMS2; AtMS2; 5-methyltetrahydropteroyltriglutam... 30.0 6.3
hsa:79828 METTL8, FLJ13334, FLJ13984, FLJ31054, FLJ42098, FLJ7... 30.0 6.5
xla:444613 trim16, MGC84103, ebbp; tripartite motif containing... 29.6 7.6
ath:AT5G42880 hypothetical protein 29.3 9.6
> tgo:TGME49_078490 hypothetical protein ; K13125 nitric oxide
synthase-interacting protein
Length=322
Score = 130 bits (327), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 87/249 (34%), Positives = 119/249 (47%), Gaps = 43/249 (17%)
Query 7 IQKETKAWEALQAQKEQEAAAAAEANATAAQRAFLEAEASLTLETSAAPAQTKNSPQSAE 66
+ KE K WEA Q K++ A A+A + FLE E + A+ + P AE
Sbjct 79 MAKELKEWEAQQIAKKEADAVKKMEEASAEKNKFLEEENKV-----ASYYAKQRKPTVAE 133
Query 67 EFRQTLEQPTI----NSKEEARAKNFWVSE------------------------------ 92
+ P + K AR K+FWV E
Sbjct 134 LELAPKKNPHLLIGEGGKAAAREKSFWVREAQEENSLVKNFSETVTLYDVRAGRIDGKPP 193
Query 93 ---NTPDEPEAKLKEPQKGFCCPITGAPLRIKQLITVKPVLASKEDTENTRWLCALSGRE 149
NTP P + K P++ CPIT PL++KQL+ + P L ++TE RW C +S R
Sbjct 194 AVENTPSAPPEEAKPPKRSLECPITRKPLKLKQLVYLNPELLDDKETEFNRWSCKISKRA 253
Query 150 ISHQKAAVVKSTGQVILLEYLEKLVFGKKGALTSG-VIDRKDTVALIPGGTGFSAHNNVE 208
I++QKAA V TG VIL+E +EK V KKG VI ++D + L+PGGTG+S+HN VE
Sbjct 254 ITNQKAAAVIPTGDVILMECIEKYVLNKKGGFYGDKVITKQDIIPLVPGGTGYSSHNTVE 313
Query 209 VAVARPNVQ 217
V R +Q
Sbjct 314 GVVRRACIQ 322
> pfa:PFE0900w conserved protein, unknown function; K13125 nitric
oxide synthase-interacting protein
Length=270
Score = 113 bits (283), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 62/157 (39%), Positives = 88/157 (56%), Gaps = 12/157 (7%)
Query 68 FRQTLEQ-PTIN-SKEEAR-----AKNFWVSENTP----DEPEAKLKEPQKGFCCPITGA 116
F Q LE T+N KEE + + NFW+S NT D + KLK P K CPIT
Sbjct 111 FVQDLENLNTVNVQKEEEKNLLDISNNFWLSCNTSKVKKDTIQKKLKPPSKNLICPITKK 170
Query 117 PLRIKQLITVKPVLASKEDTENTRWLCALSGREISHQKAAVVKSTGQVILLEYLEKLVFG 176
PL++ +LIT+ P + D+EN W+C+ S + I H KA ++K TG++IL + E ++G
Sbjct 171 PLKMNELITINPEVIKNGDSENGGWVCSFSKKNIDHHKAVLLKKTGKIILKSFFENFIYG 230
Query 177 KKGALTSGVIDRKDTVALIPGGTGFSAHNNVEVAVAR 213
KK + V D + L GGT F +H+NVE + R
Sbjct 231 KKNSYDITV-GEDDFINLEAGGTAFCSHSNVEKTLYR 266
> bbo:BBOV_II003220 18.m06270; hypothetical protein; K13125 nitric
oxide synthase-interacting protein
Length=262
Score = 92.4 bits (228), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 50/128 (39%), Positives = 72/128 (56%), Gaps = 20/128 (15%)
Query 87 NFWVS----ENTPDEPEAKLKEPQKGFC-CPITGAPLRIKQLITVKPVLASKEDTENTRW 141
+FWV+ + DE L EP K CPITG PL+IK+L+ + P L+ D + W
Sbjct 138 SFWVAGVSGNSKGDETSTSLPEPPKKILRCPITGKPLKIKELVDIHPDLSENTDNGDPIW 197
Query 142 LCALSGREISHQKAAVVKSTGQVILLEYLEKLVFGKKGALTSGVIDRKDT--VALIPGGT 199
+C++S R I H++ + +STG+++L Y+E V + DT + LIPGGT
Sbjct 198 ICSVSQRPIGHKEVMLNRSTGRLVLRRYIE-------------VCNDMDTKYIKLIPGGT 244
Query 200 GFSAHNNV 207
FSAHNNV
Sbjct 245 AFSAHNNV 252
> tpv:TP04_0271 hypothetical protein; K13125 nitric oxide synthase-interacting
protein
Length=241
Score = 82.8 bits (203), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 42/116 (36%), Positives = 66/116 (56%), Gaps = 11/116 (9%)
Query 104 EPQKGFCCPITGAPLRIKQLITVKPVLASKEDTENTR--WLCALSGREISHQKAAVVKST 161
+P+ CPI+G PL++K LI + P S D+ ++ WLC++S + I H +A K
Sbjct 135 KPKNLLSCPISGRPLKVKDLIDLDPDTRSTSDSPSSEVVWLCSVSKKPILHNQAYAYKKN 194
Query 162 GQVILLEYLEKLVFGKKGALTSGVIDRKDTVALIPGGTGFSAHNNVEVAVARPNVQ 217
G++++ +Y+ GA T D + V+L+P GT FS+HNNVE RP +Q
Sbjct 195 GKIVMKQYV--------GA-TDNPQDTESFVSLVPAGTAFSSHNNVEAKKFRPCMQ 241
> hsa:51070 NOSIP; nitric oxide synthase interacting protein;
K13125 nitric oxide synthase-interacting protein
Length=301
Score = 76.3 bits (186), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 60/226 (26%), Positives = 110/226 (48%), Gaps = 21/226 (9%)
Query 7 IQKETKAWEALQAQKEQEAAAAAEANATAAQRAFLEAEASLT---LETSAAPAQTKNSPQ 63
I ++ KA+E + + +E A + R FLE E+++ L A A + SP
Sbjct 81 IARQMKAYEKQRGTRREEQKELQRAASQDHVRGFLEKESAIVSRPLNPFTAKALSGTSPD 140
Query 64 SAEEFRQTLEQPTINSKEEARAK---NFWVSENTPDEPEAKLKEPQKGFCCPITGAPLRI 120
+ P++ + + K +FW+ TP+ KL++P + CP++G PLR+
Sbjct 141 DVQP------GPSVGPPSKDKDKVLPSFWIPSLTPEAKATKLEKPSRTVTCPMSGKPLRM 194
Query 121 KQLITVK--PVLASKED----TENTRWLCALSGREISH-QKAAVVKSTGQVILLEYLEKL 173
L V P+ +S + T + R++CA++ +S+ AV++ +G V+ LE +EKL
Sbjct 195 SDLTPVHFTPLDSSVDRVGLITRSERYVCAVTRDSLSNATPCAVLRPSGAVVTLECVEKL 254
Query 174 VFGKK-GALTSGVIDRKDTVALIPGGTGFSAHN-NVEVAVARPNVQ 217
+ +T + +D + L GGTGF+ ++ +RP +Q
Sbjct 255 IRKDMVDPVTGDKLTDRDIIVLQRGGTGFAGSGVKLQAEKSRPVMQ 300
> cel:R05G6.4 hypothetical protein; K13125 nitric oxide synthase-interacting
protein
Length=310
Score = 69.3 bits (168), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 38/134 (28%), Positives = 73/134 (54%), Gaps = 3/134 (2%)
Query 87 NFWVSENTPDEPEAKLKEPQKGFCCPITGAPLRIKQLITVKPV-LASKEDTENTRWLCAL 145
+FW+ E P KL++P CP++G P+++K+L+ VK + E + ++LC +
Sbjct 175 SFWIPELNPTAVATKLEKPSSKVLCPVSGKPIKLKELLEVKFTPMPGTETAAHRKFLCPV 234
Query 146 SGREISH-QKAAVVKSTGQVILLEYLEKLVFGKKGALTSG-VIDRKDTVALIPGGTGFSA 203
+ E+++ + A +K + V+ + +EKL+ G +G + D + L GGTG+SA
Sbjct 235 TRDELTNTTRCAYLKKSKSVVKYDVVEKLIKGDGIDPINGEPMSEDDIIELQRGGTGYSA 294
Query 204 HNNVEVAVARPNVQ 217
N + + RP ++
Sbjct 295 TNETKAKLIRPQLE 308
> xla:414557 nosip, MGC78783; nitric oxide synthase interacting
protein; K13125 nitric oxide synthase-interacting protein
Length=298
Score = 68.9 bits (167), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 58/225 (25%), Positives = 110/225 (48%), Gaps = 22/225 (9%)
Query 7 IQKETKAWEALQAQKEQEAAAAAEANATAAQRAFLEAEASLTLETSAAPAQTKNSPQSAE 66
I ++ KA+E + K+ E +A + +AFL+ E +T+ + T+ S +
Sbjct 81 IARQMKAYEKQKNSKKAEMEELNKAAKESKMKAFLDKE--MTIVSKPLNPFTRKSDSGID 138
Query 67 EFRQTLEQPTINSKEEARAKNFWVSENTPDEPEAKLKEPQKGFCCPITGAPLRIKQLITV 126
+ Q + K + + +FW+ TP+ + +K+P K CP++G PL++K L+
Sbjct 139 AAEPSCSQQSSEEKNK-QLPSFWIPSLTPEAKTSLVKKPDKTVYCPMSGRPLKMKDLM-- 195
Query 127 KPVLASKED---------TENTRWLCALSGREISHQ-KAAVVKSTGQVILLEYLEKLVFG 176
PV ++ D R++CA++ + + AV++ +G V+ +E +EKL+
Sbjct 196 -PVNFTRVDEKVDRVGLINRQDRYVCAVTRDMLGNSVPCAVLRPSGAVVTMECVEKLI-- 252
Query 177 KKGA---LTSGVIDRKDTVALIPGGTGFSAHNN-VEVAVARPNVQ 217
KK ++ + +D + L GGTGFS ++ ARP +Q
Sbjct 253 KKDMIDPISGDKLSERDIIMLQRGGTGFSGSGVLLQAKEARPVMQ 297
> dre:492793 nosip, wu:fb80a11, zgc:101669; nitric oxide synthase
interacting protein; K13125 nitric oxide synthase-interacting
protein
Length=304
Score = 68.9 bits (167), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 45/142 (31%), Positives = 78/142 (54%), Gaps = 13/142 (9%)
Query 87 NFWVSENTPDEPEAKLKEPQKGFCCPITGAPLRIKQLITVK--PVLASKED----TENTR 140
+FW+ TP+ LK+P K CP++G PL++ LITV+ P+ S + T R
Sbjct 164 SFWIPSLTPEAKPTLLKKPSKTVSCPMSGRPLKMSDLITVRFTPLDPSLDRVALLTRQDR 223
Query 141 WLCALSGREISHQ-KAAVVKSTGQVILLEYLEKLVFGKKGA---LTSGVIDRKDTVALIP 196
++CA++ + + AV++ +G V+ +E +EKL+ KK +T + KD + +
Sbjct 224 YVCAVTKDTLGNSVPCAVLRPSGVVVTMECVEKLI--KKDMVDPITGDKLKEKDIIPIQR 281
Query 197 GGTGFSAHN-NVEVAVARPNVQ 217
GGTGF+ +++ ARP +Q
Sbjct 282 GGTGFAGSGVDLKAKEARPVMQ 303
> mmu:66394 Nosip, 2310061K06Rik, CGI-25, MGC115777; nitric oxide
synthase interacting protein; K13125 nitric oxide synthase-interacting
protein
Length=276
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 52/221 (23%), Positives = 98/221 (44%), Gaps = 32/221 (14%)
Query 5 RGIQKETKAWEALQAQKEQEAAAAAEANATAAQRAFLEAEASLTLETSAAPAQTKNSPQS 64
R I ++ KA+E + + +E A A R FLE EA++ + P++
Sbjct 79 REIARQVKAYEKQRGARREEQKELQRAAAQDQVRGFLEKEAAIV-----SRPLNPFMPKA 133
Query 65 AEEFRQTLEQP-----TINSKEEARAKNFWVSENTPDEPEAKLKEPQKGFCCPITGAPLR 119
A EQP + ++ +FW+ TP+ KL++ P+ + R
Sbjct 134 ATLPNTEGEQPGPSVGPVGKDKDKALPSFWIPSLTPEAKATKLEK-------PLDDSVDR 186
Query 120 IKQLITVKPVLASKEDTENTRWLCALSGREISH-QKAAVVKSTGQVILLEYLEKLVFGKK 178
+ + T + R++CA++ +S+ AV++ +G V+ LE +EKL+
Sbjct 187 VGLI------------TRSERYVCAVTRDSLSNATPCAVLRPSGAVVTLECVEKLIRKDM 234
Query 179 GALTSG-VIDRKDTVALIPGGTGFSAHN-NVEVAVARPNVQ 217
+G + +D + L GGTGF+ ++ ++RP +Q
Sbjct 235 VDPVNGDTLTERDIIVLQRGGTGFAGSGVKLQAEMSRPVMQ 275
> dre:100332064 nitric oxide synthase-interacting protein-like
Length=115
Score = 44.3 bits (103), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 35/116 (30%), Positives = 64/116 (55%), Gaps = 13/116 (11%)
Query 113 ITGAPLRIKQLITVK--PVLASKED----TENTRWLCALSGREISHQ-KAAVVKSTGQVI 165
++G PL++ LITV+ P+ S + T R++CA++ + + AV++ +G V+
Sbjct 1 MSGRPLKMSDLITVRFTPLDPSLDRVALLTRQDRYVCAVTKDTLGNSVPCAVLRPSGVVV 60
Query 166 LLEYLEKLVFGKKGA---LTSGVIDRKDTVALIPGGTGFSAHN-NVEVAVARPNVQ 217
+E +EKL+ KK +T + KD + + GGTGF+ +++ ARP +Q
Sbjct 61 TMECVEKLI--KKDMVDPITGDKLKEKDIIPIQRGGTGFAGSGVDLKAKEARPVMQ 114
> dre:559280 MGC171819; zgc:171819
Length=797
Score = 35.0 bits (79), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 19/38 (50%), Gaps = 0/38 (0%)
Query 108 GFCCPITGAPLRIKQLITVKPVLASKEDTENTRWLCAL 145
G C P APL QLI P+ A ED E W CA+
Sbjct 510 GLCLPKLLAPLIRHQLIGWNPLQAESEDFEALPWYCAV 547
> eco:b3440 yhhX, ECK3425, JW3403; predicted oxidoreductase with
NAD(P)-binding Rossmann-fold domain
Length=345
Score = 34.3 bits (77), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 36/57 (63%), Gaps = 4/57 (7%)
Query 156 AVVKSTGQVILLEYLEKLVFGKKGALTSGVIDRKDT---VALIPGGTGFSAHNNVEV 209
A+VK T ++ ++Y + +V GKKG+ ID+++T ++PG GF+A ++V V
Sbjct 221 AIVK-TSHLVKIDYPKFIVHGKKGSFIKYGIDQQETSLKANIMPGEPGFAADDSVGV 276
> ath:AT1G61620 phosphoinositide binding; K13125 nitric oxide
synthase-interacting protein
Length=310
Score = 32.0 bits (71), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 42/174 (24%), Positives = 71/174 (40%), Gaps = 23/174 (13%)
Query 52 SAAPAQT-KNSPQSAEEFRQTLEQPTINSKEEA--RAKNFWVSENTPDEPEAKLKEPQKG 108
SA P + KN + F T + +EEA K FW+ TP ++ P+
Sbjct 117 SAMPRNSDKNHNEDKNGFHGANSVKTTSFEEEALRTMKAFWLPSATP-AASVRVDAPETH 175
Query 109 FCCPITGAPLRIKQLITVKPVLASKEDTEN-----------TRWLCALSGREISHQKAAV 157
CP L++K L ++ + E+ E ++C +++ + V
Sbjct 176 TVCPEGKEKLKLKNLFAIRFTEDNSEEEETKTKSASSSSYDKSYICPSCKVTLTNTMSLV 235
Query 158 -VKSTGQVILLEYLEKLVFGKKGALTSGVIDR----KDTVALIPGGTGFSAHNN 206
+ S G V + EK + K L V D+ ++ V L GGTGF+ H++
Sbjct 236 ALSSCGHVFCKKCAEKFMPVDKVCL---VCDKPCKDRNLVGLKKGGTGFAEHDD 286
> sce:YMR167W MLH1, PMS2; Mlh1p; K08734 DNA mismatch repair protein
MLH1
Length=769
Score = 32.0 bits (71), Expect = 1.9, Method: Composition-based stats.
Identities = 23/93 (24%), Positives = 35/93 (37%), Gaps = 0/93 (0%)
Query 11 TKAWEALQAQKEQEAAAAAEANATAAQRAFLEAEASLTLETSAAPAQTKNSPQSAEEFRQ 70
T A L+ K QE + A +FL + E S+ Q + Q
Sbjct 383 TTANSQLRKAKRQENKLVRIDASQAKITSFLSSSQQFNFEGSSTKRQLSEPKVTNVSHSQ 442
Query 71 TLEQPTINSKEEARAKNFWVSENTPDEPEAKLK 103
E+ T+N E+ R N + D+P+ K K
Sbjct 443 EAEKLTLNESEQPRDANTINDNDLKDQPKKKQK 475
> mmu:76608 Hectd3, 1700064K09Rik, AI467540, AW743312; HECT domain
containing 3; K12233 E3 ubiquitin-protein ligase HECTD3
[EC:6.3.2.19]
Length=861
Score = 31.6 bits (70), Expect = 2.0, Method: Composition-based stats.
Identities = 32/96 (33%), Positives = 44/96 (45%), Gaps = 19/96 (19%)
Query 113 ITGAPLRIKQ-LITVKPVLASKEDTENTRWLCALSGREISHQKAAVVKSTGQVILLEYLE 171
+ GA LR K+ L+ P K+ LSG E+S K + V LLE +E
Sbjct 604 LMGAALRGKEFLVLALPGFVWKQ----------LSGEEVSWSKDFPAVDSVLVKLLEVME 653
Query 172 -------KLVFGKKGALTSGVIDRKDTVALIPGGTG 200
+ FGK+ T+ V+ + V LIPGGTG
Sbjct 654 GVDKETFEFKFGKELTFTT-VLSDQQVVELIPGGTG 688
> hsa:23072 HECW1, KIAA0322, NEDL1; HECT, C2 and WW domain containing
E3 ubiquitin protein ligase 1; K12167 E3 ubiquitin-protein
ligase HECW1 [EC:6.3.2.19]
Length=1606
Score = 30.4 bits (67), Expect = 4.5, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 25/55 (45%), Gaps = 0/55 (0%)
Query 17 LQAQKEQEAAAAAEANATAAQRAFLEAEASLTLETSAAPAQTKNSPQSAEEFRQT 71
L AQ +++ A A A Q E ++L LE APA TK P E Q+
Sbjct 441 LLAQVQKDIQPAPSAEELAEQLDLGEEASALLLEDGEAPASTKEEPLEEEATTQS 495
> ath:AT3G03780 AtMS2; AtMS2; 5-methyltetrahydropteroyltriglutamate-homocysteine
S-methyltransferase/ methionine synthase
(EC:2.1.1.14); K00549 5-methyltetrahydropteroyltriglutamate--homocysteine
methyltransferase [EC:2.1.1.14]
Length=765
Score = 30.0 bits (66), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 26/59 (44%), Gaps = 0/59 (0%)
Query 7 IQKETKAWEALQAQKEQEAAAAAEANATAAQRAFLEAEASLTLETSAAPAQTKNSPQSA 65
+ E K+W A AQK E A A+A A +F A A ++P T S Q A
Sbjct 343 LDAEIKSWLAFAAQKVVEVDALAKALAGQTNESFFTANADALSSRRSSPRVTNESVQKA 401
> hsa:79828 METTL8, FLJ13334, FLJ13984, FLJ31054, FLJ42098, FLJ77788,
TIP; methyltransferase like 8
Length=407
Score = 30.0 bits (66), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 10 ETKAWEALQAQKEQEAAAAAEANATAAQRAFLEAEASLTLETS 52
E W+ +Q KE+EAAA + +A R LE + E S
Sbjct 44 EHNMWDHMQWSKEEEAAARKKVKENSAVRVLLEEQVKYEREAS 86
> xla:444613 trim16, MGC84103, ebbp; tripartite motif containing
16; K12006 tripartite motif-containing protein 16
Length=563
Score = 29.6 bits (65), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 26/49 (53%), Gaps = 0/49 (0%)
Query 30 EANATAAQRAFLEAEASLTLETSAAPAQTKNSPQSAEEFRQTLEQPTIN 78
EAN T+ Q + LEA+ ++ L+ + K S EF +T E+ +N
Sbjct 195 EANTTSIQNSVLEAKRAIDLQFADVHGAVKKSHDKVREFLETRERAALN 243
> ath:AT5G42880 hypothetical protein
Length=751
Score = 29.3 bits (64), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 41/74 (55%), Gaps = 11/74 (14%)
Query 17 LQAQKEQEAAAAAEANATAAQRAFLEAEASLTLETSAAPAQTKNSPQ----SAEEFRQTL 72
++A+KE EAA A+E A AA +A E E+S E + NSP+ S EE+ + L
Sbjct 533 VEAKKEMEAARASEKLALAAIKALQETESSQRFE------EINNSPRSIIISVEEYYE-L 585
Query 73 EQPTINSKEEARAK 86
+ + S+EEA +
Sbjct 586 SKQALESEEEANTR 599
Lambda K H
0.310 0.124 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7009334380
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40