bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0838_orf3
Length=208
Score E
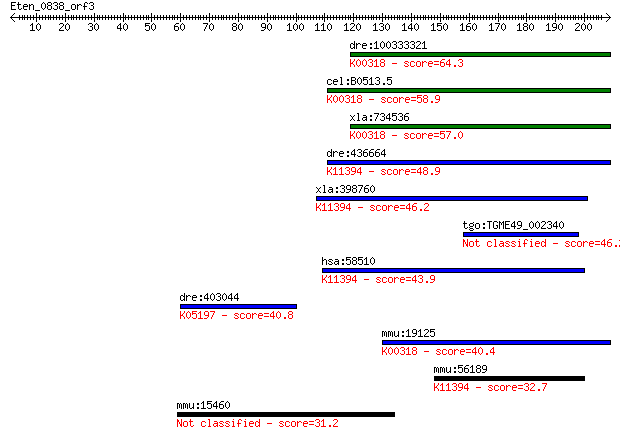
Sequences producing significant alignments: (Bits) Value
dre:100333321 prodha, im:7136007, wu:fc02a12; proline dehydrog... 64.3 3e-10
cel:B0513.5 hypothetical protein; K00318 proline dehydrogenase... 58.9 1e-08
xla:734536 prodh, MGC115247; proline dehydrogenase (oxidase) 1... 57.0 5e-08
dre:436664 wu:fb24e01; zgc:92040 (EC:1.5.99.8); K11394 hydroxy... 48.9 1e-05
xla:398760 prodh2, MGC68483; proline dehydrogenase (oxidase) 2... 46.2 9e-05
tgo:TGME49_002340 proline oxidase, putative (EC:1.5.99.8) 46.2 9e-05
hsa:58510 PRODH2, HSPOX1; proline dehydrogenase (oxidase) 2 (E... 43.9 4e-04
dre:403044 gria1b, Gria1, glur1b, gria1.2, sb:eu293; glutamate... 40.8 0.003
mmu:19125 Prodh, MGC159030, Pro-1, Pro1, Ym24d07; proline dehy... 40.4 0.004
mmu:56189 Prodh2, 2510028N04Rik, 2510038B11Rik, MmPOX, MmPOX1,... 32.7 0.88
mmu:15460 Hr, ALUNC, AU, N, ba, bldy, rh, rh-bmh, rhino; hairless 31.2 2.4
> dre:100333321 prodha, im:7136007, wu:fc02a12; proline dehydrogenase
(oxidase) 1a; K00318 proline dehydrogenase [EC:1.5.99.8]
Length=617
Score = 64.3 bits (155), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 51/90 (56%), Gaps = 0/90 (0%)
Query 119 DLLRSLFVFSLCRHHWLVDNWESLLNVSCRIFGSRAVFAFIRFSFFKVFCGGEALEEVVQ 178
+LLRSL VF LC +LVD + L+++S +I G R ++ +F+ F GE +
Sbjct 85 ELLRSLLVFKLCTFDFLVDKNKELIDLSKKILGQRLFEKLMKMTFYGQFVAGEDQNSIKP 144
Query 179 TLDKLKQRNLGAILDYAAEQQLPARAAARK 208
++K + +G++LDY+ E+ L A +K
Sbjct 145 LIEKNQAFGVGSVLDYSVEEDLTQEEAEKK 174
> cel:B0513.5 hypothetical protein; K00318 proline dehydrogenase
[EC:1.5.99.8]
Length=616
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 31/98 (31%), Positives = 52/98 (53%), Gaps = 0/98 (0%)
Query 111 ALRLYSMSDLLRSLFVFSLCRHHWLVDNWESLLNVSCRIFGSRAVFAFIRFSFFKVFCGG 170
A + S ++L+R+L V LC LV+ + +LN R+ G ++ +FF F G
Sbjct 67 AFKSKSNTELVRALVVLRLCGIQTLVNQNQIILNTMRRVLGKNLFKKTLKNTFFGHFVAG 126
Query 171 EALEEVVQTLDKLKQRNLGAILDYAAEQQLPARAAARK 208
E EEV ++KL+ + +ILDY+ E + ++ A K
Sbjct 127 ETEEEVRHVVEKLRNYGVKSILDYSVEADITSQEATDK 164
> xla:734536 prodh, MGC115247; proline dehydrogenase (oxidase)
1; K00318 proline dehydrogenase [EC:1.5.99.8]
Length=617
Score = 57.0 bits (136), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 27/90 (30%), Positives = 50/90 (55%), Gaps = 0/90 (0%)
Query 119 DLLRSLFVFSLCRHHWLVDNWESLLNVSCRIFGSRAVFAFIRFSFFKVFCGGEALEEVVQ 178
+LLRS+ VF LC LVD + +L++S ++ G + ++ +F+ F GE E +
Sbjct 85 ELLRSIMVFWLCSIDTLVDRNQQILSLSKKVLGQKLFEKMMKMTFYGQFVAGEDQESIKP 144
Query 179 TLDKLKQRNLGAILDYAAEQQLPARAAARK 208
+ + + +G++LDY+ E+ L A +K
Sbjct 145 LIRRNQAFGVGSVLDYSVEEDLTQEEAEKK 174
> dre:436664 wu:fb24e01; zgc:92040 (EC:1.5.99.8); K11394 hydroxyproline
oxidase [EC:1.5.-.-]
Length=465
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 48/98 (48%), Gaps = 0/98 (0%)
Query 111 ALRLYSMSDLLRSLFVFSLCRHHWLVDNWESLLNVSCRIFGSRAVFAFIRFSFFKVFCGG 170
A R+ S+ +L+R+L VF LC LV+N L++ S R+ G R +R S + F G
Sbjct 31 AFRVKSLWELIRALGVFRLCSFPVLVNNCGKLMSFSRRVLGKRCFCMVLRPSVYAQFVAG 90
Query 171 EALEEVVQTLDKLKQRNLGAILDYAAEQQLPARAAARK 208
E E+ ++ K+ L +L E+ L ++
Sbjct 91 ETEGEIADSMQKMSSLGLHPMLAVPIEEDLGESTGEKR 128
> xla:398760 prodh2, MGC68483; proline dehydrogenase (oxidase)
2 (EC:1.5.99.8); K11394 hydroxyproline oxidase [EC:1.5.-.-]
Length=466
Score = 46.2 bits (108), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 47/94 (50%), Gaps = 0/94 (0%)
Query 107 AGAAALRLYSMSDLLRSLFVFSLCRHHWLVDNWESLLNVSCRIFGSRAVFAFIRFSFFKV 166
+ +L S ++ R L +F +C LV + E +L+VS R+ G R ++ S +
Sbjct 45 SDGGVFKLKSSWEVARGLLIFRMCSFPSLVKHSEKMLSVSRRLLGRRLFEWGMKGSVYGQ 104
Query 167 FCGGEALEEVVQTLDKLKQRNLGAILDYAAEQQL 200
F GE L E+ +D+L+Q + +L E+ L
Sbjct 105 FVAGETLPEIRVCVDRLRQLGIHPMLAVPIEEDL 138
> tgo:TGME49_002340 proline oxidase, putative (EC:1.5.99.8)
Length=485
Score = 46.2 bits (108), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 28/40 (70%), Gaps = 0/40 (0%)
Query 158 FIRFSFFKVFCGGEALEEVVQTLDKLKQRNLGAILDYAAE 197
++ S +KVFCGGE +E++ T+ KL+ R + +LDYA E
Sbjct 3 LLKKSVYKVFCGGETEDELLATMQKLQSRGVKIVLDYAVE 42
> hsa:58510 PRODH2, HSPOX1; proline dehydrogenase (oxidase) 2
(EC:1.5.99.8); K11394 hydroxyproline oxidase [EC:1.5.-.-]
Length=536
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 43/91 (47%), Gaps = 0/91 (0%)
Query 109 AAALRLYSMSDLLRSLFVFSLCRHHWLVDNWESLLNVSCRIFGSRAVFAFIRFSFFKVFC 168
A L +L R+L V LC LV + L S R+ GSR AF+R S + F
Sbjct 102 GGAFHLKGTGELTRALLVLRLCAWPPLVTHGLLLQAWSRRLLGSRLSGAFLRASVYGQFV 161
Query 169 GGEALEEVVQTLDKLKQRNLGAILDYAAEQQ 199
GE EEV + +L+ +L +L E++
Sbjct 162 AGETAEEVKGCVQQLRTLSLRPLLAVPTEEE 192
> dre:403044 gria1b, Gria1, glur1b, gria1.2, sb:eu293; glutamate
receptor, ionotropic, AMPA 1b; K05197 glutamate receptor,
ionotropic, AMPA 1
Length=917
Score = 40.8 bits (94), Expect = 0.003, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 25/44 (56%), Gaps = 4/44 (9%)
Query 60 FAP----GQQRQQTHPQEESHQPPSPAPRAPPNQRQQQKQQLQQ 99
F+P ++T Q+ +QPPSP P PNQ Q Q+++ Q+
Sbjct 558 FSPYEWHADDEEETADQQNQNQPPSPEPNQSPNQSQNQREEKQE 601
> mmu:19125 Prodh, MGC159030, Pro-1, Pro1, Ym24d07; proline dehydrogenase
(EC:1.5.99.8); K00318 proline dehydrogenase [EC:1.5.99.8]
Length=599
Score = 40.4 bits (93), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 36/79 (45%), Gaps = 0/79 (0%)
Query 130 CRHHWLVDNWESLLNVSCRIFGSRAVFAFIRFSFFKVFCGGEALEEVVQTLDKLKQRNLG 189
C L+ + E L V+ ++ G R ++ +F+ F GE E + + K +G
Sbjct 79 CASPVLLAHHEQLFQVARKLLGQRMFERLMKMTFYGHFVAGEDQESIRPLIRHNKAFGVG 138
Query 190 AILDYAAEQQLPARAAARK 208
ILDY E+ L A RK
Sbjct 139 FILDYGVEEDLSPEEAERK 157
> mmu:56189 Prodh2, 2510028N04Rik, 2510038B11Rik, MmPOX, MmPOX1,
POX1; proline dehydrogenase (oxidase) 2 (EC:1.5.99.8); K11394
hydroxyproline oxidase [EC:1.5.-.-]
Length=456
Score = 32.7 bits (73), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 148 RIFGSRAVFAFIRFSFFKVFCGGEALEEVVQTLDKLKQRNLGAILDYAAEQQ 199
R+ GSR A +R S + F GE EEV + +L+ L +L E++
Sbjct 65 RLLGSRLSGALLRASIYGQFVAGETAEEVRNCVGQLQALGLQPLLAVPTEEE 116
> mmu:15460 Hr, ALUNC, AU, N, ba, bldy, rh, rh-bmh, rhino; hairless
Length=1182
Score = 31.2 bits (69), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 36/77 (46%), Gaps = 3/77 (3%)
Query 59 IFAPGQQRQQTHPQEESHQPPSPAPRAPPNQRQQQKQQLQQLVLTG--DAAGAAALRLYS 116
++APG QQ P E++ P P+ N+ + K++ + D AG + L S
Sbjct 693 VWAPGDGGQQKEPTEKTPPTPQPSCNGDSNRTKDIKEETPDSTESPAEDGAGRSPLPCPS 752
Query 117 MSDLLRSLFVFSLCRHH 133
+ +LL S V LC H
Sbjct 753 LCELLASTAV-KLCLGH 768
Lambda K H
0.324 0.134 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6511501916
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40