bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0882_orf2
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
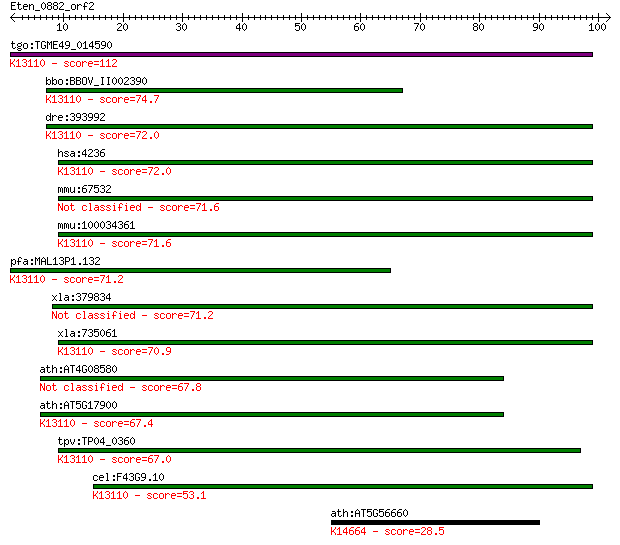
tgo:TGME49_014590 microfibrillar-associated protein 1, putativ... 112 3e-25
bbo:BBOV_II002390 18.m06195; micro-fibrillar-associated protei... 74.7 7e-14
dre:393992 mfap1, MGC56551, zgc:56551; microfibrillar-associat... 72.0 4e-13
hsa:4236 MFAP1; microfibrillar-associated protein 1; K13110 mi... 72.0 5e-13
mmu:67532 Mfap1a, 4432409M24Rik, Mfap1; microfibrillar-associa... 71.6 5e-13
mmu:100034361 Mfap1b; microfibrillar-associated protein 1B; K1... 71.6 5e-13
pfa:MAL13P1.132 microfibril-associated protein homologue, puta... 71.2 7e-13
xla:379834 mfap1, MGC53463; microfibrillar-associated protein 1 71.2
xla:735061 hypothetical protein MGC85054; K13110 microfibrilla... 70.9 1e-12
ath:AT4G08580 hypothetical protein 67.8 9e-12
ath:AT5G17900 hypothetical protein; K13110 microfibrillar-asso... 67.4 1e-11
tpv:TP04_0360 hypothetical protein; K13110 microfibrillar-asso... 67.0 2e-11
cel:F43G9.10 hypothetical protein; K13110 microfibrillar-assoc... 53.1 2e-07
ath:AT5G56660 ILL2; ILL2; IAA-Ala conjugate hydrolase/ IAA-ami... 28.5 4.8
> tgo:TGME49_014590 microfibrillar-associated protein 1, putative
; K13110 microfibrillar-associated protein 1
Length=438
Score = 112 bits (280), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 57/98 (58%), Positives = 70/98 (71%), Gaps = 2/98 (2%)
Query 1 QDLARSGEEPLYLWDFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDL 60
QDLARSGEE LYL D+NAPVGEDK DKK LP +++RRGE R G++KHTHLVDVDT+D
Sbjct 343 QDLARSGEEALYLRDYNAPVGEDKWDKKILPSAMRVRRGEFGRQGQTKHTHLVDVDTTDF 402
Query 61 SHPWAQATRDLQGQKLLKKAAGIKGANDFERPSLRKPK 98
+ WA T+ +K K G KGA +F+RP+ R K
Sbjct 403 TSAWAFNTK--IKEKTDSKMGGRKGAKEFDRPAARPKK 438
> bbo:BBOV_II002390 18.m06195; micro-fibrillar-associated protein
1 C-terminus containing protein; K13110 microfibrillar-associated
protein 1
Length=437
Score = 74.7 bits (182), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 31/60 (51%), Positives = 41/60 (68%), Gaps = 0/60 (0%)
Query 7 GEEPLYLWDFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDLSHPWAQ 66
G EP+Y DFNAP +D VDK +PK +Q+RRG+ + GRSK+THL DT+ PW+Q
Sbjct 353 GSEPIYKRDFNAPTADDCVDKSLMPKSMQVRRGQYGKMGRSKYTHLTAEDTTKFDMPWSQ 412
> dre:393992 mfap1, MGC56551, zgc:56551; microfibrillar-associated
protein 1; K13110 microfibrillar-associated protein 1
Length=437
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 39/93 (41%), Positives = 57/93 (61%), Gaps = 7/93 (7%)
Query 7 GEEPLYLWDFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDLSHPWAQ 66
GE+ ++ DF+AP ED +K LPK++Q++ R GR+K+THLVD DT+ WAQ
Sbjct 350 GEQDVFKRDFSAPTLEDHFNKTILPKVMQVK--NFGRSGRTKYTHLVDQDTTSFDSAWAQ 407
Query 67 ATRDLQGQKLLKKAAGIKGAND-FERPSLRKPK 98
+ Q K K+ AG G D F+RP+++K K
Sbjct 408 ES--AQNSKFFKQKAG--GVRDVFDRPTVQKRK 436
> hsa:4236 MFAP1; microfibrillar-associated protein 1; K13110
microfibrillar-associated protein 1
Length=439
Score = 72.0 bits (175), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 40/91 (43%), Positives = 54/91 (59%), Gaps = 7/91 (7%)
Query 9 EPLYLWDFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDLSHPWAQAT 68
E +Y DF+AP ED +K LPK++Q++ R GR+K+THLVD DT+ W Q +
Sbjct 353 EEVYKRDFSAPTLEDHFNKTILPKVMQVK--NFGRSGRTKYTHLVDQDTTSFDSAWGQES 410
Query 69 RDLQGQKLLK-KAAGIKGANDFERPSLRKPK 98
Q K K KAAG++ FERPS +K K
Sbjct 411 --AQNTKFFKQKAAGVRDV--FERPSAKKRK 437
> mmu:67532 Mfap1a, 4432409M24Rik, Mfap1; microfibrillar-associated
protein 1A
Length=439
Score = 71.6 bits (174), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 40/91 (43%), Positives = 54/91 (59%), Gaps = 7/91 (7%)
Query 9 EPLYLWDFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDLSHPWAQAT 68
E +Y DF+AP ED +K LPK++Q++ R GR+K+THLVD DT+ W Q +
Sbjct 353 EEVYKRDFSAPTLEDHFNKTILPKVMQVK--NFGRSGRTKYTHLVDQDTTSFDSAWGQES 410
Query 69 RDLQGQKLLK-KAAGIKGANDFERPSLRKPK 98
Q K K KAAG++ FERPS +K K
Sbjct 411 --AQNTKFFKQKAAGVRDV--FERPSAKKRK 437
> mmu:100034361 Mfap1b; microfibrillar-associated protein 1B;
K13110 microfibrillar-associated protein 1
Length=439
Score = 71.6 bits (174), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 40/91 (43%), Positives = 54/91 (59%), Gaps = 7/91 (7%)
Query 9 EPLYLWDFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDLSHPWAQAT 68
E +Y DF+AP ED +K LPK++Q++ R GR+K+THLVD DT+ W Q +
Sbjct 353 EEVYKRDFSAPTLEDHFNKTILPKVMQVK--NFGRSGRTKYTHLVDQDTTSFDSAWGQES 410
Query 69 RDLQGQKLLK-KAAGIKGANDFERPSLRKPK 98
Q K K KAAG++ FERPS +K K
Sbjct 411 --AQNTKFFKQKAAGVRDV--FERPSAKKRK 437
> pfa:MAL13P1.132 microfibril-associated protein homologue, putative;
K13110 microfibrillar-associated protein 1
Length=492
Score = 71.2 bits (173), Expect = 7e-13, Method: Composition-based stats.
Identities = 35/64 (54%), Positives = 47/64 (73%), Gaps = 0/64 (0%)
Query 1 QDLARSGEEPLYLWDFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDL 60
QDL G+E +YL D+N PV EDKVD++ LPK+LQ+RRG+ + G+SK+THL+D DTS
Sbjct 403 QDLFEEGKEEIYLRDYNEPVYEDKVDRQNLPKVLQVRRGKFGKQGQSKYTHLLDNDTSRK 462
Query 61 SHPW 64
W
Sbjct 463 DSLW 466
> xla:379834 mfap1, MGC53463; microfibrillar-associated protein
1
Length=441
Score = 71.2 bits (173), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 40/92 (43%), Positives = 56/92 (60%), Gaps = 7/92 (7%)
Query 8 EEPLYLWDFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDLSHPWAQA 67
+E +Y DF+AP ED +K LPK++Q++ R GR+K+THLVD DT+ W Q
Sbjct 354 DENVYKRDFSAPTLEDHFNKTILPKVMQVK--NFGRSGRTKYTHLVDQDTTSFDSAWGQD 411
Query 68 TRDLQGQKLLK-KAAGIKGANDFERPSLRKPK 98
+ Q K K KAAG++ FERPS++K K
Sbjct 412 --NPQNTKFFKQKAAGVRDV--FERPSVQKRK 439
> xla:735061 hypothetical protein MGC85054; K13110 microfibrillar-associated
protein 1
Length=442
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 40/91 (43%), Positives = 55/91 (60%), Gaps = 7/91 (7%)
Query 9 EPLYLWDFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDLSHPWAQAT 68
E +Y DF+AP ED +K LPK++Q++ R GR+K+THLVD DT+ W Q
Sbjct 356 ENVYKRDFSAPTLEDHFNKTILPKVMQVK--NFGRSGRTKYTHLVDQDTTSFDSAWGQD- 412
Query 69 RDLQGQKLLK-KAAGIKGANDFERPSLRKPK 98
+ Q K K KAAG++ FERPS++K K
Sbjct 413 -NPQNTKFFKQKAAGVRDV--FERPSVQKRK 440
> ath:AT4G08580 hypothetical protein
Length=435
Score = 67.8 bits (164), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 34/78 (43%), Positives = 52/78 (66%), Gaps = 4/78 (5%)
Query 6 SGEEPLYLWDFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDLSHPWA 65
+G + ++ DF+AP GED++DK LPK++Q++ R GR+K THLV+ DT+D S+PW
Sbjct 344 AGTDGIFQRDFSAPTGEDRLDKSILPKVMQVK--HFGRSGRTKWTHLVNEDTTDWSNPW- 400
Query 66 QATRDLQGQKLLKKAAGI 83
+ D +K KK AG+
Sbjct 401 -TSNDPLREKYNKKMAGM 417
> ath:AT5G17900 hypothetical protein; K13110 microfibrillar-associated
protein 1
Length=435
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 34/78 (43%), Positives = 52/78 (66%), Gaps = 4/78 (5%)
Query 6 SGEEPLYLWDFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDLSHPWA 65
+G + ++ DF+AP GED++DK LPK++Q++ R GR+K THLV+ DT+D S+PW
Sbjct 344 AGTDGIFQRDFSAPTGEDRLDKSILPKVMQVK--HFGRSGRTKWTHLVNEDTTDWSNPW- 400
Query 66 QATRDLQGQKLLKKAAGI 83
+ D +K KK AG+
Sbjct 401 -TSNDPLREKYNKKMAGM 417
> tpv:TP04_0360 hypothetical protein; K13110 microfibrillar-associated
protein 1
Length=408
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 39/88 (44%), Positives = 50/88 (56%), Gaps = 11/88 (12%)
Query 9 EPLYLWDFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDLSHPWAQAT 68
EPLY DFNAP ED VDK LPK +++RRG + G+ KHTHL DVDT+ W++
Sbjct 332 EPLYARDFNAPTAEDCVDKSLLPKPMRVRRGLYGKQGQVKHTHLKDVDTTQFD-AWSKTD 390
Query 69 RDLQGQKLLKKAAGIKGANDFERPSLRK 96
+ K G K F+RPS +K
Sbjct 391 K--------YKLTGTKQV--FDRPSKKK 408
> cel:F43G9.10 hypothetical protein; K13110 microfibrillar-associated
protein 1
Length=466
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/84 (35%), Positives = 47/84 (55%), Gaps = 5/84 (5%)
Query 15 DFNAPVGEDKVDKKTLPKILQLRRGELARGGRSKHTHLVDVDTSDLSHPWAQATRDLQGQ 74
+F +D+ DK LPK++Q++ + R+K+THL + DT+D WA +T L Q
Sbjct 387 NFAEATNDDQFDKTILPKVMQVK--NFGKASRTKYTHLTEEDTTDHQGVWA-STNQLNSQ 443
Query 75 KLLKKAAGIKGANDFERPSLRKPK 98
K+A G + FERP+ +K K
Sbjct 444 FSTKRAGGSRPV--FERPATKKRK 465
> ath:AT5G56660 ILL2; ILL2; IAA-Ala conjugate hydrolase/ IAA-amino
acid conjugate hydrolase/ metallopeptidase; K14664 IAA-amino
acid hydrolase [EC:3.5.1.-]
Length=439
Score = 28.5 bits (62), Expect = 4.8, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 55 VDTSDLSHPWAQATRDLQGQKLLKKAAGIKGANDF 89
V+ DL + + RDL GQ+ +AA + G+ DF
Sbjct 336 VNNKDLYKQFKKVVRDLLGQEAFVEAAPVMGSEDF 370
Lambda K H
0.313 0.133 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2036602604
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40