bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0903_orf1
Length=162
Score E
Sequences producing significant alignments: (Bits) Value
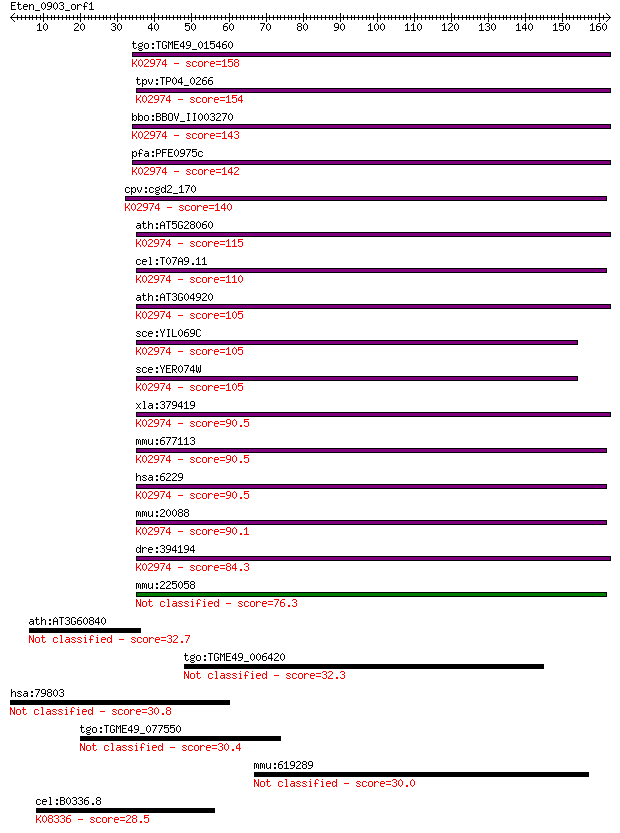
tgo:TGME49_015460 40S ribosomal protein S24, putative ; K02974... 158 7e-39
tpv:TP04_0266 40S ribosomal protein S24; K02974 small subunit ... 154 1e-37
bbo:BBOV_II003270 18.m06275; 40S ribosomal protein S24 protein... 143 3e-34
pfa:PFE0975c 40S ribosomal protein S24, putative; K02974 small... 142 6e-34
cpv:cgd2_170 40s ribosomal protein s24 ; K02974 small subunit ... 140 2e-33
ath:AT5G28060 40S ribosomal protein S24 (RPS24B); K02974 small... 115 5e-26
cel:T07A9.11 rps-24; Ribosomal Protein, Small subunit family m... 110 2e-24
ath:AT3G04920 40S ribosomal protein S24 (RPS24A); K02974 small... 105 7e-23
sce:YIL069C RPS24B, RPS24EB; Rps24bp; K02974 small subunit rib... 105 8e-23
sce:YER074W RPS24A, RPS24EA; Rps24ap; K02974 small subunit rib... 105 8e-23
xla:379419 rps24, MGC64320; ribosomal protein S24; K02974 smal... 90.5 2e-18
mmu:677113 40S ribosomal protein S24-like; K02974 small subuni... 90.5 2e-18
hsa:6229 RPS24, DBA3, DKFZp686N1586; ribosomal protein S24; K0... 90.5 2e-18
mmu:20088 Rps24, MGC107549; ribosomal protein S24; K02974 smal... 90.1 3e-18
dre:394194 rps24, wu:fa66b02, zgc:103596, zgc:109740; ribosoma... 84.3 2e-16
mmu:225058 Gm4832, EG225058; predicted gene 4832 76.3
ath:AT3G60840 MAP65-4 (MICROTUBULE-ASSOCIATED PROTEIN 65-4) 32.7 0.61
tgo:TGME49_006420 myosin head motor domain-containing protein 32.3 0.72
hsa:79803 HPS6, FLJ22501, MGC20522, RP11-302K17.1; Hermansky-P... 30.8 2.2
tgo:TGME49_077550 uvrD/REP helicase domain-containing protein ... 30.4 2.7
mmu:619289 Rfx8, 4933400N17Rik; regulatory factor X 8 30.0
cel:B0336.8 lgg-3; LC3, GABARAP and GATE-16 family member (lgg... 28.5 8.9
> tgo:TGME49_015460 40S ribosomal protein S24, putative ; K02974
small subunit ribosomal protein S24e
Length=135
Score = 158 bits (400), Expect = 7e-39, Method: Compositional matrix adjust.
Identities = 77/129 (59%), Positives = 100/129 (77%), Gaps = 0/129 (0%)
Query 34 FTLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTA 93
F+LR RKL++NPLLQRKQ VD+LH R +VS+KEL +++A+QF+V D + +V+ +TA
Sbjct 7 FSLRFRKLKTNPLLQRKQFGVDILHPSRGSVSKKELVEKIAKQFRVSDSQSIVVFGLKTA 66
Query 94 FGGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEK 153
FGG RS G A+IY++ AA++FE RFRLVR+G EA+P KGRRA KELKNRRKKVRG EK
Sbjct 67 FGGGRSSGFAMIYDNPNAAKKFENRFRLVRLGYVEAKPTKGRRAYKELKNRRKKVRGKEK 126
Query 154 AKTQAGKQK 162
AK +K
Sbjct 127 AKCSGAAKK 135
> tpv:TP04_0266 40S ribosomal protein S24; K02974 small subunit
ribosomal protein S24e
Length=135
Score = 154 bits (389), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 75/128 (58%), Positives = 98/128 (76%), Gaps = 0/128 (0%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
+R++K SNP+L RKQ ++VLH GR+NVS+ EL++RLA+QFKV D + +VL +T F
Sbjct 8 VVRVKKFLSNPILSRKQFSIEVLHPGRSNVSKTELKERLAKQFKVKDSKTVVLFGLKTLF 67
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEKA 154
GG RS G LIY++ + +R+ER +RLVR+G EA+PK GRRA KELKNRRKKVRG EKA
Sbjct 68 GGGRSTGFGLIYDNLASLKRYERSYRLVRLGIEEAKPKLGRRAKKELKNRRKKVRGKEKA 127
Query 155 KTQAGKQK 162
K AGK+K
Sbjct 128 KCTAGKKK 135
> bbo:BBOV_II003270 18.m06275; 40S ribosomal protein S24 protein;
K02974 small subunit ribosomal protein S24e
Length=135
Score = 143 bits (360), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 70/129 (54%), Positives = 93/129 (72%), Gaps = 0/129 (0%)
Query 34 FTLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTA 93
F++R++K +NPLL RKQ V+VLH GRANVS+ +L++R+A+QFKV DP+ +VL F+T
Sbjct 7 FSIRVKKFLTNPLLSRKQFGVEVLHPGRANVSKNDLRERIAKQFKVKDPKTIVLFGFKTY 66
Query 94 FGGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEK 153
FGG S G LIY+ +++E +R VR+G E GRRA KELKNRRKKVRG +K
Sbjct 67 FGGGMSSGYCLIYDTLANLKKYELHYRQVRLGLEEPSKGMGRRAKKELKNRRKKVRGKDK 126
Query 154 AKTQAGKQK 162
AK AGK+K
Sbjct 127 AKCSAGKKK 135
> pfa:PFE0975c 40S ribosomal protein S24, putative; K02974 small
subunit ribosomal protein S24e
Length=133
Score = 142 bits (357), Expect = 6e-34, Method: Compositional matrix adjust.
Identities = 65/129 (50%), Positives = 97/129 (75%), Gaps = 0/129 (0%)
Query 34 FTLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTA 93
FT+R++K SNPLL+RKQ +++LH + +V++KE+++RLA+ +K+ + +VL F+T
Sbjct 5 FTIRVKKYMSNPLLRRKQFALEILHPNKGSVAKKEVKERLAKMYKLNNVNTIVLFGFKTL 64
Query 94 FGGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEK 153
FGG R++G LIY++ A ++FE+++RLVR G + E K GRRA+KELKNRRKKVRGTEK
Sbjct 65 FGGGRTKGFGLIYKNVDAVKKFEKKYRLVREGLIDKETKAGRRASKELKNRRKKVRGTEK 124
Query 154 AKTQAGKQK 162
K K+K
Sbjct 125 TKVSGAKKK 133
> cpv:cgd2_170 40s ribosomal protein s24 ; K02974 small subunit
ribosomal protein S24e
Length=159
Score = 140 bits (352), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 68/130 (52%), Positives = 89/130 (68%), Gaps = 0/130 (0%)
Query 32 SPFTLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFR 91
+ FT+R+R+ +NPLL RKQ VDV+H VS+ +L+ +LA+ +KV D C+VL F+
Sbjct 30 NNFTVRVRQFLNNPLLARKQFVVDVIHPSLGGVSKSDLKAKLAKLYKVQDANCIVLFGFK 89
Query 92 TAFGGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGT 151
TAFGG RS G +IY + A ++FE R+R VRMG AE GR+ KE KNRRKKVRGT
Sbjct 90 TAFGGGRSSGFCVIYNNISALKKFEHRYRQVRMGIAEKVTATGRKGRKETKNRRKKVRGT 149
Query 152 EKAKTQAGKQ 161
EKAK GK+
Sbjct 150 EKAKISGGKK 159
> ath:AT5G28060 40S ribosomal protein S24 (RPS24B); K02974 small
subunit ribosomal protein S24e
Length=133
Score = 115 bits (289), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 59/128 (46%), Positives = 84/128 (65%), Gaps = 1/128 (0%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R R +N LL RKQ +DVLH GRANVS+ EL+++LA+ ++V DP + FRT F
Sbjct 7 TIRTRNFMTNRLLARKQFVIDVLHPGRANVSKAELKEKLARMYEVKDPNAIFCFKFRTHF 66
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEKA 154
GG +S G LIY+ A++FE ++RL+R G + + +K R+ KE KNR KK+RG +K
Sbjct 67 GGGKSSGYGLIYDTVENAKKFEPKYRLIRNG-LDTKIEKSRKQIKERKNRAKKIRGVKKT 125
Query 155 KTQAGKQK 162
K K+K
Sbjct 126 KAGDTKKK 133
> cel:T07A9.11 rps-24; Ribosomal Protein, Small subunit family
member (rps-24); K02974 small subunit ribosomal protein S24e
Length=131
Score = 110 bits (276), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 57/127 (44%), Positives = 86/127 (67%), Gaps = 1/127 (0%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R RK+ +N LL RKQ+ V+V+H GR V + ++++++A+ +K P ++ F +
Sbjct 6 TIRTRKVLTNKLLYRKQMVVEVIHPGRPTVPKADIREKIAKLYKT-TPDTVIPFGFESKI 64
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEKA 154
GG +S+G AL+Y+ A++FE ++RLVRMG A K GR+ KE KNR+KKVRGT KA
Sbjct 65 GGGKSKGFALVYDTIDFAKKFEPKYRLVRMGLATKVEKPGRKQRKERKNRQKKVRGTAKA 124
Query 155 KTQAGKQ 161
K AGK+
Sbjct 125 KVSAGKK 131
> ath:AT3G04920 40S ribosomal protein S24 (RPS24A); K02974 small
subunit ribosomal protein S24e
Length=133
Score = 105 bits (262), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 60/128 (46%), Positives = 87/128 (67%), Gaps = 1/128 (0%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R RK +N LL RKQ +DVLH GRANVS+ EL+++LA+ ++V DP + + FRT F
Sbjct 7 TIRTRKFMTNRLLSRKQFVIDVLHPGRANVSKAELKEKLARMYEVKDPNAIFVFKFRTHF 66
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEKA 154
GG +S G LIY+ +A++FE ++RL+R G + + +K R+ KE KNR KK+RG +K
Sbjct 67 GGGKSSGFGLIYDTVESAKKFEPKYRLIRNG-LDTKIEKSRKQIKERKNRAKKIRGVKKT 125
Query 155 KTQAGKQK 162
K K+K
Sbjct 126 KAGDAKKK 133
> sce:YIL069C RPS24B, RPS24EB; Rps24bp; K02974 small subunit ribosomal
protein S24e
Length=135
Score = 105 bits (261), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 58/119 (48%), Positives = 77/119 (64%), Gaps = 1/119 (0%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R RK+ SNPLL RKQ VDVLH RANVS+ EL+++LA+ +K + + + FRT F
Sbjct 6 TIRTRKVISNPLLARKQFVVDVLHPNRANVSKDELREKLAEVYK-AEKDAVSVFGFRTQF 64
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEK 153
GG +S G L+Y A++FE +RLVR G AE K R+ K+ KNR KK+ GT K
Sbjct 65 GGGKSVGFGLVYNSVAEAKKFEPTYRLVRYGLAEKVEKASRQQRKQKKNRDKKIFGTGK 123
> sce:YER074W RPS24A, RPS24EA; Rps24ap; K02974 small subunit ribosomal
protein S24e
Length=135
Score = 105 bits (261), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 58/119 (48%), Positives = 77/119 (64%), Gaps = 1/119 (0%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R RK+ SNPLL RKQ VDVLH RANVS+ EL+++LA+ +K + + + FRT F
Sbjct 6 TIRTRKVISNPLLARKQFVVDVLHPNRANVSKDELREKLAEVYK-AEKDAVSVFGFRTQF 64
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEK 153
GG +S G L+Y A++FE +RLVR G AE K R+ K+ KNR KK+ GT K
Sbjct 65 GGGKSVGFGLVYNSVAEAKKFEPTYRLVRYGLAEKVEKASRQQRKQKKNRDKKIFGTGK 123
> xla:379419 rps24, MGC64320; ribosomal protein S24; K02974 small
subunit ribosomal protein S24e
Length=132
Score = 90.5 bits (223), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 57/128 (44%), Positives = 82/128 (64%), Gaps = 2/128 (1%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R RK +N LLQRKQ+ +DVLH G+A V + E++++LA+ +K P + + FRT F
Sbjct 6 TIRTRKFMTNRLLQRKQMVIDVLHPGKATVPKTEIREKLAKMYKTT-PDVIFVFGFRTHF 64
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEKA 154
GG ++ G +IY+ A++ E + RL + G E + K R+ KE KNR KKVRGT KA
Sbjct 65 GGGKTTGFGMIYDSLDYAKKNEPKHRLAKHGLYEKK-KTSRKQRKERKNRMKKVRGTAKA 123
Query 155 KTQAGKQK 162
AGK+K
Sbjct 124 NVGAGKKK 131
> mmu:677113 40S ribosomal protein S24-like; K02974 small subunit
ribosomal protein S24e
Length=133
Score = 90.5 bits (223), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 57/127 (44%), Positives = 81/127 (63%), Gaps = 2/127 (1%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R RK +N LLQRKQ+ +DVLH G+A V + E++++LA+ +K P + + FRT F
Sbjct 6 TIRTRKFMTNRLLQRKQMVIDVLHPGKATVPKTEIREKLAKMYKTT-PDVIFVFGFRTHF 64
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEKA 154
GG ++ G +IY+ A++ E + RL R G E + K R+ KE KNR KKVRGT KA
Sbjct 65 GGGKTTGFGMIYDSLDYAKKNEPKHRLARHGLYEKK-KTSRKQRKERKNRMKKVRGTAKA 123
Query 155 KTQAGKQ 161
AGK+
Sbjct 124 NVGAGKK 130
> hsa:6229 RPS24, DBA3, DKFZp686N1586; ribosomal protein S24;
K02974 small subunit ribosomal protein S24e
Length=133
Score = 90.5 bits (223), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 57/127 (44%), Positives = 81/127 (63%), Gaps = 2/127 (1%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R RK +N LLQRKQ+ +DVLH G+A V + E++++LA+ +K P + + FRT F
Sbjct 6 TIRTRKFMTNRLLQRKQMVIDVLHPGKATVPKTEIREKLAKMYKTT-PDVIFVFGFRTHF 64
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEKA 154
GG ++ G +IY+ A++ E + RL R G E + K R+ KE KNR KKVRGT KA
Sbjct 65 GGGKTTGFGMIYDSLDYAKKNEPKHRLARHGLYEKK-KTSRKQRKERKNRMKKVRGTAKA 123
Query 155 KTQAGKQ 161
AGK+
Sbjct 124 NVGAGKK 130
> mmu:20088 Rps24, MGC107549; ribosomal protein S24; K02974 small
subunit ribosomal protein S24e
Length=130
Score = 90.1 bits (222), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 57/127 (44%), Positives = 81/127 (63%), Gaps = 2/127 (1%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R RK +N LLQRKQ+ +DVLH G+A V + E++++LA+ +K P + + FRT F
Sbjct 6 TIRTRKFMTNRLLQRKQMVIDVLHPGKATVPKTEIREKLAKMYKTT-PDVIFVFGFRTHF 64
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEKA 154
GG ++ G +IY+ A++ E + RL R G E + K R+ KE KNR KKVRGT KA
Sbjct 65 GGGKTTGFGMIYDSLDYAKKNEPKHRLARHGLYEKK-KTSRKQRKERKNRMKKVRGTAKA 123
Query 155 KTQAGKQ 161
AGK+
Sbjct 124 NVGAGKK 130
> dre:394194 rps24, wu:fa66b02, zgc:103596, zgc:109740; ribosomal
protein S24; K02974 small subunit ribosomal protein S24e
Length=131
Score = 84.3 bits (207), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 55/128 (42%), Positives = 81/128 (63%), Gaps = 2/128 (1%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
T+R RK +N LLQRKQ+ VDVLH G+A V + E++++LA+ +K P + + F+T F
Sbjct 6 TVRTRKFMTNRLLQRKQMIVDVLHPGKATVPKTEIREKLAKMYKTT-PDVVFVFGFKTQF 64
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEKA 154
GG ++ G +IY+ A++ E ++RL R G E + K R+ KE KNR KKVRG +KA
Sbjct 65 GGGKTTGFGMIYDSLDYAKKNEPKYRLQRHGLYEKK-KTSRKQRKERKNRMKKVRGIKKA 123
Query 155 KTQAGKQK 162
A +K
Sbjct 124 SVGASGKK 131
> mmu:225058 Gm4832, EG225058; predicted gene 4832
Length=133
Score = 76.3 bits (186), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 52/127 (40%), Positives = 78/127 (61%), Gaps = 2/127 (1%)
Query 35 TLRLRKLQSNPLLQRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAF 94
TLR RK +N LLQRKQ+ + VLH G+A V + E++++L++ +K P + + +RT F
Sbjct 6 TLRTRKFMTNRLLQRKQVVIGVLHPGKATVLKTEIREKLSRMYKTT-PDVIFVFGYRTHF 64
Query 95 GGARSRGQALIYEDFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNRRKKVRGTEKA 154
GG ++ G + Y+ A++ E + L R G E + K R+ KE KNR KKV+GT KA
Sbjct 65 GGGKTTGFGMSYDSLDYAKKNEPKHGLARHGFYE-KKKTSRKQRKERKNRIKKVKGTTKA 123
Query 155 KTQAGKQ 161
AGK+
Sbjct 124 NDGAGKK 130
> ath:AT3G60840 MAP65-4 (MICROTUBULE-ASSOCIATED PROTEIN 65-4)
Length=648
Score = 32.7 bits (73), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 6 APQTHKAVRKEAPAPPAAAAAMSEPKSPFT 35
P+T++ ++ P PAA+ AM+E +PFT
Sbjct 593 TPKTNEEKKRAVPTTPAASVAMTEATTPFT 622
> tgo:TGME49_006420 myosin head motor domain-containing protein
Length=1387
Score = 32.3 bits (72), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 36/97 (37%), Gaps = 6/97 (6%)
Query 48 QRKQLWVDVLHLGRANVSRKELQQRLAQQFKVPDPRCLVLCCFRTAFGGARSRGQALIYE 107
QR+ LW D + S QQR A P+ C LC R GGA ++
Sbjct 1295 QRRALWTDSREQTKTQTSHNNQQQRSAAPMPTPETWC-TLCWLRMQSGGAGGSSAMCVWA 1353
Query 108 DFRAAQRFERRFRLVRMGAAEAEPKKGRRATKELKNR 144
A+ R +G E + RR TK NR
Sbjct 1354 PNSASPS-----RSKEVGHEERRSQHKRRKTKLQTNR 1385
> hsa:79803 HPS6, FLJ22501, MGC20522, RP11-302K17.1; Hermansky-Pudlak
syndrome 6
Length=775
Score = 30.8 bits (68), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 33/70 (47%), Gaps = 11/70 (15%)
Query 1 GGTRAAPQTHKAVR-----KEAPAPPAAAAAMSEPKSPF------TLRLRKLQSNPLLQR 49
GGT + Q+H R +EAP P +AA+ + TL L + S LL+R
Sbjct 286 GGTVSLLQSHGGTRAVGTLQEAPVGPWGSAALGTFQGTLACVLGSTLELLDMGSGQLLER 345
Query 50 KQLWVDVLHL 59
K L D +HL
Sbjct 346 KVLSTDRVHL 355
> tgo:TGME49_077550 uvrD/REP helicase domain-containing protein
(EC:3.1.11.5 3.4.21.72)
Length=2851
Score = 30.4 bits (67), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 22/55 (40%), Positives = 29/55 (52%), Gaps = 3/55 (5%)
Query 20 PPAAAAAMSEPKSPFTLRLRKLQSNPLLQ-RKQLWVDVLHLGRANVSRKELQQRL 73
PPA A +S PK+ LR + + L Q RK W D + L R N KEL++ L
Sbjct 1449 PPAMCAVLSSPKAEAAYILRFIVA--LKQTRKLCWGDFVILSRTNRGLKELEELL 1501
> mmu:619289 Rfx8, 4933400N17Rik; regulatory factor X 8
Length=596
Score = 30.0 bits (66), Expect = 3.3, Method: Composition-based stats.
Identities = 27/100 (27%), Positives = 40/100 (40%), Gaps = 10/100 (10%)
Query 67 KELQQRLAQQFKVPDPRCLVLCCFRTAFGGARSRGQALIYED-----FRAAQRFERRFRL 121
K QQ +PD R L+ C + G ED ++ + F +RF+L
Sbjct 207 KSRQQDTVMLMSLPDVRQLLGCYDMQLYKGLEDMLLPDFLEDVSIQYLKSVRLFSKRFKL 266
Query 122 VRMGAAEAEP-----KKGRRATKELKNRRKKVRGTEKAKT 156
+GA E P K + T +K R+K + AKT
Sbjct 267 WLLGALEGFPAILQISKLKEVTGFVKRLRRKTYLSNMAKT 306
> cel:B0336.8 lgg-3; LC3, GABARAP and GATE-16 family member (lgg-3);
K08336 autophagy-related protein 12
Length=118
Score = 28.5 bits (62), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 8 QTHKAVRKEAPAPPAAAAAMSEPKSP-FTLRLRKLQSNPLLQRKQLWVD 55
+T A P AAA+ PKS T+RLR + P+L+ K++ V+
Sbjct 2 ETETATTPTGNTEPTAAASAEPPKSDKVTVRLRNIADAPVLKNKKMVVN 50
Lambda K H
0.320 0.132 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3767900632
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40