bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0904_orf1
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
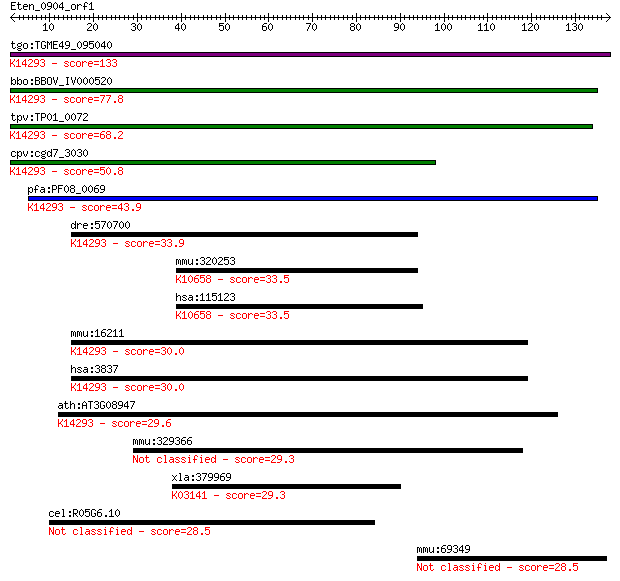
tgo:TGME49_095040 importin subunit beta-1, putative ; K14293 i... 133 2e-31
bbo:BBOV_IV000520 21.m02834; importin beta subunit; K14293 imp... 77.8 1e-14
tpv:TP01_0072 importin beta; K14293 importin subunit beta-1 68.2 8e-12
cpv:cgd7_3030 importin/karyopherin ; K14293 importin subunit b... 50.8 1e-06
pfa:PF08_0069 importin beta, putative; K14293 importin subunit... 43.9 2e-04
dre:570700 kpnb1, MGC123184, wu:fc12b10, zgc:123184; karyopher... 33.9 0.17
mmu:320253 March3, 6330411I15Rik, A530081L18Rik, AI843639, MAR... 33.5 0.21
hsa:115123 MARCH3, FLJ20096, MARCH-III, MGC48332, RNF173; memb... 33.5 0.21
mmu:16211 Kpnb1, AA409963, IPOB, Impnb, MGC8315; karyopherin (... 30.0 2.2
hsa:3837 KPNB1, IMB1, IPO1, IPOB, Impnb, MGC2155, MGC2156, MGC... 30.0 2.2
ath:AT3G08947 binding / protein transporter; K14293 importin s... 29.6 2.9
mmu:329366 4932418E24Rik; RIKEN cDNA 4932418E24 gene 29.3 3.4
xla:379969 gtf2h1, MGC53309; general transcription factor IIH,... 29.3 3.7
cel:R05G6.10 hypothetical protein 28.5 6.1
mmu:69349 1700008O03Rik; RIKEN cDNA 1700008O03 gene 28.5 6.6
> tgo:TGME49_095040 importin subunit beta-1, putative ; K14293
importin subunit beta-1
Length=971
Score = 133 bits (334), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 70/139 (50%), Positives = 89/139 (64%), Gaps = 2/139 (1%)
Query 1 TPYDAGPPNWQLNEEWIWYVHELRSATLEALASCVYALKEAQLQEVLRKEVNGMLQVVKA 60
T YD GP NWQ+NEEW+WY+H+LR L+A S VY+ KE +QE L+ VN ML VVKA
Sbjct 833 TTYDVGPSNWQVNEEWLWYIHDLREGVLQAYMSIVYSFKEKCMQEQLKLYVNAMLDVVKA 892
Query 61 VAEAPTWVHGSAQMVQQALELTGDLLSTFGRTLGGHLSTASFVLQLQREAERLAT--EEG 118
VA + G + V+QA+EL GDL+ST+G L HL A F+ QL + A+ L T + G
Sbjct 893 VAATSPKMRGVDENVKQAIELVGDLISTYGGDLTLHLQRAPFMEQLLQLAQVLGTVKDAG 952
Query 119 NAAVGAKAQWLAQLLNHYS 137
A KAQWL QL+ YS
Sbjct 953 GEACLQKAQWLRQLMARYS 971
> bbo:BBOV_IV000520 21.m02834; importin beta subunit; K14293 importin
subunit beta-1
Length=859
Score = 77.8 bits (190), Expect = 1e-14, Method: Composition-based stats.
Identities = 48/136 (35%), Positives = 71/136 (52%), Gaps = 6/136 (4%)
Query 1 TPYDAGPPNWQLNEEWIWYVHELRSATLEALASCVYALKEAQLQEVLRKEVNGMLQVVKA 60
T +D GP + NEEWIWY+++LR TL + +Y K+ + LR V+ +L ++
Sbjct 726 TSFDLGPVD---NEEWIWYINDLREGTLLSFTGILYGQKDINQVDSLRGYVSSILYFIQQ 782
Query 61 VAEAPTWVHGSAQMVQQALELTGDLLSTFGRTLGGHLSTASFVLQLQREAERLATEEGNA 120
V E P + SA + A+ LTGDL++ FG L HL + V ++ + L EE
Sbjct 783 VVETPA-EYFSAGNFRLAVALTGDLITAFGGDLSVHLVNSPLVEKIYERQKALEAEEHPM 841
Query 121 A--VGAKAQWLAQLLN 134
A K WL +LLN
Sbjct 842 ASECREKVNWLYKLLN 857
> tpv:TP01_0072 importin beta; K14293 importin subunit beta-1
Length=873
Score = 68.2 bits (165), Expect = 8e-12, Method: Composition-based stats.
Identities = 41/135 (30%), Positives = 70/135 (51%), Gaps = 6/135 (4%)
Query 1 TPYDAGPPNWQLNEEWIWYVHELRSATLEALASCVYALKEAQLQEVLRKEVNGMLQVVKA 60
T Y+ GP + NE+WI++V++L+ ++L+ VY LKE +LR V+ +LQ +
Sbjct 740 TTYEMGPID---NEDWIYFVNQLQESSLQCFTGIVYGLKEGGALHLLRPYVSSLLQFAQQ 796
Query 61 VAEAPTWVHGSAQMVQQALELTGDLLSTFGRTLGGHLSTASFVLQLQREAER--LATEEG 118
V E P + + + A+ L GDL S+FG L HL ++ + ++ ++ LA + G
Sbjct 797 VVETPDPFFDT-NLYKLAVSLIGDLSSSFGSDLSRHLVDSNLIRGIESRLKQLELAQDPG 855
Query 119 NAAVGAKAQWLAQLL 133
+ WL L
Sbjct 856 IKDCRDRVGWLHSTL 870
> cpv:cgd7_3030 importin/karyopherin ; K14293 importin subunit
beta-1
Length=882
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 30/97 (30%), Positives = 50/97 (51%), Gaps = 3/97 (3%)
Query 1 TPYDAGPPNWQLNEEWIWYVHELRSATLEALASCVYALKEAQLQEVLRKEVNGMLQVVKA 60
T Y+ GP N +E+WI Y+ ELR A L+ VY +K+A+ E+L V ++Q +
Sbjct 752 TQYNDGPVN---SEDWIEYLGELREAVLQGYTGIVYGMKDAKRLEILGPYVPSIIQFIDN 808
Query 61 VAEAPTWVHGSAQMVQQALELTGDLLSTFGRTLGGHL 97
+ + + ++ A L GDL++ F L +L
Sbjct 809 IVNDYSGEFPNDSNLKNATALVGDLITAFNGQLIQYL 845
> pfa:PF08_0069 importin beta, putative; K14293 importin subunit
beta-1
Length=877
Score = 43.9 bits (102), Expect = 2e-04, Method: Composition-based stats.
Identities = 31/132 (23%), Positives = 65/132 (49%), Gaps = 6/132 (4%)
Query 5 AGPPNWQLNEEWIWYVHELRSATLEALASCVYALKEAQLQEVLRKEVNGMLQVVKAVAEA 64
+G P+ +++W+ YV ELR A L ++ +YAL + L+ + +L ++ +
Sbjct 744 SGSPD---SDDWVNYVFELRDAILLTYSNIIYALIDGNEINKLKPYIPNILDFIELIL-I 799
Query 65 PTWVHGSAQMVQQALELTGDLLSTFGRTLGGHLSTASFVLQLQREAERLATE--EGNAAV 122
H +AQ Q ++ L GDL+ +G L + ++ + + + L+++ E
Sbjct 800 KEINHFNAQNFQNSVSLLGDLVHAYGYELIENSKLTDLIISVYGKIDILSSQGDEKCETC 859
Query 123 GAKAQWLAQLLN 134
+K +WL ++ N
Sbjct 860 VSKIKWLKRICN 871
> dre:570700 kpnb1, MGC123184, wu:fc12b10, zgc:123184; karyopherin
(importin) beta 1; K14293 importin subunit beta-1
Length=876
Score = 33.9 bits (76), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 40/84 (47%), Gaps = 8/84 (9%)
Query 15 EWIWYVHELRSATLEALASCVYALKEAQLQ-----EVLRKEVNGMLQVVKAVAEAPTWVH 69
+ + Y++ELR LEA + LK Q +++ V +L + +AE
Sbjct 753 DMVDYLNELREGCLEAYTGIIQGLKGDQENVHPDVMLVQPRVEFILSFIHHIAEDEDRSD 812
Query 70 GSAQMVQQALELTGDLLSTFGRTL 93
G +V A+ L GDL +TFG+ +
Sbjct 813 G---VVANAVGLIGDLCTTFGKDV 833
> mmu:320253 March3, 6330411I15Rik, A530081L18Rik, AI843639, MARCH-III,
MGC130167, MGC130168; membrane-associated ring finger
(C3HC4) 3; K10658 E3 ubiquitin-protein ligase MARCH3 [EC:6.3.2.19]
Length=218
Score = 33.5 bits (75), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 21/55 (38%), Positives = 27/55 (49%), Gaps = 0/55 (0%)
Query 39 KEAQLQEVLRKEVNGMLQVVKAVAEAPTWVHGSAQMVQQALELTGDLLSTFGRTL 93
+ + L EVL + VVK V + + V+G Q V Q G LLST RTL
Sbjct 5 RCSHLPEVLPDCTSSAAPVVKTVEDCGSLVNGQPQYVMQVSAKDGQLLSTVVRTL 59
> hsa:115123 MARCH3, FLJ20096, MARCH-III, MGC48332, RNF173; membrane-associated
ring finger (C3HC4) 3; K10658 E3 ubiquitin-protein
ligase MARCH3 [EC:6.3.2.19]
Length=253
Score = 33.5 bits (75), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 27/56 (48%), Gaps = 0/56 (0%)
Query 39 KEAQLQEVLRKEVNGMLQVVKAVAEAPTWVHGSAQMVQQALELTGDLLSTFGRTLG 94
+ + L EVL + VVK V + + V+G Q V Q G LLST RTL
Sbjct 5 RCSHLPEVLPDCTSSAAPVVKTVEDCGSLVNGQPQYVMQVSAKDGQLLSTVVRTLA 60
> mmu:16211 Kpnb1, AA409963, IPOB, Impnb, MGC8315; karyopherin
(importin) beta 1; K14293 importin subunit beta-1
Length=876
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 27/109 (24%), Positives = 46/109 (42%), Gaps = 8/109 (7%)
Query 15 EWIWYVHELRSATLEALASCVYALKEAQLQ-----EVLRKEVNGMLQVVKAVAEAPTWVH 69
+ + Y++ELR + LEA V LK Q +++ V +L + +A
Sbjct 753 DMVDYLNELRESCLEAYTGIVQGLKGDQENVHPDVMLVQPRVEFILSFIDHIAGDEDHTD 812
Query 70 GSAQMVQQALELTGDLLSTFGRTLGGHLSTASFVLQLQREAERLATEEG 118
G +V A L GDL + FG+ + + + +L E R T +
Sbjct 813 G---VVACAAGLIGDLCTAFGKDVLKLVEARPMIHELLTEGRRSKTNKA 858
> hsa:3837 KPNB1, IMB1, IPO1, IPOB, Impnb, MGC2155, MGC2156, MGC2157,
NTF97; karyopherin (importin) beta 1; K14293 importin
subunit beta-1
Length=876
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 27/109 (24%), Positives = 46/109 (42%), Gaps = 8/109 (7%)
Query 15 EWIWYVHELRSATLEALASCVYALKEAQLQ-----EVLRKEVNGMLQVVKAVAEAPTWVH 69
+ + Y++ELR + LEA V LK Q +++ V +L + +A
Sbjct 753 DMVDYLNELRESCLEAYTGIVQGLKGDQENVHPDVMLVQPRVEFILSFIDHIAGDEDHTD 812
Query 70 GSAQMVQQALELTGDLLSTFGRTLGGHLSTASFVLQLQREAERLATEEG 118
G +V A L GDL + FG+ + + + +L E R T +
Sbjct 813 G---VVACAAGLIGDLCTAFGKDVLKLVEARPMIHELLTEGRRSKTNKA 858
> ath:AT3G08947 binding / protein transporter; K14293 importin
subunit beta-1
Length=873
Score = 29.6 bits (65), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 26/114 (22%), Positives = 50/114 (43%), Gaps = 6/114 (5%)
Query 12 LNEEWIWYVHELRSATLEALASCVYALKEAQLQEVLRKEVNGMLQVVKAVAEAPTWVHGS 71
L+EE + Y ++LR + EA + + K+ + E++ +LQ V+ V++ P
Sbjct 753 LDEELMDYANQLRRSIFEAYSGILQGFKDTK-AELMMPYAQHLLQFVELVSKDPLR---D 808
Query 72 AQMVQQALELTGDLLSTFGRTLGGHLSTASFVLQLQREAERLATEEGNAAVGAK 125
+ + A+ GDL G +F + E L +E+ + V A+
Sbjct 809 ESVTKAAVAAMGDLADVVGENTKQLFQNFTFFGEFLNEC--LESEDEDLKVTAR 860
> mmu:329366 4932418E24Rik; RIKEN cDNA 4932418E24 gene
Length=958
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 44/99 (44%), Gaps = 10/99 (10%)
Query 29 EALASCVYALKEAQLQEVLRKEVNGML-QVVKAVAEAPTWVHGSAQMVQQALELTGDLLS 87
+ALA+ L+ +LQEV RK+ +L + + V++ + M +E G L S
Sbjct 611 KALAAHTLELRNQRLQEVYRKQREAVLGKDIPVVSQRRPGIVTFVPMQSGGMEAPGSLGS 670
Query 88 ---------TFGRTLGGHLSTASFVLQLQREAERLATEE 117
T G LG + SF L L + R+ T++
Sbjct 671 PREQTWSKVTSGMVLGDQEAPDSFCLCLNKPWNRIETQD 709
> xla:379969 gtf2h1, MGC53309; general transcription factor IIH,
polypeptide 1, 62kDa; K03141 transcription initiation factor
TFIIH subunit 1
Length=532
Score = 29.3 bits (64), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 29/56 (51%), Gaps = 4/56 (7%)
Query 38 LKEAQLQEVLRKE----VNGMLQVVKAVAEAPTWVHGSAQMVQQALELTGDLLSTF 89
+K+ +LQE + E NGM +V + ++ + HG + Q + D+LS+F
Sbjct 348 VKKVKLQEAIEYEDLDQTNGMKPIVLNLKKSDRYYHGPTPIQSQQYASSQDILSSF 403
> cel:R05G6.10 hypothetical protein
Length=461
Score = 28.5 bits (62), Expect = 6.1, Method: Composition-based stats.
Identities = 20/78 (25%), Positives = 34/78 (43%), Gaps = 4/78 (5%)
Query 10 WQLNEEWIWYVHELRSATLEALASC----VYALKEAQLQEVLRKEVNGMLQVVKAVAEAP 65
W N + + +R+ E L C Y K +LQ LR +N + + KAVA
Sbjct 113 WATNIPYDFKTEYMRTRLNELLRLCAVDKTYQQKTTELQSTLRSSLNKLDRYEKAVANLQ 172
Query 66 TWVHGSAQMVQQALELTG 83
+ + + +Q+ +TG
Sbjct 173 KALSENTNLPEQSDSMTG 190
> mmu:69349 1700008O03Rik; RIKEN cDNA 1700008O03 gene
Length=196
Score = 28.5 bits (62), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 24/43 (55%), Gaps = 1/43 (2%)
Query 94 GGHLSTASFVLQLQREAERLATEEGNAAVGAKAQWLAQLLNHY 136
G H T SF++ L+ E ++ G A +K Q+L Q+++ Y
Sbjct 17 GSHRETGSFLVDLESMEESMSRSLGKPAKSSK-QYLRQVISEY 58
Lambda K H
0.315 0.128 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2428006156
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40