bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0942_orf3
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
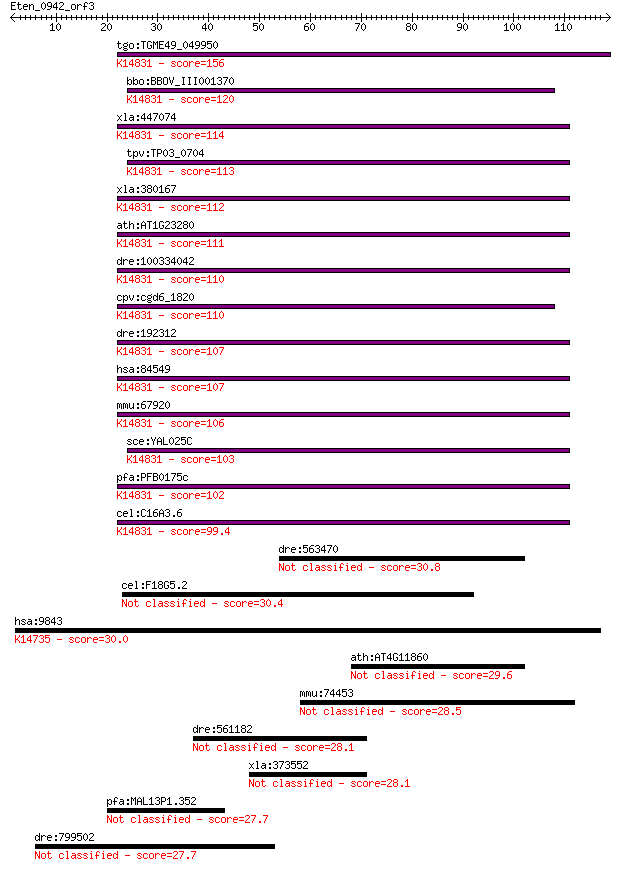
tgo:TGME49_049950 MAK16 protein, putative ; K14831 protein MAK16 156 2e-38
bbo:BBOV_III001370 17.m07141; Mak16 family protein; K14831 pro... 120 1e-27
xla:447074 mak16-a, MGC85217, mak16l, rbm13; MAK16 homolog; K1... 114 1e-25
tpv:TP03_0704 hypothetical protein; K14831 protein MAK16 113 1e-25
xla:380167 mak16-b, MGC53057, mak16, mak16l, rbm13; MAK16 homo... 112 3e-25
ath:AT1G23280 MAK16 protein-related; K14831 protein MAK16 111 5e-25
dre:100334042 Protein MAK16 homolog; K14831 protein MAK16 110 9e-25
cpv:cgd6_1820 hypothetical protein ; K14831 protein MAK16 110 1e-24
dre:192312 mak16, chunp6925, fb34e01, mak16l, wu:fb34e01; MAK1... 107 1e-23
hsa:84549 MAK16, MAK16L, RBM13; MAK16 homolog (S. cerevisiae);... 107 1e-23
mmu:67920 Mak16, 2600016B03Rik, AI314911, Rbm13; MAK16 homolog... 106 2e-23
sce:YAL025C MAK16; Essential nuclear protein, constituent of 6... 103 2e-22
pfa:PFB0175c nucleolar preribosomal assembly protein, putative... 102 3e-22
cel:C16A3.6 hypothetical protein; K14831 protein MAK16 99.4 3e-21
dre:563470 fam63a, si:ch211-210h11.5; family with sequence sim... 30.8 1.1
cel:F18G5.2 pes-8; Patterned Expression Site family member (pe... 30.4 1.5
hsa:9843 HEPH, CPL, KIAA0698; hephaestin; K14735 hephaestin 30.0 2.0
ath:AT4G11860 hypothetical protein 29.6 2.3
mmu:74453 Ccdc11, 4933415I03Rik, MGC117752; coiled-coil domain... 28.5 5.5
dre:561182 fj68a07; wu:fj68a07 28.1 6.6
xla:373552 crb2, Xerl, erl-a; crumbs homolog 2 28.1
pfa:MAL13P1.352 conserved Plasmodium protein, unknown function 27.7 9.1
dre:799502 similar to novel NACHT domain containing protein 27.7 9.2
> tgo:TGME49_049950 MAK16 protein, putative ; K14831 protein MAK16
Length=333
Score = 156 bits (394), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 67/97 (69%), Positives = 82/97 (84%), Gaps = 0/97 (0%)
Query 22 MQNDELIWCVVNKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGKL 81
MQNDE+IW V+NK FCAFK KT +EDMCRN YNVTGKCNRVSCPLANS+Y TV+E QG L
Sbjct 1 MQNDEVIWNVINKDFCAFKVKTLREDMCRNEYNVTGKCNRVSCPLANSNYGTVVEHQGSL 60
Query 82 YLCLKTVERAHLPSRLWEKLKLPQSLREAKRFVQLRM 118
+LCLKT+ERAHLP +LWEK++LP ++++AK + M
Sbjct 61 FLCLKTIERAHLPKKLWEKIRLPLNIKQAKEVIDREM 97
> bbo:BBOV_III001370 17.m07141; Mak16 family protein; K14831 protein
MAK16
Length=226
Score = 120 bits (300), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 51/84 (60%), Positives = 66/84 (78%), Gaps = 0/84 (0%)
Query 24 NDELIWCVVNKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGKLYL 83
+D+++W +V + FCAF+K TD +D C N YNVTG CNR SCPLANS+YATV+E QG L+L
Sbjct 2 SDDVVWQLVREGFCAFRKCTDTKDFCSNAYNVTGLCNRASCPLANSNYATVIEDQGTLFL 61
Query 84 CLKTVERAHLPSRLWEKLKLPQSL 107
C+KT ER H PS WE+LKL ++L
Sbjct 62 CMKTAERCHTPSEQWERLKLSKNL 85
> xla:447074 mak16-a, MGC85217, mak16l, rbm13; MAK16 homolog;
K14831 protein MAK16
Length=301
Score = 114 bits (284), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 49/90 (54%), Positives = 70/90 (77%), Gaps = 1/90 (1%)
Query 22 MQNDELIWCVV-NKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGK 80
MQ+D++IW VV NK FC+FK KT ++ CRN YN+TG CNR +CPLANS YAT+ E++G
Sbjct 1 MQHDDVIWDVVGNKQFCSFKIKTKTQNFCRNEYNITGLCNRSACPLANSQYATIKEEKGI 60
Query 81 LYLCLKTVERAHLPSRLWEKLKLPQSLREA 110
YL +KT+ERA P+R+WE+++L ++ +A
Sbjct 61 CYLYMKTIERAAFPARMWERVRLSKNYEQA 90
> tpv:TP03_0704 hypothetical protein; K14831 protein MAK16
Length=232
Score = 113 bits (283), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 49/87 (56%), Positives = 64/87 (73%), Gaps = 0/87 (0%)
Query 24 NDELIWCVVNKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGKLYL 83
+D+++W ++ FC+F+K TD + C NVYNVTG CN SCPLANS+YATV+E QG L L
Sbjct 2 SDDVVWQLIKNGFCSFRKTTDNKAFCTNVYNVTGLCNHASCPLANSNYATVIEDQGDLLL 61
Query 84 CLKTVERAHLPSRLWEKLKLPQSLREA 110
CLKTVER H P W+K++L +S EA
Sbjct 62 CLKTVERCHTPRDQWQKIRLSKSKTEA 88
> xla:380167 mak16-b, MGC53057, mak16, mak16l, rbm13; MAK16 homolog;
K14831 protein MAK16
Length=304
Score = 112 bits (280), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 48/90 (53%), Positives = 70/90 (77%), Gaps = 1/90 (1%)
Query 22 MQNDELIWCVV-NKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGK 80
MQ+D++IW VV NK FC+FK KT ++ CRN +N+TG CNR +CPLANS YAT+ E++G
Sbjct 1 MQHDDVIWDVVGNKQFCSFKIKTKTQNFCRNEFNITGLCNRSACPLANSQYATIKEEKGI 60
Query 81 LYLCLKTVERAHLPSRLWEKLKLPQSLREA 110
YL +KT+ERA P+R+WE+++L ++ +A
Sbjct 61 CYLYMKTIERAAFPARMWERVRLSKNYEQA 90
> ath:AT1G23280 MAK16 protein-related; K14831 protein MAK16
Length=303
Score = 111 bits (278), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 47/89 (52%), Positives = 64/89 (71%), Gaps = 0/89 (0%)
Query 22 MQNDELIWCVVNKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGKL 81
MQ+DE+IW V+ C++ K + CRN YNVTG CNR SCPLANS YAT+ + G
Sbjct 1 MQHDEVIWQVIRHKHCSYMAKIETGIFCRNQYNVTGICNRSSCPLANSRYATIRDHDGVF 60
Query 82 YLCLKTVERAHLPSRLWEKLKLPQSLREA 110
YL +KT+ERAH+P++LWE++KLP + +A
Sbjct 61 YLYMKTIERAHMPNKLWERVKLPVNYEKA 89
> dre:100334042 Protein MAK16 homolog; K14831 protein MAK16
Length=305
Score = 110 bits (276), Expect = 9e-25, Method: Compositional matrix adjust.
Identities = 48/90 (53%), Positives = 69/90 (76%), Gaps = 1/90 (1%)
Query 22 MQNDELIWCVV-NKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGK 80
MQ+D++IW ++ NK FC++K KT + CRN YN+TG CNR SCPLANS YAT+ E++G+
Sbjct 1 MQHDDVIWDLIGNKSFCSYKVKTKTQQFCRNEYNITGLCNRSSCPLANSQYATIREEKGQ 60
Query 81 LYLCLKTVERAHLPSRLWEKLKLPQSLREA 110
+L +K +ERA PSR+WEK+KL ++ +A
Sbjct 61 CFLYMKVIERAAFPSRMWEKVKLDRNYAKA 90
> cpv:cgd6_1820 hypothetical protein ; K14831 protein MAK16
Length=258
Score = 110 bits (275), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 50/86 (58%), Positives = 60/86 (69%), Gaps = 0/86 (0%)
Query 22 MQNDELIWCVVNKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGKL 81
M + ELIW V+ +FC+FK K DK CRN YNVTG CNR SCPL+N YATV+E +G
Sbjct 1 MNDPELIWNSVSNNFCSFKVKCDKTSFCRNEYNVTGLCNRTSCPLSNLLYATVIEHEGDC 60
Query 82 YLCLKTVERAHLPSRLWEKLKLPQSL 107
YL +KT ERAH P LWEK+KL +
Sbjct 61 YLYMKTAERAHTPRYLWEKIKLSNNF 86
> dre:192312 mak16, chunp6925, fb34e01, mak16l, wu:fb34e01; MAK16
homolog (S. cerevisiae); K14831 protein MAK16
Length=303
Score = 107 bits (266), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 47/90 (52%), Positives = 68/90 (75%), Gaps = 1/90 (1%)
Query 22 MQNDELIWCVV-NKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGK 80
MQ+D++IW ++ NK FC++K KT + RN YN+TG CNR SCPLANS YAT+ E++G+
Sbjct 1 MQHDDVIWDLIGNKSFCSYKVKTKTQQFWRNEYNITGLCNRSSCPLANSQYATIREEKGQ 60
Query 81 LYLCLKTVERAHLPSRLWEKLKLPQSLREA 110
+L +K +ERA PSR+WEK+KL ++ +A
Sbjct 61 CFLYMKVIERAAFPSRMWEKVKLDRNYAKA 90
> hsa:84549 MAK16, MAK16L, RBM13; MAK16 homolog (S. cerevisiae);
K14831 protein MAK16
Length=300
Score = 107 bits (266), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 46/90 (51%), Positives = 67/90 (74%), Gaps = 1/90 (1%)
Query 22 MQNDELIWCVV-NKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGK 80
MQ+D++IW + NK FC+FK +T + CRN Y++TG CNR SCPLANS YAT+ E++G+
Sbjct 1 MQSDDVIWDTLGNKQFCSFKIRTKTQSFCRNEYSLTGLCNRSSCPLANSQYATIKEEKGQ 60
Query 81 LYLCLKTVERAHLPSRLWEKLKLPQSLREA 110
YL +K +ERA P RLWE+++L ++ +A
Sbjct 61 CYLYMKVIERAAFPRRLWERVRLSKNYEKA 90
> mmu:67920 Mak16, 2600016B03Rik, AI314911, Rbm13; MAK16 homolog
(S. cerevisiae); K14831 protein MAK16
Length=296
Score = 106 bits (264), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 46/90 (51%), Positives = 67/90 (74%), Gaps = 1/90 (1%)
Query 22 MQNDELIWCVV-NKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGK 80
MQ+D++IW + NK FC+FK +T + CRN Y++TG CNR SCPLANS YAT+ E++G+
Sbjct 1 MQSDDVIWNTLGNKQFCSFKIRTKTQGFCRNEYSLTGLCNRSSCPLANSQYATIKEEKGQ 60
Query 81 LYLCLKTVERAHLPSRLWEKLKLPQSLREA 110
YL +K +ERA P RLWE+++L ++ +A
Sbjct 61 CYLYMKVIERAAFPRRLWERVRLSKNYEKA 90
> sce:YAL025C MAK16; Essential nuclear protein, constituent of
66S pre-ribosomal particles; required for maturation of 25S
and 5.8S rRNAs; required for maintenance of M1 satellite double-stranded
RNA of the L-A virus; K14831 protein MAK16
Length=306
Score = 103 bits (256), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 47/88 (53%), Positives = 65/88 (73%), Gaps = 1/88 (1%)
Query 24 NDELIWCVVNKHFCAFK-KKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGKLY 82
+DE++W V+N+ FC+ + K + ++ CRN YNVTG C R SCPLANS YATV GKLY
Sbjct 2 SDEIVWQVINQSFCSHRIKAPNGQNFCRNEYNVTGLCTRQSCPLANSKYATVKCDNGKLY 61
Query 83 LCLKTVERAHLPSRLWEKLKLPQSLREA 110
L +KT ERAH P++LWE++KL ++ +A
Sbjct 62 LYMKTPERAHTPAKLWERIKLSKNYTKA 89
> pfa:PFB0175c nucleolar preribosomal assembly protein, putative;
K14831 protein MAK16
Length=442
Score = 102 bits (254), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 44/89 (49%), Positives = 63/89 (70%), Gaps = 0/89 (0%)
Query 22 MQNDELIWCVVNKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGKL 81
M ND + W ++ K C+FKKK D E C N YNVTG C + +CPL+NS Y+T++ +G++
Sbjct 1 MNNDSVTWEILGKGKCSFKKKVDTEIFCLNEYNVTGLCTKANCPLSNSVYSTIILDKGEI 60
Query 82 YLCLKTVERAHLPSRLWEKLKLPQSLREA 110
YL +K+VERAHLPS LW ++ L + +EA
Sbjct 61 YLYMKSVERAHLPSALWSRVLLSLNKKEA 89
> cel:C16A3.6 hypothetical protein; K14831 protein MAK16
Length=323
Score = 99.4 bits (246), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 47/89 (52%), Positives = 60/89 (67%), Gaps = 0/89 (0%)
Query 22 MQNDELIWCVVNKHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVLEQQGKL 81
MQ D++ W ++NK CA+K T + C+N N+TG CNR SCPLANS YATV E+ G
Sbjct 1 MQCDDVTWNILNKGQCAYKAWTKPKMFCKNEMNLTGLCNRASCPLANSQYATVREENGVC 60
Query 82 YLCLKTVERAHLPSRLWEKLKLPQSLREA 110
YL K VER+H P RLWEK KL + + +A
Sbjct 61 YLYAKVVERSHYPRRLWEKTKLSKDMNKA 89
> dre:563470 fam63a, si:ch211-210h11.5; family with sequence similarity
63, member A
Length=520
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 15/48 (31%), Positives = 25/48 (52%), Gaps = 0/48 (0%)
Query 54 NVTGKCNRVSCPLANSHYATVLEQQGKLYLCLKTVERAHLPSRLWEKL 101
N T K +S N+H++T+++ +G LYL + S +WE L
Sbjct 355 NTTAKEGELSVFFRNNHFSTMIKHKGHLYLLVTDQGFLQEESVVWESL 402
> cel:F18G5.2 pes-8; Patterned Expression Site family member (pes-8)
Length=467
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 20/76 (26%), Positives = 31/76 (40%), Gaps = 7/76 (9%)
Query 23 QNDELI-WCVVN------KHFCAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSHYATVL 75
QND +I WC +N C + E + + YN G R P +H
Sbjct 41 QNDYIINWCNMNGDQLIGARGCVNCNQCGYESVETHSYNRPGAIKRTLVPFVAAHTTINF 100
Query 76 EQQGKLYLCLKTVERA 91
E + ++Y+C ER+
Sbjct 101 ECEIEMYICSGCAERS 116
> hsa:9843 HEPH, CPL, KIAA0698; hephaestin; K14735 hephaestin
Length=1160
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 32/122 (26%), Positives = 51/122 (41%), Gaps = 25/122 (20%)
Query 2 LKGQAKARKLDLRRRGELAKMQNDEL----IWCVVNKHFCAFKKKTDKEDMCRNVYNVTG 57
L+G K + +A MQ D L I+C H +E R +YNV+
Sbjct 670 LQGMRKGAAMLFPHTFVMAIMQPDNLGTFEIYCQAGSH---------REAGMRAIYNVS- 719
Query 58 KCNRVSCPLANSHYATVLE--QQGKLYLCL-KTVERAHLPSRLWEKLKLPQSLREAKRFV 114
CP H AT + Q ++Y + + VE + P R WE+ QS +++ ++
Sbjct 720 -----QCP---GHQATPRQRYQAARIYYIMAEEVEWDYCPDRSWEREWHNQSEKDSYGYI 771
Query 115 QL 116
L
Sbjct 772 FL 773
> ath:AT4G11860 hypothetical protein
Length=682
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 68 NSHYATVLEQQGKLYLCLKTVERAHLPSRLWEKL 101
N+H+ T+ + +G+LYL + P +WEKL
Sbjct 521 NNHFCTMFKYEGELYLLATDQGYLNQPDLVWEKL 554
> mmu:74453 Ccdc11, 4933415I03Rik, MGC117752; coiled-coil domain
containing 11
Length=514
Score = 28.5 bits (62), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 31/59 (52%), Gaps = 5/59 (8%)
Query 58 KCNRVSCPLANSHYATVLEQQGKLYLCLKTVER-----AHLPSRLWEKLKLPQSLREAK 111
+C + LA+ H V+E++ K ++R HL +RLWE+ +L + REA+
Sbjct 160 RCEELRTKLASIHEKKVVEERNAQIEFNKELKRQKLVEEHLFARLWEEDRLAKERREAQ 218
> dre:561182 fj68a07; wu:fj68a07
Length=929
Score = 28.1 bits (61), Expect = 6.6, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 17/34 (50%), Gaps = 0/34 (0%)
Query 37 CAFKKKTDKEDMCRNVYNVTGKCNRVSCPLANSH 70
C F K KE M Y + G CN SCP ++ +
Sbjct 741 CPFSHKVAKEKMPVCSYFLKGICNNSSCPYSHVY 774
> xla:373552 crb2, Xerl, erl-a; crumbs homolog 2
Length=778
Score = 28.1 bits (61), Expect = 7.3, Method: Composition-based stats.
Identities = 10/23 (43%), Positives = 14/23 (60%), Gaps = 0/23 (0%)
Query 48 MCRNVYNVTGKCNRVSCPLANSH 70
+C+ +N T +C SCP NSH
Sbjct 94 VCQPGFNCTNECQSNSCPFLNSH 116
> pfa:MAL13P1.352 conserved Plasmodium protein, unknown function
Length=1106
Score = 27.7 bits (60), Expect = 9.1, Method: Composition-based stats.
Identities = 11/23 (47%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 20 AKMQNDELIWCVVNKHFCAFKKK 42
+K+ N+ LI C+VNK C+F +K
Sbjct 383 SKLYNNILIECIVNKLLCSFNEK 405
> dre:799502 similar to novel NACHT domain containing protein
Length=748
Score = 27.7 bits (60), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 13/47 (27%), Positives = 23/47 (48%), Gaps = 0/47 (0%)
Query 6 AKARKLDLRRRGELAKMQNDELIWCVVNKHFCAFKKKTDKEDMCRNV 52
A ++L + EL + + E WC++N H C K + + D + V
Sbjct 2 ANVKQLLKKSLDELEEAELKEFQWCLINDHGCVSKAEMENADRLKTV 48
Lambda K H
0.323 0.135 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2027872200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40