bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0943_orf1
Length=140
Score E
Sequences producing significant alignments: (Bits) Value
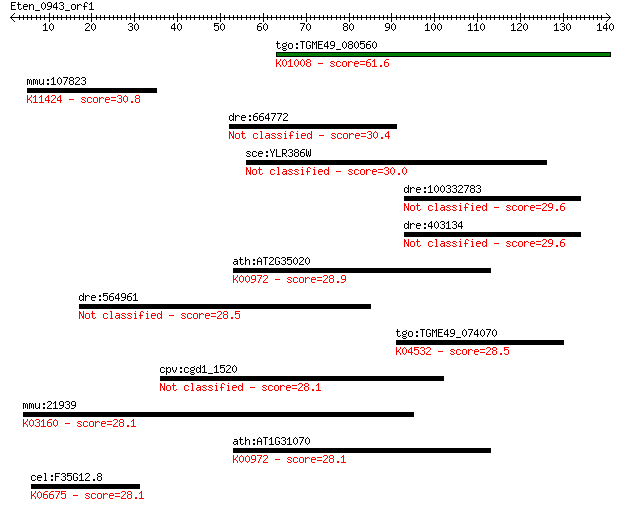
tgo:TGME49_080560 selenophosphate synthetase, putative (EC:2.7... 61.6 7e-10
mmu:107823 Whsc1, 5830445G22Rik, 9430010A17Rik, AW555663, C130... 30.8 1.5
dre:664772 MGC136804; zgc:136804 30.4 1.9
sce:YLR386W VAC14; Protein involved in regulated synthesis of ... 30.0 2.5
dre:100332783 toll-like receptor 19-like 29.6 2.7
dre:403134 tlr19; toll-like receptor 19 29.6
ath:AT2G35020 UTP--glucose-1-phosphate uridylyltransferase fam... 28.9 5.8
dre:564961 dnttip2, cb88, sb:cb1016, sb:cb88; deoxynucleotidyl... 28.5 6.3
tgo:TGME49_074070 thiF family domain-containing protein ; K045... 28.5 7.0
cpv:cgd1_1520 extracellular membrane associated protein with a... 28.1 7.9
mmu:21939 Cd40, AI326936, Bp50, GP39, HIGM1, IGM, IMD3, T-BAM,... 28.1 9.1
ath:AT1G31070 UDP-N-acetylglucosamine pyrophosphorylase-relate... 28.1 9.9
cel:F35G12.8 smc-4; SMC (structural maintenance of chromosomes... 28.1 9.9
> tgo:TGME49_080560 selenophosphate synthetase, putative (EC:2.7.9.3
1.8.1.4); K01008 selenide, water dikinase [EC:2.7.9.3]
Length=1189
Score = 61.6 bits (148), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 28/78 (35%), Positives = 42/78 (53%), Gaps = 0/78 (0%)
Query 63 PVSSVSLLLVTETTEVLYSAMVPGYITGEFNLPDLTVDLAPLCSFAGASLITARAVGLLP 122
P + L LV+ T+ Y ++PG + G + VDL LC AG + A +G+
Sbjct 41 PEKGMRLTLVSADTDTPYKGLLPGLLAGAYTRDQAHVDLLQLCQVAGGRFVRASVLGVDR 100
Query 123 QQKLLLLDGSRPPLRFDV 140
++KL+ D RPPLR+DV
Sbjct 101 ERKLIFCDDGRPPLRYDV 118
> mmu:107823 Whsc1, 5830445G22Rik, 9430010A17Rik, AW555663, C130020C13Rik,
D030027O06Rik, D930023B08Rik, Whsc1l, mKIAA1090;
Wolf-Hirschhorn syndrome candidate 1 (human) (EC:2.1.1.43);
K11424 histone-lysine N-methyltransferase NSD1/2 [EC:2.1.1.43]
Length=1366
Score = 30.8 bits (68), Expect = 1.5, Method: Composition-based stats.
Identities = 19/32 (59%), Positives = 21/32 (65%), Gaps = 2/32 (6%)
Query 5 LRKQRETTADKTTPCSS--AAGAASTDKTAAA 34
LRKQRET DKT SS A AAS+ K+ AA
Sbjct 556 LRKQRETITDKTARTSSYKAIEAASSLKSQAA 587
> dre:664772 MGC136804; zgc:136804
Length=449
Score = 30.4 bits (67), Expect = 1.9, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 52 CSNNSGCINNLPVSSVSLLLVTETTEVLYSAMVPGYITG 90
CS+ C+ P +SV L L E ++ A +PG ITG
Sbjct 364 CSSQGRCVRRDPSASVYLHLNPEMWSIIPRAQLPGPITG 402
> sce:YLR386W VAC14; Protein involved in regulated synthesis of
PtdIns(3,5)P(2), in control of trafficking of some proteins
to the vacuole lumen via the MVB, and in maintenance of vacuole
size and acidity; interacts with Fig4p; activator of Fab1p
Length=880
Score = 30.0 bits (66), Expect = 2.5, Method: Composition-based stats.
Identities = 26/70 (37%), Positives = 33/70 (47%), Gaps = 4/70 (5%)
Query 56 SGCINNLPVSSVSLLLVTETTEVLYSAMVPGYITGEFNLPDLTVDLAPLCSFAGASLITA 115
S C N PVS +SL V E E+ Y+ + Y E L DL V L L + + T
Sbjct 651 SWCPN--PVSVISLCFVAENYELAYTVL-QTYANYELKLNDL-VQLDILIQLFESPVFTR 706
Query 116 RAVGLLPQQK 125
+ LL QQK
Sbjct 707 MRLQLLEQQK 716
> dre:100332783 toll-like receptor 19-like
Length=735
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 25/48 (52%), Gaps = 7/48 (14%)
Query 93 NLPDLTVDLA-------PLCSFAGASLITARAVGLLPQQKLLLLDGSR 133
N+ DL+ DL+ LC F + I A+A LP ++L +DG R
Sbjct 20 NITDLSTDLSQVGFEVRTLCVFGDITSIPAKAFSHLPSLEVLHIDGIR 67
> dre:403134 tlr19; toll-like receptor 19
Length=735
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 25/48 (52%), Gaps = 7/48 (14%)
Query 93 NLPDLTVDLA-------PLCSFAGASLITARAVGLLPQQKLLLLDGSR 133
N+ DL+ DL+ LC F + I A+A LP ++L +DG R
Sbjct 20 NITDLSTDLSQVGFEVRTLCVFGDITSIPAKAFSHLPSLEVLHIDGIR 67
> ath:AT2G35020 UTP--glucose-1-phosphate uridylyltransferase family
protein (EC:2.7.7.23); K00972 UDP-N-acetylglucosamine
pyrophosphorylase [EC:2.7.7.23]
Length=502
Score = 28.9 bits (63), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 32/60 (53%), Gaps = 3/60 (5%)
Query 53 SNNSGCINNLPVSSVSLLLVTETTEVLYSAMVPGYITGEFNLPDLTVDLAPLCSFAGASL 112
N +G + P S+ L+L T V+ + G++T L V+++PLCS+AG +L
Sbjct 429 KNANGSNYDTPESARLLVLRLHTRWVIAAG---GFLTHSVPLYATGVEVSPLCSYAGENL 485
> dre:564961 dnttip2, cb88, sb:cb1016, sb:cb88; deoxynucleotidyltransferase,
terminal, interacting protein 2
Length=941
Score = 28.5 bits (62), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 20/73 (27%), Positives = 31/73 (42%), Gaps = 5/73 (6%)
Query 17 TPCSSAAGAASTDKTAAAVLRSILGKDAAVAADEECSNNSGCINNL-----PVSSVSLLL 71
TPCSS G+AS+++ A K A EE S + + + P + +
Sbjct 320 TPCSSRTGSASSNRAVPASRTRSKAKVITSAMSEEQSEDEQLVTHHEGIQAPEEQLDQTM 379
Query 72 VTETTEVLYSAMV 84
ET+E S M+
Sbjct 380 TMETSEAQESTMI 392
> tgo:TGME49_074070 thiF family domain-containing protein ; K04532
amyloid beta precursor protein binding protein 1
Length=779
Score = 28.5 bits (62), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 26/44 (59%), Gaps = 5/44 (11%)
Query 91 EFNLPDLTVD---LAPLCSFAGASLITARAV--GLLPQQKLLLL 129
E N+PDLTVD + + +F GA + T A+ G+ Q+ + L+
Sbjct 711 EINVPDLTVDERIIRQMVAFGGAEIHTTAAIVGGVAAQEAVKLI 754
> cpv:cgd1_1520 extracellular membrane associated protein with
a signal peptide, EGF domain, 9x transmembrane domains and
a pentraxin domain
Length=3008
Score = 28.1 bits (61), Expect = 7.9, Method: Composition-based stats.
Identities = 22/67 (32%), Positives = 34/67 (50%), Gaps = 4/67 (5%)
Query 36 LRSILGKDAAVAADEECSNNSGCINNLPVSSVSLLLVTETTEVLYSAMVPGYITGEFNLP 95
LR L + + E N+G I N+ ++ L+ +T + Y+ +PGYI E+N P
Sbjct 1029 LRLKLNRGTGLLFSEPNLENNG-IENMELNHS--LINEDTKDKKYNVPIPGYIEEEYNEP 1085
Query 96 -DLTVDL 101
L VDL
Sbjct 1086 RSLDVDL 1092
> mmu:21939 Cd40, AI326936, Bp50, GP39, HIGM1, IGM, IMD3, T-BAM,
TRAP, Tnfrsf5, p50; CD40 antigen; K03160 tumor necrosis factor
receptor superfamily member 5
Length=289
Score = 28.1 bits (61), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 22/91 (24%), Positives = 37/91 (40%), Gaps = 21/91 (23%)
Query 4 GLRKQRETTADKTTPCSSAAGAASTDKTAAAVLRSILGKDAAVAADEECSNNSGCINNLP 63
GLR ++E TA+ T C+ G T K A C+ ++ CI
Sbjct 88 GLRVKKEGTAESDTVCTCKEGQHCTSKDCEA-----------------CAQHTPCIPGFG 130
Query 64 VSSVSLLLVTETTEVLYSAMVPGYITGEFNL 94
V + + TETT+ + G+ + + +L
Sbjct 131 V----MEMATETTDTVCHPCPVGFFSNQSSL 157
> ath:AT1G31070 UDP-N-acetylglucosamine pyrophosphorylase-related
(EC:2.7.7.23); K00972 UDP-N-acetylglucosamine pyrophosphorylase
[EC:2.7.7.23]
Length=505
Score = 28.1 bits (61), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 32/60 (53%), Gaps = 3/60 (5%)
Query 53 SNNSGCINNLPVSSVSLLLVTETTEVLYSAMVPGYITGEFNLPDLTVDLAPLCSFAGASL 112
N +G + P S+ L+L T V+ + G++T L V+++PLCS+AG +L
Sbjct 432 KNVNGSNFDTPESARLLVLRLHTRWVIAAG---GFLTHSVPLYATGVEVSPLCSYAGENL 488
> cel:F35G12.8 smc-4; SMC (structural maintenance of chromosomes)
family member (smc-4); K06675 structural maintenance of
chromosome 4
Length=1549
Score = 28.1 bits (61), Expect = 9.9, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 6 RKQRETTADKTTPCSSAAGAASTDK 30
RK R AD+TTP S + +AST K
Sbjct 1499 RKARNIEADETTPPSKRSNSASTPK 1523
Lambda K H
0.316 0.131 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2552834388
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40