bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0947_orf1
Length=238
Score E
Sequences producing significant alignments: (Bits) Value
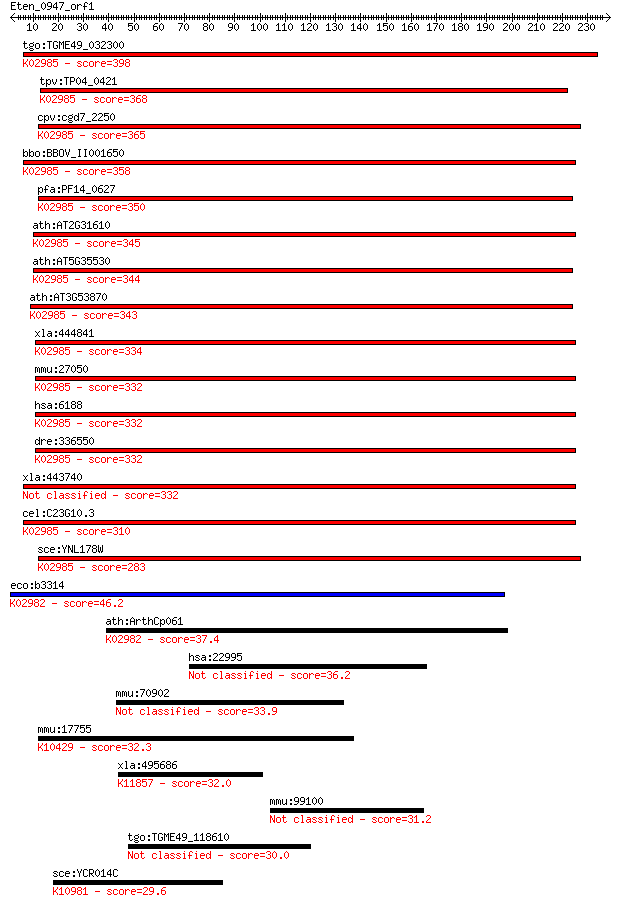
tgo:TGME49_032300 40S ribosomal protein S3, putative ; K02985 ... 398 1e-110
tpv:TP04_0421 40S ribosomal protein S3; K02985 small subunit r... 368 8e-102
cpv:cgd7_2250 40S ribosomal protein S3, KH domain ; K02985 sma... 365 7e-101
bbo:BBOV_II001650 18.m06128; 40S ribosomal protein S3 protein;... 358 7e-99
pfa:PF14_0627 40S ribosomal protein S3, putative; K02985 small... 350 3e-96
ath:AT2G31610 40S ribosomal protein S3 (RPS3A); K02985 small s... 345 1e-94
ath:AT5G35530 40S ribosomal protein S3 (RPS3C); K02985 small s... 344 2e-94
ath:AT3G53870 40S ribosomal protein S3 (RPS3B); K02985 small s... 343 3e-94
xla:444841 rps3-a, MGC53327, rps3; ribosomal protein S3; K0298... 334 2e-91
mmu:27050 Rps3, D7Ertd795e, Rs_3; ribosomal protein S3; K02985... 332 6e-91
hsa:6188 RPS3, FLJ26283, FLJ27450, MGC87870; ribosomal protein... 332 8e-91
dre:336550 rps3, MGC56088, fb13d09, wu:fb13d09, zgc:56088; rib... 332 9e-91
xla:443740 rps3-b, rpls1b, rpls3-b; ribosomal protein S3 332 9e-91
cel:C23G10.3 rps-3; Ribosomal Protein, Small subunit family me... 310 3e-84
sce:YNL178W RPS3, SUF14; Rps3p; K02985 small subunit ribosomal... 283 3e-76
eco:b3314 rpsC, ECK3301, JW3276; 30S ribosomal subunit protein... 46.2 1e-04
ath:ArthCp061 rps3; 30S ribosomal protein S3; K02982 small sub... 37.4 0.047
hsa:22995 CEP152, FLJ21594, KIAA0912, MCPH4; centrosomal prote... 36.2 0.12
mmu:70902 Lpcat2b, 4921521K07Rik, Aytl1b; lysophosphatidylchol... 33.9 0.57
mmu:17755 Mtap1b, A230055D22, AI843217, LC1, MAP1B, MAP5, MGC1... 32.3 1.4
xla:495686 usp47; ubiquitin specific peptidase 47; K11857 ubiq... 32.0 2.0
mmu:99100 Cep152, AI851464, mKIAA0912; centrosomal protein 152 31.2
tgo:TGME49_118610 hypothetical protein 30.0 8.6
sce:YCR014C POL4; DNA polymerase IV, undergoes pair-wise inter... 29.6 9.4
> tgo:TGME49_032300 40S ribosomal protein S3, putative ; K02985
small subunit ribosomal protein S3e
Length=235
Score = 398 bits (1022), Expect = 1e-110, Method: Compositional matrix adjust.
Identities = 199/228 (87%), Positives = 210/228 (92%), Gaps = 1/228 (0%)
Query 6 MAANTNISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREV 65
MA+ N KKRKFV DGVFQAELNEFLS TL+EDGYSGVEVRVTPIRTEIIIRATRTREV
Sbjct 1 MASTGNTCKKRKFVKDGVFQAELNEFLSCTLSEDGYSGVEVRVTPIRTEIIIRATRTREV 60
Query 66 LGEKGRRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRA 125
LG+KGRRIRELTS+VQKRFGF PDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRA
Sbjct 61 LGDKGRRIRELTSVVQKRFGFAPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRA 120
Query 126 CYGVLRHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLR 185
CYGVLRHIMESGAKGCEVVVSGKLRAQRAKSMKF+DGY+ISTGEP + FV QAIRSVQLR
Sbjct 121 CYGVLRHIMESGAKGCEVVVSGKLRAQRAKSMKFKDGYLISTGEPSKMFVDQAIRSVQLR 180
Query 186 QGVLGVRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPELPVAVQDPEL 233
QGVLGVRV IMLP DPEGK GP+ PLPDTIIV +PKPE+PV VQ E+
Sbjct 181 QGVLGVRVKIMLPHDPEGKRGPANPLPDTIIVMDPKPEIPV-VQPEEM 227
> tpv:TP04_0421 40S ribosomal protein S3; K02985 small subunit
ribosomal protein S3e
Length=216
Score = 368 bits (945), Expect = 8e-102, Method: Compositional matrix adjust.
Identities = 174/209 (83%), Positives = 196/209 (93%), Gaps = 0/209 (0%)
Query 13 SKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEKGRR 72
SK+++F+NDGVF AELNEFLSRTLAEDGYSGVE+R+TP+RTEIIIRATRTREVLGEK RR
Sbjct 8 SKQKRFINDGVFNAELNEFLSRTLAEDGYSGVELRITPVRTEIIIRATRTREVLGEKARR 67
Query 73 IRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYGVLRH 132
IRELTSLVQKRFGF DSVEL+AERVENRGLCAMAQAESLRYKLLKGLA+RRA YGVLR
Sbjct 68 IRELTSLVQKRFGFSSDSVELYAERVENRGLCAMAQAESLRYKLLKGLAIRRAAYGVLRQ 127
Query 133 IMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLRQGVLGVR 192
IMESGAKGCEV+VSGKLRAQRAKSMKF+DGYIISTG+P Q +V+ A+RSVQLRQGVLG++
Sbjct 128 IMESGAKGCEVIVSGKLRAQRAKSMKFKDGYIISTGQPAQEYVNTAVRSVQLRQGVLGIK 187
Query 193 VSIMLPFDPEGKMGPSTPLPDTIIVSEPK 221
V IMLP+DPEG++GPST LPDT+ V +PK
Sbjct 188 VKIMLPYDPEGRLGPSTLLPDTVTVLDPK 216
> cpv:cgd7_2250 40S ribosomal protein S3, KH domain ; K02985 small
subunit ribosomal protein S3e
Length=221
Score = 365 bits (937), Expect = 7e-101, Method: Compositional matrix adjust.
Identities = 176/215 (81%), Positives = 199/215 (92%), Gaps = 1/215 (0%)
Query 12 ISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEKGR 71
ISKKRKF+NDGVF+AELNEFLS TLAEDGY+GVEVRVTPI+TEIIIRATRTREVLG KGR
Sbjct 6 ISKKRKFINDGVFEAELNEFLSCTLAEDGYAGVEVRVTPIKTEIIIRATRTREVLGVKGR 65
Query 72 RIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYGVLR 131
RIRELTSL+QKRF FP SVELFAER+ENRGLCAMAQAESLRYKLLKG+AVRRACYGVLR
Sbjct 66 RIRELTSLIQKRFNFPEGSVELFAERIENRGLCAMAQAESLRYKLLKGVAVRRACYGVLR 125
Query 132 HIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLRQGVLGV 191
H+MESGAKGCEV+VSGKLRAQRAKSMKFRDGY+I+TG+P + FV +A+R+V ++QGVLGV
Sbjct 126 HVMESGAKGCEVIVSGKLRAQRAKSMKFRDGYMIATGQPFRDFVDKAVRNVMMKQGVLGV 185
Query 192 RVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPELPV 226
+V IMLP+DPEGK GPS PLPD ++V EPK E P+
Sbjct 186 QVKIMLPYDPEGKTGPSKPLPDAVMVREPK-EYPI 219
> bbo:BBOV_II001650 18.m06128; 40S ribosomal protein S3 protein;
K02985 small subunit ribosomal protein S3e
Length=223
Score = 358 bits (920), Expect = 7e-99, Method: Compositional matrix adjust.
Identities = 171/219 (78%), Positives = 194/219 (88%), Gaps = 0/219 (0%)
Query 6 MAANTNISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREV 65
M T S RKF+ DGVF AELNEFLSR LAEDGYSGVE+R+TP+RTEIIIRATR REV
Sbjct 1 MRGFTRKSMCRKFIGDGVFNAELNEFLSRVLAEDGYSGVELRITPVRTEIIIRATRAREV 60
Query 66 LGEKGRRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRA 125
+GEK RRIRELTSL+QKRFGF PD+VEL+AERVE++GLCAMAQAESLRYKLLKGLAVRRA
Sbjct 61 VGEKARRIRELTSLIQKRFGFSPDTVELYAERVEHKGLCAMAQAESLRYKLLKGLAVRRA 120
Query 126 CYGVLRHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLR 185
YGVLRHIMESGAKGCEV+VSGKLRAQRAKSMKF+DGY+ISTG+P + FV+ A RSVQLR
Sbjct 121 AYGVLRHIMESGAKGCEVIVSGKLRAQRAKSMKFKDGYLISTGQPAKTFVTSATRSVQLR 180
Query 186 QGVLGVRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPEL 224
QGVLG++V IM P DPEG++GPST +PDTI V EP P++
Sbjct 181 QGVLGIKVKIMHPHDPEGRLGPSTVMPDTITVKEPSPDM 219
> pfa:PF14_0627 40S ribosomal protein S3, putative; K02985 small
subunit ribosomal protein S3e
Length=221
Score = 350 bits (897), Expect = 3e-96, Method: Compositional matrix adjust.
Identities = 173/213 (81%), Positives = 191/213 (89%), Gaps = 1/213 (0%)
Query 12 ISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEKGR 71
ISKKRKF+NDGVFQAELNEFL+R LAEDGYSGVEVRVTPIRTE+IIRATRTREVLG+KGR
Sbjct 5 ISKKRKFINDGVFQAELNEFLARILAEDGYSGVEVRVTPIRTEVIIRATRTREVLGDKGR 64
Query 72 RIRELTSLVQKRF-GFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYGVL 130
RIRELTSLVQKRF +SVELFAERVE+RGLCAMAQAESLRYKLLKGLAVRRACYGVL
Sbjct 65 RIRELTSLVQKRFFNKSTNSVELFAERVEHRGLCAMAQAESLRYKLLKGLAVRRACYGVL 124
Query 131 RHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLRQGVLG 190
RHIMESGAKGCEV+VSGKLRAQRAKSMKFRDGY+ISTGEP +RFV+ A RS QL+QGVLG
Sbjct 125 RHIMESGAKGCEVIVSGKLRAQRAKSMKFRDGYLISTGEPSKRFVNTATRSAQLKQGVLG 184
Query 191 VRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPE 223
++V IMLP + + G ++ LPD I V EPK +
Sbjct 185 IKVKIMLPTAIDTRTGLTSILPDNISVLEPKTD 217
> ath:AT2G31610 40S ribosomal protein S3 (RPS3A); K02985 small
subunit ribosomal protein S3e
Length=250
Score = 345 bits (884), Expect = 1e-94, Method: Compositional matrix adjust.
Identities = 164/215 (76%), Positives = 188/215 (87%), Gaps = 0/215 (0%)
Query 10 TNISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEK 69
T ISKKRKFV DGVF AELNE L+R LAEDGYSGVEVRVTP+RTEIIIRATRT+ VLGEK
Sbjct 3 TQISKKRKFVADGVFYAELNEVLTRELAEDGYSGVEVRVTPMRTEIIIRATRTQNVLGEK 62
Query 70 GRRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYGV 129
GRRIRELTSLVQKRF FP DSVEL+AE+V NRGLCA+AQAESLRYKLL GLAVRRACYGV
Sbjct 63 GRRIRELTSLVQKRFKFPVDSVELYAEKVNNRGLCAIAQAESLRYKLLGGLAVRRACYGV 122
Query 130 LRHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLRQGVL 189
LR +MESGAKGCEV+VSGKLRA RAKSMKF+DGY++S+G+P + ++ A+R V LRQGVL
Sbjct 123 LRFVMESGAKGCEVIVSGKLRAARAKSMKFKDGYMVSSGQPTKEYIDAAVRHVLLRQGVL 182
Query 190 GVRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPEL 224
G++V IML +DP GK GP TPLPD +I+ PK ++
Sbjct 183 GIKVKIMLDWDPTGKSGPKTPLPDVVIIHAPKDDV 217
> ath:AT5G35530 40S ribosomal protein S3 (RPS3C); K02985 small
subunit ribosomal protein S3e
Length=248
Score = 344 bits (883), Expect = 2e-94, Method: Compositional matrix adjust.
Identities = 164/214 (76%), Positives = 188/214 (87%), Gaps = 0/214 (0%)
Query 10 TNISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEK 69
T ISKKRKFV DGVF AELNE L+R LAEDGYSGVEVRVTP+RTEIIIRATRT+ VLGEK
Sbjct 3 TQISKKRKFVADGVFYAELNEVLTRELAEDGYSGVEVRVTPMRTEIIIRATRTQNVLGEK 62
Query 70 GRRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYGV 129
GRRIRELTSLVQKRF FP DSVEL+AE+V NRGLCA+AQAESLRYKLL GLAVRRACYGV
Sbjct 63 GRRIRELTSLVQKRFKFPQDSVELYAEKVANRGLCAIAQAESLRYKLLGGLAVRRACYGV 122
Query 130 LRHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLRQGVL 189
LR +MESGAKGCEV+VSGKLRA RAKSMKF+DGY++S+G+P + ++ A+R V LRQGVL
Sbjct 123 LRFVMESGAKGCEVIVSGKLRAARAKSMKFKDGYMVSSGQPTKEYIDAAVRHVLLRQGVL 182
Query 190 GVRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPE 223
G++V IML +DP+GK GP TPLPD +I+ PK +
Sbjct 183 GLKVKIMLDWDPKGKQGPMTPLPDVVIIHTPKED 216
> ath:AT3G53870 40S ribosomal protein S3 (RPS3B); K02985 small
subunit ribosomal protein S3e
Length=249
Score = 343 bits (880), Expect = 3e-94, Method: Compositional matrix adjust.
Identities = 163/215 (75%), Positives = 187/215 (86%), Gaps = 0/215 (0%)
Query 9 NTNISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGE 68
T ISKKRKFV DGVF AELNE L+R LAEDGYSGVEVRVTP+RTEIIIRATRT+ VLGE
Sbjct 2 TTQISKKRKFVADGVFYAELNEVLTRELAEDGYSGVEVRVTPMRTEIIIRATRTQNVLGE 61
Query 69 KGRRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYG 128
KGRRIRELTSLVQKRF FP DSVEL+AE+V NRGLCA+AQAESLRYKLL GLAVRRACYG
Sbjct 62 KGRRIRELTSLVQKRFKFPVDSVELYAEKVNNRGLCAIAQAESLRYKLLGGLAVRRACYG 121
Query 129 VLRHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLRQGV 188
VLR +MESGAKGCEV+VSGKLRA RAKSMKF+DGY++S+G+P + ++ A+R V LRQGV
Sbjct 122 VLRFVMESGAKGCEVIVSGKLRAARAKSMKFKDGYMVSSGQPTKEYIDSAVRHVLLRQGV 181
Query 189 LGVRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPE 223
LG++V +ML +DP+G GP TPLPD +I+ PK E
Sbjct 182 LGIKVKVMLDWDPKGISGPKTPLPDVVIIHSPKEE 216
> xla:444841 rps3-a, MGC53327, rps3; ribosomal protein S3; K02985
small subunit ribosomal protein S3e
Length=246
Score = 334 bits (857), Expect = 2e-91, Method: Compositional matrix adjust.
Identities = 162/214 (75%), Positives = 183/214 (85%), Gaps = 0/214 (0%)
Query 11 NISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEKG 70
ISKKRKFV DG+F+AELNEFL+R LAEDGYSGVEVRVTP RTEIII ATRT+ VLGEKG
Sbjct 4 QISKKRKFVADGIFKAELNEFLTRELAEDGYSGVEVRVTPTRTEIIILATRTQNVLGEKG 63
Query 71 RRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYGVL 130
RRIRELT++VQKRFGFP SVEL+AE+V RGLCA+AQAESLRYKLL GLAVRRACYGVL
Sbjct 64 RRIRELTAVVQKRFGFPEGSVELYAEKVATRGLCAIAQAESLRYKLLGGLAVRRACYGVL 123
Query 131 RHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLRQGVLG 190
R IMESGAKGCEVVVSGKLR QRAKSMKF DG +I +G+P +V A+R V LRQGVLG
Sbjct 124 RFIMESGAKGCEVVVSGKLRGQRAKSMKFVDGLMIHSGDPVNYYVDTAVRHVLLRQGVLG 183
Query 191 VRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPEL 224
++V IMLP+DP GK+GP PLPD + + EPK E+
Sbjct 184 IKVKIMLPWDPSGKIGPKKPLPDHVSIVEPKDEI 217
> mmu:27050 Rps3, D7Ertd795e, Rs_3; ribosomal protein S3; K02985
small subunit ribosomal protein S3e
Length=243
Score = 332 bits (852), Expect = 6e-91, Method: Compositional matrix adjust.
Identities = 162/214 (75%), Positives = 183/214 (85%), Gaps = 0/214 (0%)
Query 11 NISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEKG 70
ISKKRKFV DG+F+AELNEFL+R LAEDGYSGVEVRVTP RTEIII ATRT+ VLGEKG
Sbjct 4 QISKKRKFVADGIFKAELNEFLTRELAEDGYSGVEVRVTPTRTEIIILATRTQNVLGEKG 63
Query 71 RRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYGVL 130
RRIRELT++VQKRFGFP SVEL+AE+V RGLCA+AQAESLRYKLL GLAVRRACYGVL
Sbjct 64 RRIRELTAVVQKRFGFPEGSVELYAEKVATRGLCAIAQAESLRYKLLGGLAVRRACYGVL 123
Query 131 RHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLRQGVLG 190
R IMESGAKGCEVVVSGKLR QRAKSMKF DG +I +G+P +V A+R V LRQGVLG
Sbjct 124 RFIMESGAKGCEVVVSGKLRGQRAKSMKFVDGLMIHSGDPVNYYVDTAVRHVLLRQGVLG 183
Query 191 VRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPEL 224
++V IMLP+DP GK+GP PLPD + + EPK E+
Sbjct 184 IKVKIMLPWDPSGKIGPKKPLPDHVSIVEPKDEI 217
> hsa:6188 RPS3, FLJ26283, FLJ27450, MGC87870; ribosomal protein
S3; K02985 small subunit ribosomal protein S3e
Length=243
Score = 332 bits (850), Expect = 8e-91, Method: Compositional matrix adjust.
Identities = 162/214 (75%), Positives = 183/214 (85%), Gaps = 0/214 (0%)
Query 11 NISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEKG 70
ISKKRKFV DG+F+AELNEFL+R LAEDGYSGVEVRVTP RTEIII ATRT+ VLGEKG
Sbjct 4 QISKKRKFVADGIFKAELNEFLTRELAEDGYSGVEVRVTPTRTEIIILATRTQNVLGEKG 63
Query 71 RRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYGVL 130
RRIRELT++VQKRFGFP SVEL+AE+V RGLCA+AQAESLRYKLL GLAVRRACYGVL
Sbjct 64 RRIRELTAVVQKRFGFPEGSVELYAEKVATRGLCAIAQAESLRYKLLGGLAVRRACYGVL 123
Query 131 RHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLRQGVLG 190
R IMESGAKGCEVVVSGKLR QRAKSMKF DG +I +G+P +V A+R V LRQGVLG
Sbjct 124 RFIMESGAKGCEVVVSGKLRGQRAKSMKFVDGLMIHSGDPVNYYVDTAVRHVLLRQGVLG 183
Query 191 VRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPEL 224
++V IMLP+DP GK+GP PLPD + + EPK E+
Sbjct 184 IKVKIMLPWDPTGKIGPKKPLPDHVSIVEPKDEI 217
> dre:336550 rps3, MGC56088, fb13d09, wu:fb13d09, zgc:56088; ribosomal
protein S3; K02985 small subunit ribosomal protein
S3e
Length=245
Score = 332 bits (850), Expect = 9e-91, Method: Compositional matrix adjust.
Identities = 161/214 (75%), Positives = 183/214 (85%), Gaps = 0/214 (0%)
Query 11 NISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEKG 70
ISKKRKFV+DG+F+AELNEFL+R LAEDGYSGVEVRVTP RTEIII ATRT+ VLGEKG
Sbjct 4 QISKKRKFVSDGIFKAELNEFLTRELAEDGYSGVEVRVTPTRTEIIILATRTQNVLGEKG 63
Query 71 RRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYGVL 130
RRIRELT++VQKRFGFP VEL+AE+V RGLCA+AQAESLRYKLL GLAVRRACYGVL
Sbjct 64 RRIRELTAVVQKRFGFPEGGVELYAEKVATRGLCAIAQAESLRYKLLGGLAVRRACYGVL 123
Query 131 RHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLRQGVLG 190
R IMESGAKGCEVVVSGKLR QRAKSMKF DG +I +G+P +V A+R V LRQGVLG
Sbjct 124 RFIMESGAKGCEVVVSGKLRGQRAKSMKFVDGLMIHSGDPVNYYVDTAVRHVLLRQGVLG 183
Query 191 VRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPEL 224
++V IMLP+DP GK+GP PLPD + + EPK E+
Sbjct 184 IKVKIMLPWDPSGKIGPKKPLPDHVSIVEPKEEV 217
> xla:443740 rps3-b, rpls1b, rpls3-b; ribosomal protein S3
Length=246
Score = 332 bits (850), Expect = 9e-91, Method: Compositional matrix adjust.
Identities = 163/219 (74%), Positives = 186/219 (84%), Gaps = 2/219 (0%)
Query 6 MAANTNISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREV 65
MAA +SKKRKFV DG+F+AELNEFL+R LAEDGYSGVEVRVTP +TEIII ATRT+ V
Sbjct 1 MAAQ--MSKKRKFVADGIFKAELNEFLTRELAEDGYSGVEVRVTPTQTEIIILATRTQNV 58
Query 66 LGEKGRRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRA 125
LGEKGRRIRELT++VQKRFGFP SVEL+AE+V RGLCA+AQAESLRYKLL GLAVRRA
Sbjct 59 LGEKGRRIRELTAVVQKRFGFPEGSVELYAEKVATRGLCAIAQAESLRYKLLGGLAVRRA 118
Query 126 CYGVLRHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLR 185
CYGVLR IMESGAKGCEVVVSGKLR QRAKSMKF DG +I +G+P +V A+R V LR
Sbjct 119 CYGVLRFIMESGAKGCEVVVSGKLRGQRAKSMKFVDGLMIHSGDPVNYYVDTAVRHVLLR 178
Query 186 QGVLGVRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPEL 224
QGVLG++V IMLP+DP GK+GP PLPD + + EPK E+
Sbjct 179 QGVLGIKVKIMLPWDPSGKIGPKKPLPDHVSIVEPKDEI 217
> cel:C23G10.3 rps-3; Ribosomal Protein, Small subunit family
member (rps-3); K02985 small subunit ribosomal protein S3e
Length=247
Score = 310 bits (794), Expect = 3e-84, Method: Compositional matrix adjust.
Identities = 147/219 (67%), Positives = 179/219 (81%), Gaps = 0/219 (0%)
Query 6 MAANTNISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREV 65
MAAN N++KK+K V G+F+AELN FL + LAEDGYSGVEVR TP R E+II ATRT+ V
Sbjct 1 MAANQNVTKKKKAVIGGIFKAELNNFLMKELAEDGYSGVEVRSTPARAEVIIMATRTQNV 60
Query 66 LGEKGRRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRA 125
LGE+GRRI+ELTS+VQKRFGF SVEL+AE+V NRGLCA+AQ ESLRYKL+ GLAVRRA
Sbjct 61 LGERGRRIKELTSVVQKRFGFEEGSVELYAEKVSNRGLCAVAQCESLRYKLVGGLAVRRA 120
Query 126 CYGVLRHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLR 185
CYGVLR IMESGA+G EV+VSGKLR QRAK+MKF DG +I +G P ++ QA+R VQLR
Sbjct 121 CYGVLRFIMESGAQGVEVIVSGKLRGQRAKAMKFVDGLMIHSGHPVNDYIQQAVRHVQLR 180
Query 186 QGVLGVRVSIMLPFDPEGKMGPSTPLPDTIIVSEPKPEL 224
QGV+G++V IMLP+DP G+ GP LPD + + EP+ E+
Sbjct 181 QGVIGIKVKIMLPYDPRGQNGPRNALPDHVQIVEPQEEV 219
> sce:YNL178W RPS3, SUF14; Rps3p; K02985 small subunit ribosomal
protein S3e
Length=240
Score = 283 bits (725), Expect = 3e-76, Method: Compositional matrix adjust.
Identities = 140/216 (64%), Positives = 174/216 (80%), Gaps = 4/216 (1%)
Query 12 ISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLGEKGR 71
ISKKRK V DGVF AELNEF +R LAE+GYSGVEVRVTP +TE+IIRATRT++VLGE GR
Sbjct 5 ISKKRKLVADGVFYAELNEFFTRELAEEGYSGVEVRVTPTKTEVIIRATRTQDVLGENGR 64
Query 72 RIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGLAVRRACYGVLR 131
RI ELT LVQKRF + P ++ L+AERV++RGL A+AQAES+++KLL GLA+RRA YGV+R
Sbjct 65 RINELTLLVQKRFKYAPGTIVLYAERVQDRGLSAVAQAESMKFKLLNGLAIRRAAYGVVR 124
Query 132 HIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYIISTGEPRQRFVSQAIRSVQLRQGVLGV 191
++MESGAKGCEVVVSGKLRA RAK+MKF DG++I +G+P F+ A R V +RQGVLG+
Sbjct 125 YVMESGAKGCEVVVSGKLRAARAKAMKFADGFLIHSGQPVNDFIDTATRHVLMRQGVLGI 184
Query 192 RVSIMLPFDP-EGKMGPSTPLPDTIIVSEPKPELPV 226
+V IM DP + + GP LPD + + EPK E P+
Sbjct 185 KVKIMR--DPAKSRTGPKA-LPDAVTIIEPKEEEPI 217
> eco:b3314 rpsC, ECK3301, JW3276; 30S ribosomal subunit protein
S3; K02982 small subunit ribosomal protein S3
Length=233
Score = 46.2 bits (108), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 48/200 (24%), Positives = 91/200 (45%), Gaps = 12/200 (6%)
Query 1 LSLAKMAANTNISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRAT 60
L + K +T + ++F ++ ++ ++L++ LA+ S + + + I
Sbjct 12 LGIVKPWNSTWFANTKEFADNLDSDFKVRQYLTKELAKASVSRIVIERPAKSIRVTIHTA 71
Query 61 RTREVLGEKGRRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMAQAESLRYKLLKGL 120
R V+G+KG + +L +V G P ++ V L A A+S+ +L + +
Sbjct 72 RPGIVIGKKGEDVEKLRKVVADIAGVP---AQINIAEVRKPELDAKLVADSITSQLERRV 128
Query 121 AVRRACYGVLRHIMESGAKGCEVVVSGKL-RAQRAKSMKFRDGYIISTGEPRQRF---VS 176
RRA +++ M GAKG +V VSG+L A+ A++ +R+G + P +
Sbjct 129 MFRRAMKRAVQNAMRLGAKGIKVEVSGRLGGAEIARTEWYREGRV-----PLHTLRADID 183
Query 177 QAIRSVQLRQGVLGVRVSIM 196
GV+GV+V I
Sbjct 184 YNTSEAHTTYGVIGVKVWIF 203
> ath:ArthCp061 rps3; 30S ribosomal protein S3; K02982 small subunit
ribosomal protein S3
Length=218
Score = 37.4 bits (85), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 35/161 (21%), Positives = 78/161 (48%), Gaps = 5/161 (3%)
Query 39 DGYSGVEVRVTPIRTEIIIRATRTREVLGEKGRRIRELTSLVQKRFGFPPDSVELFAERV 98
+G + +E++ +III + ++ +K RR+ EL VQK + + R+
Sbjct 58 EGIARIEIQKRIDLIQIIIYMGFPKLLIEDKPRRVEELQMNVQKELNCVNRKLNIAITRI 117
Query 99 ENRGLCAMAQAESLRYKLLKGLAVRRACYGVLRHIMESGAKGCEVVVSGKLRAQRAKSMK 158
N AE + +L ++ R+A + ++ KG +V ++G++ + ++
Sbjct 118 SNPYGDPNILAEFIAGQLKNRVSFRKAMKKAIELTEQANTKGIQVQIAGRIDGKEIARVE 177
Query 159 F-RDGYI-ISTGEPRQRFVSQAIRSVQLRQGVLGVRVSIML 197
+ R+G + + T E + + S +R++ GVLG+++ I +
Sbjct 178 WIREGRVPLQTIEAKIDYCSYTVRTI---YGVLGIKIWIFV 215
> hsa:22995 CEP152, FLJ21594, KIAA0912, MCPH4; centrosomal protein
152kDa
Length=1710
Score = 36.2 bits (82), Expect = 0.12, Method: Composition-based stats.
Identities = 31/95 (32%), Positives = 50/95 (52%), Gaps = 9/95 (9%)
Query 72 RIRELTSLVQKRFGFPPDSVELFAERVENRGL-CAMAQAESLRYKLLKGLAVRRACYGVL 130
++ EL +L + PP S L A +EN L C+ E LR + +K AV++ +L
Sbjct 1231 KLEELQTLCKT----PPRS--LSAGAIENACLPCSGGALEELRGQYIK--AVKKIKCDML 1282
Query 131 RHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGYII 165
R+I ES + E+V + LR ++ + K R Y+I
Sbjct 1283 RYIQESKERAAEMVKAEVLRERQETARKMRKYYLI 1317
> mmu:70902 Lpcat2b, 4921521K07Rik, Aytl1b; lysophosphatidylcholine
acyltransferase 2B
Length=516
Score = 33.9 bits (76), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 23/91 (25%), Positives = 41/91 (45%), Gaps = 7/91 (7%)
Query 43 GVEVRVTPIRTEIIIRATRTREVLGEKGRRI-RELTSLVQKRFGFPPDSVELFAERVENR 101
G+ V P TE I++ + L E G RELT+++Q FG P V +++ +
Sbjct 415 GLTVLCNPSNTEKILQMSFKLFDLDEDGYVTERELTTMLQAAFGVPDLDVSTLFQQMAGK 474
Query 102 GLCAMAQAESLRYKLLKGLAVRRACYGVLRH 132
++ + Y+ + A++ Y L H
Sbjct 475 ------DSDQVSYRTFRRFALKHPAYAKLFH 499
> mmu:17755 Mtap1b, A230055D22, AI843217, LC1, MAP1B, MAP5, MGC169657,
MGC169658, Mtap-5, Mtap5; microtubule-associated protein
1B; K10429 microtubule-associated protein 1
Length=2464
Score = 32.3 bits (72), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 34/136 (25%), Positives = 49/136 (36%), Gaps = 21/136 (15%)
Query 12 ISKKRKFVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEIIIRATRTREVLG---E 68
I K K DG + E+ E +RT E GYS EI + TRT EV G E
Sbjct 1922 IEKTIKTPEDGGYTCEITEKTTRTPEEGGYS----------YEISEKTTRTPEVSGYTYE 1971
Query 69 KGRRIRELTSLVQKRFGFPPDSVELFAE----RVENRGLCAMAQAESLRY----KLLKGL 120
K R R L + + D + + + ++ES Y K +
Sbjct 1972 KTERSRRLLDDISNGYDDTEDGGHTLGDCSYSYETTEKITSFPESESYSYETSTKTTRSP 2031
Query 121 AVRRACYGVLRHIMES 136
CY + I ++
Sbjct 2032 DTSAYCYETMEKITKT 2047
> xla:495686 usp47; ubiquitin specific peptidase 47; K11857 ubiquitin
carboxyl-terminal hydrolase 47 [EC:3.1.2.15]
Length=1350
Score = 32.0 bits (71), Expect = 2.0, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 26/57 (45%), Gaps = 3/57 (5%)
Query 44 VEVRVTPIRTEIIIRATRTREVLGEKGRRIRELTSLVQKRFGFPPDSVELFAERVEN 100
V+V V ++TE + R L + I E L+ K PPDS+ + ER N
Sbjct 700 VKVYVVDLKTETVASPVSVRAYLSQT---IIEFKQLISKSVDLPPDSMRVILERCYN 753
> mmu:99100 Cep152, AI851464, mKIAA0912; centrosomal protein 152
Length=1736
Score = 31.2 bits (69), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 34/61 (55%), Gaps = 2/61 (3%)
Query 104 CAMAQAESLRYKLLKGLAVRRACYGVLRHIMESGAKGCEVVVSGKLRAQRAKSMKFRDGY 163
C+ E LR + +K AVR+ +LR+I ES + E+V + LR ++ + K R+ Y
Sbjct 1294 CSRQALEELRGQYIK--AVRKIKRDMLRYIQESKERAAEMVKAEVLRERQETARKMRNYY 1351
Query 164 I 164
+
Sbjct 1352 L 1352
> tgo:TGME49_118610 hypothetical protein
Length=1369
Score = 30.0 bits (66), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 30/72 (41%), Gaps = 12/72 (16%)
Query 48 VTPIRTEIIIRATRTREVLGEKGRRIRELTSLVQKRFGFPPDSVELFAERVENRGLCAMA 107
V P RTE I R T V+ E+ R + L +L K F P V ++G C
Sbjct 1206 VAPDRTECITRGTGDENVISERERVLNVLQNL--KEFALP----------VASKGKCQKI 1253
Query 108 QAESLRYKLLKG 119
Q S L+ G
Sbjct 1254 QCASQSLVLIAG 1265
> sce:YCR014C POL4; DNA polymerase IV, undergoes pair-wise interactions
with Dnl4p-Lif1p and Rad27p to mediate repair of DNA
double-strand breaks by non-homologous end joining (NHEJ);
homologous to mammalian DNA polymerase beta (EC:2.7.7.7);
K10981 DNA polymerase IV [EC:2.7.7.7]
Length=582
Score = 29.6 bits (65), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 37/71 (52%), Gaps = 5/71 (7%)
Query 18 FVNDGVFQAELNEFLSRTLAEDGYSGVEVRVTPIRTEI----IIRATRTREVLGEKGRRI 73
F ND A++ E L L +DGY +++TP ++ I+ RT +++G G R
Sbjct 376 FCNDTTELAKIMETLCIKLYKDGYIHCFLQLTPNLEKLFLKRIVERFRTAKIVG-YGERK 434
Query 74 RELTSLVQKRF 84
R +S + K+F
Sbjct 435 RWYSSEIIKKF 445
Lambda K H
0.320 0.137 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8274230796
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40