bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0951_orf1
Length=105
Score E
Sequences producing significant alignments: (Bits) Value
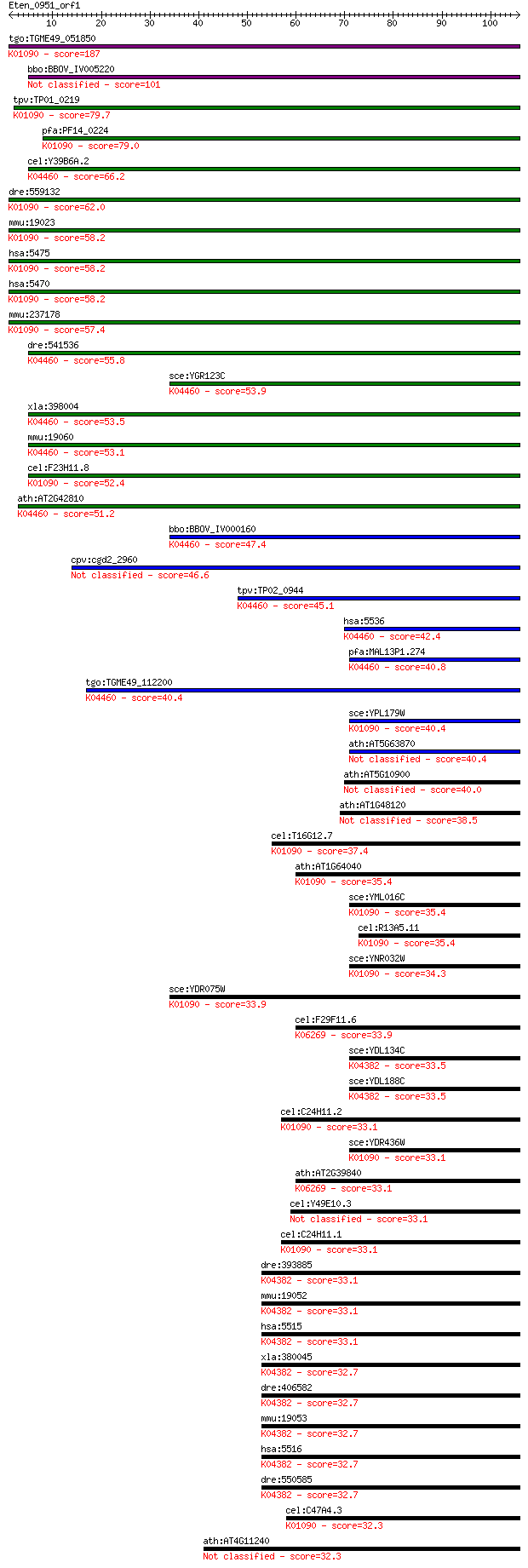
tgo:TGME49_051850 serine/threonine protein phosphatase 5, puta... 187 7e-48
bbo:BBOV_IV005220 23.m06074; serine/threonin protein phosphata... 101 7e-22
tpv:TP01_0219 serine/threonine protein phosphatase; K01090 pro... 79.7 2e-15
pfa:PF14_0224 PP7; serine/threonine protein phosphatase; K0109... 79.0 3e-15
cel:Y39B6A.2 pph-5; Protein PHosphatase family member (pph-5);... 66.2 2e-11
dre:559132 ppef1, MGC171580, zgc:171580; protein phosphatase, ... 62.0 4e-10
mmu:19023 Ppef2; protein phosphatase, EF hand calcium-binding ... 58.2 6e-09
hsa:5475 PPEF1, PP7, PPEF, PPP7C, PPP7CA; protein phosphatase,... 58.2 6e-09
hsa:5470 PPEF2, PPP7CB; protein phosphatase, EF-hand calcium b... 58.2 7e-09
mmu:237178 Ppef1, PPEF-1; protein phosphatase with EF hand cal... 57.4 1e-08
dre:541536 fc83f08, im:7146608, ppp5c, wu:fc83f08; zgc:110801 ... 55.8 3e-08
sce:YGR123C PPT1; Ppt1p (EC:3.1.3.16); K04460 protein phosphat... 53.9 1e-07
xla:398004 ppp5c, pp5; protein phosphatase 5, catalytic subuni... 53.5 1e-07
mmu:19060 Ppp5c, AU020526, PP5; protein phosphatase 5, catalyt... 53.1 2e-07
cel:F23H11.8 pef-1; Phosphatase with EF hands family member (p... 52.4 3e-07
ath:AT2G42810 PP5.2; PP5.2 (PROTEIN PHOSPHATASE 5.2); phosphop... 51.2 9e-07
bbo:BBOV_IV000160 21.m02802; serine/threonine protein phosphat... 47.4 1e-05
cpv:cgd2_2960 phosphoprotein phosphatase related 46.6 2e-05
tpv:TP02_0944 serine/threonine protein phosphatase; K04460 pro... 45.1 5e-05
hsa:5536 PPP5C, FLJ36922, FLJ55954, PP5, PPP5, PPT; protein ph... 42.4 4e-04
pfa:MAL13P1.274 PfPP5; serine/threonine protein phosphatase (E... 40.8 0.001
tgo:TGME49_112200 serine/threonine protein phosphatase, putati... 40.4 0.001
sce:YPL179W PPQ1, SAL6; Ppq1p (EC:3.1.3.16); K01090 protein ph... 40.4 0.001
ath:AT5G63870 PP7; PP7 (SERINE/THREONINE PHOSPHATASE 7); prote... 40.4 0.002
ath:AT5G10900 calcineurin-like phosphoesterase family protein 40.0 0.002
ath:AT1G48120 calcineurin-like phosphoesterase family protein 38.5 0.006
cel:T16G12.7 hypothetical protein; K01090 protein phosphatase ... 37.4 0.012
ath:AT1G64040 TOPP3; TOPP3; protein serine/threonine phosphata... 35.4 0.040
sce:YML016C PPZ1; Ppz1p (EC:3.1.3.16); K01090 protein phosphat... 35.4 0.048
cel:R13A5.11 hypothetical protein; K01090 protein phosphatase ... 35.4 0.050
sce:YNR032W PPG1; Ppg1p (EC:3.1.3.16); K01090 protein phosphat... 34.3 0.094
sce:YDR075W PPH3; Catalytic subunit of an evolutionarily conse... 33.9 0.11
cel:F29F11.6 gsp-1; yeast Glc Seven-like Phosphatases family m... 33.9 0.14
sce:YDL134C PPH21, PPH1; Catalytic subunit of protein phosphat... 33.5 0.18
sce:YDL188C PPH22, PPH2; Catalytic subunit of protein phosphat... 33.5 0.18
cel:C24H11.2 hypothetical protein; K01090 protein phosphatase ... 33.1 0.20
sce:YDR436W PPZ2; Ppz2p (EC:3.1.3.16); K01090 protein phosphat... 33.1 0.22
ath:AT2G39840 TOPP4; TOPP4; protein serine/threonine phosphata... 33.1 0.22
cel:Y49E10.3 pph-4.2; Protein PHosphatase family member (pph-4.2) 33.1 0.22
cel:C24H11.1 hypothetical protein; K01090 protein phosphatase ... 33.1 0.23
dre:393885 MGC56064, Ppp2ca; zgc:56064 (EC:3.1.3.16); K04382 p... 33.1 0.24
mmu:19052 Ppp2ca, PP2A, R75353; protein phosphatase 2 (formerl... 33.1 0.25
hsa:5515 PPP2CA, PP2Ac, PP2CA, PP2Calpha, RP-C; protein phosph... 33.1 0.25
xla:380045 ppp2ca, MGC53449, pp2ac; protein phosphatase 2, cat... 32.7 0.26
dre:406582 ppp2cb, fb17h07, fc05c02, fk24a01, wu:fb17h07, wu:f... 32.7 0.27
mmu:19053 Ppp2cb, AI115466, D8Ertd766e, PP2Ac; protein phospha... 32.7 0.27
hsa:5516 PPP2CB, PP2Abeta, PP2CB; protein phosphatase 2, catal... 32.7 0.27
dre:550585 ppp2ca, zgc:110641; protein phosphatase 2 (formerly... 32.7 0.30
cel:C47A4.3 hypothetical protein; K01090 protein phosphatase [... 32.3 0.34
ath:AT4G11240 TOPP7; TOPP7; protein serine/threonine phosphatase 32.3 0.34
> tgo:TGME49_051850 serine/threonine protein phosphatase 5, putative
(EC:3.1.2.15 3.1.3.16); K01090 protein phosphatase [EC:3.1.3.16]
Length=1086
Score = 187 bits (475), Expect = 7e-48, Method: Composition-based stats.
Identities = 79/105 (75%), Positives = 92/105 (87%), Gaps = 0/105 (0%)
Query 1 LPPGYKGPHLRWPITREFALNLIYHFRDHSDVPLPRKYALQLVMAVEEHYKQTMKGAVAE 60
+P Y GP ++WPITREF L+L+ H+RD+ DVPLPRKYAL+L+MA+ +HY++TMKGAV E
Sbjct 256 IPDDYAGPRVKWPITREFCLDLVDHYRDNPDVPLPRKYALELIMAITDHYRETMKGAVVE 315
Query 61 VEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
VEIPKK G RL LVGDTHGQLNDVLWI YKFGPPSATNVYLFNGD
Sbjct 316 VEIPKKGGSRLVLVGDTHGQLNDVLWIFYKFGPPSATNVYLFNGD 360
> bbo:BBOV_IV005220 23.m06074; serine/threonin protein phosphatase
(EC:3.1.3.16)
Length=952
Score = 101 bits (251), Expect = 7e-22, Method: Composition-based stats.
Identities = 46/101 (45%), Positives = 65/101 (64%), Gaps = 0/101 (0%)
Query 5 YKGPHLRWPITREFALNLIYHFRDHSDVPLPRKYALQLVMAVEEHYKQTMKGAVAEVEIP 64
Y GP L +T +FA L+ ++ +P +YA ++++ + E Y+ GA+ VE+P
Sbjct 173 YNGPKLGSKVTSKFANELLDAYKSGQISTVPIEYAQRILVDMLEWYRSNDNGAINTVEVP 232
Query 65 KKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
G L +VGD HGQLND+LWI +KFGPPS+ NVYLFNGD
Sbjct 233 PHENGNLIVVGDLHGQLNDLLWIFFKFGPPSSRNVYLFNGD 273
> tpv:TP01_0219 serine/threonine protein phosphatase; K01090 protein
phosphatase [EC:3.1.3.16]
Length=935
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 40/104 (38%), Positives = 53/104 (50%), Gaps = 20/104 (19%)
Query 2 PPGYKGPHLRWPITREFALNLIYHFRDHSDVPLPRKYALQLVMAVEEHYKQTMKGAVAEV 61
P Y GP L +T +FA L+ ++ ++ +G + +
Sbjct 173 PSDYNGPKLGKVVTLKFAHELLDMYKSGAN--------------------NNDEGVINHI 212
Query 62 EIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+IP L +VGD HGQLND+LWI YKFGPPS NVYLFNGD
Sbjct 213 DIPTHENSNLIIVGDLHGQLNDLLWIFYKFGPPSRRNVYLFNGD 256
> pfa:PF14_0224 PP7; serine/threonine protein phosphatase; K01090
protein phosphatase [EC:3.1.3.16]
Length=959
Score = 79.0 bits (193), Expect = 3e-15, Method: Composition-based stats.
Identities = 35/99 (35%), Positives = 62/99 (62%), Gaps = 1/99 (1%)
Query 8 PHLRWPITREFALNLIYHFRDHSDVPLPRKYALQLVMAVEEHYKQTMKGAVAEVEIPKKP 67
P L+ I R FA + + F ++ LP ++++ ++ ++ +K +V +++ KK
Sbjct 177 PRLKDKIDRTFATEMFHFFLTSKEIILPYNLVHKVLIKTKKMLEENIKSSVINLDMSKKS 236
Query 68 GG-RLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+L ++GD HGQL+DVLW+ +FG PS+TN+Y+FNGD
Sbjct 237 KDTKLIILGDVHGQLHDVLWLFNRFGIPSSTNIYIFNGD 275
> cel:Y39B6A.2 pph-5; Protein PHosphatase family member (pph-5);
K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=496
Score = 66.2 bits (160), Expect = 2e-11, Method: Composition-based stats.
Identities = 39/101 (38%), Positives = 52/101 (51%), Gaps = 6/101 (5%)
Query 5 YKGPHLRWPITREFALNLIYHFRDHSDVPLPRKYALQLVMAVEEHYKQTMKGAVAEVEIP 64
Y GP L IT+EF L LI F++ L +KYA ++++ + +K VEI
Sbjct 176 YDGPRLEDKITKEFVLQLIKTFKNQQK--LHKKYAFKMLL----EFYNYVKSLPTMVEIT 229
Query 65 KKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
G + + GD HGQ D+ I G PS TN YLFNGD
Sbjct 230 VPTGKKFTICGDVHGQFYDLCNIFEINGYPSETNPYLFNGD 270
> dre:559132 ppef1, MGC171580, zgc:171580; protein phosphatase,
EF-hand calcium binding domain 1 (EC:3.1.3.16); K01090 protein
phosphatase [EC:3.1.3.16]
Length=718
Score = 62.0 bits (149), Expect = 4e-10, Method: Composition-based stats.
Identities = 37/105 (35%), Positives = 54/105 (51%), Gaps = 6/105 (5%)
Query 1 LPPGYKGPHLRWPITREFALNLIYHFRDHSDVPLPRKYALQLVMAVEEHYKQTMKGAVAE 60
+P Y GP L +P++ L+ F++ L KY LQL+ ++ KQ
Sbjct 109 VPESYSGPRLSFPLSVTDTNALLSAFKEEQT--LHGKYVLQLLYETKKFLKQMPNVIYLS 166
Query 61 VEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
K+ + + GD HG+L+D+L I YK G PSA N Y+FNGD
Sbjct 167 TSYAKE----ITICGDLHGRLDDLLLIFYKNGLPSAENPYIFNGD 207
> mmu:19023 Ppef2; protein phosphatase, EF hand calcium-binding
domain 2 (EC:3.1.3.16); K01090 protein phosphatase [EC:3.1.3.16]
Length=757
Score = 58.2 bits (139), Expect = 6e-09, Method: Composition-based stats.
Identities = 34/105 (32%), Positives = 51/105 (48%), Gaps = 6/105 (5%)
Query 1 LPPGYKGPHLRWPITREFALNLIYHFRDHSDVPLPRKYALQLVMAVEEHYKQTMKGAVAE 60
+P Y GP L +P+ + A L+ FR L +Y L L+ +H Q
Sbjct 110 VPDSYTGPRLSFPLLPDHATALVEAFRLRQQ--LHARYVLNLLYETRKHLAQLPNINRVS 167
Query 61 VEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
++ + + GD HGQL+D+++I YK G PS Y+FNGD
Sbjct 168 TCYSEE----VTVCGDLHGQLDDLIFIFYKNGLPSPERAYVFNGD 208
> hsa:5475 PPEF1, PP7, PPEF, PPP7C, PPP7CA; protein phosphatase,
EF-hand calcium binding domain 1 (EC:3.1.3.16); K01090 protein
phosphatase [EC:3.1.3.16]
Length=653
Score = 58.2 bits (139), Expect = 6e-09, Method: Composition-based stats.
Identities = 34/105 (32%), Positives = 52/105 (49%), Gaps = 6/105 (5%)
Query 1 LPPGYKGPHLRWPITREFALNLIYHFRDHSDVPLPRKYALQLVMAVEEHYKQTMKGAVAE 60
+P Y GP L++P+T L+ F++ L Y L+++ K+ +K
Sbjct 103 VPDSYNGPRLQFPLTCTDIDLLLEAFKEQQ--ILHAHYVLEVLFET----KKVLKQMPNF 156
Query 61 VEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
I P + + GD HG+L+D+ I YK G PS N Y+FNGD
Sbjct 157 THIQTSPSKEVTICGDLHGKLDDLFLIFYKNGLPSERNPYVFNGD 201
> hsa:5470 PPEF2, PPP7CB; protein phosphatase, EF-hand calcium
binding domain 2 (EC:3.1.3.16); K01090 protein phosphatase
[EC:3.1.3.16]
Length=753
Score = 58.2 bits (139), Expect = 7e-09, Method: Composition-based stats.
Identities = 34/105 (32%), Positives = 52/105 (49%), Gaps = 6/105 (5%)
Query 1 LPPGYKGPHLRWPITREFALNLIYHFRDHSDVPLPRKYALQLVMAVEEHYKQTMKGAVAE 60
+P Y GP L +P+ + A L+ FR L +Y L L+ ++H Q
Sbjct 110 VPDSYTGPRLSFPLLPDHATALVEAFRLKQQ--LHARYVLNLLYETKKHLVQLPNINRVS 167
Query 61 VEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
++ + + GD HGQL+D+++I YK G PS Y+FNGD
Sbjct 168 TCYSEE----ITVCGDLHGQLDDLIFIFYKNGLPSPERSYVFNGD 208
> mmu:237178 Ppef1, PPEF-1; protein phosphatase with EF hand calcium-binding
domain 1 (EC:3.1.3.16); K01090 protein phosphatase
[EC:3.1.3.16]
Length=650
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 33/105 (31%), Positives = 52/105 (49%), Gaps = 6/105 (5%)
Query 1 LPPGYKGPHLRWPITREFALNLIYHFRDHSDVPLPRKYALQLVMAVEEHYKQTMKGAVAE 60
+P Y GP L++P+T L+ F+ L Y L+++ + KQ +
Sbjct 106 VPDSYDGPRLQFPLTFTDIHILLQAFKQQQ--ILHAHYVLEVLFEARKVLKQMPNFS--- 160
Query 61 VEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+ P + + GD HG+L+D++ I YK G PS N Y+FNGD
Sbjct 161 -HVKTFPAKEITICGDLHGKLDDLMLIFYKNGLPSENNPYVFNGD 204
> dre:541536 fc83f08, im:7146608, ppp5c, wu:fc83f08; zgc:110801
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=481
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 39/102 (38%), Positives = 54/102 (52%), Gaps = 7/102 (6%)
Query 5 YKGPHLR-WPITREFALNLIYHFRDHSDVPLPRKYALQLVMAVEEHYKQTMKGAVAEVEI 63
Y GP L +T +F +L+ F+D L RK A Q+++ V K+ + + VEI
Sbjct 158 YAGPKLEDGKVTLKFMEDLMDWFKDQKK--LHRKCAYQILVQV----KEVLTKLPSLVEI 211
Query 64 PKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
K ++ + GDTHGQ D+L I PS TN YLFNGD
Sbjct 212 TLKETEKITICGDTHGQYYDLLNIFELNSLPSPTNPYLFNGD 253
> sce:YGR123C PPT1; Ppt1p (EC:3.1.3.16); K04460 protein phosphatase
5 [EC:3.1.3.16]
Length=513
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 26/72 (36%), Positives = 42/72 (58%), Gaps = 2/72 (2%)
Query 34 LPRKYALQLVMAVEEHYKQTMKGAVAEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGP 93
LP+KY ++ + ++Q + ++ E+E P ++++ GDTHGQ DVL + KFG
Sbjct 209 LPKKYVAAIISHADTLFRQ--EPSMVELENNSTPDVKISVCGDTHGQFYDVLNLFRKFGK 266
Query 94 PSATNVYLFNGD 105
+ YLFNGD
Sbjct 267 VGPKHTYLFNGD 278
> xla:398004 ppp5c, pp5; protein phosphatase 5, catalytic subunit
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=493
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 34/102 (33%), Positives = 54/102 (52%), Gaps = 7/102 (6%)
Query 5 YKGPHLR-WPITREFALNLIYHFRDHSDVPLPRKYALQLVMAVEEHYKQTMKGAVAEVEI 63
Y GP L+ +T +F L L+ ++D L RK Q+++ V K + + VE+
Sbjct 170 YTGPQLQDGNVTVDFMLELMQFYKDQKK--LHRKCLYQMLVQV----KDILSKLPSLVEV 223
Query 64 PKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+ ++ + GDTHGQ D++ I + G PS N Y+FNGD
Sbjct 224 SLEKSQQVTVCGDTHGQFYDLMNIFHLNGLPSENNPYIFNGD 265
> mmu:19060 Ppp5c, AU020526, PP5; protein phosphatase 5, catalytic
subunit (EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=499
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 37/102 (36%), Positives = 52/102 (50%), Gaps = 7/102 (6%)
Query 5 YKGPHLR-WPITREFALNLIYHFRDHSDVPLPRKYALQLVMAVEEHYKQTMKGAVAEVEI 63
Y GP L +T F +L+ ++D L RK A Q+++ V K+ + VE
Sbjct 176 YSGPKLEDGKVTITFMKDLMQWYKDQKK--LHRKCAYQILVQV----KEVLCKLSTLVET 229
Query 64 PKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
K ++ + GDTHGQ D+L I G PS TN Y+FNGD
Sbjct 230 TLKETEKITVCGDTHGQFYDLLNIFELNGLPSETNPYIFNGD 271
> cel:F23H11.8 pef-1; Phosphatase with EF hands family member
(pef-1); K01090 protein phosphatase [EC:3.1.3.16]
Length=707
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 32/101 (31%), Positives = 49/101 (48%), Gaps = 6/101 (5%)
Query 5 YKGPHLRWPITREFALNLIYHFRDHSDVPLPRKYALQLVMAVEEHYKQTMKGAVAEVEIP 64
YKGP L P+ + +I F+ + L KY L ++ ++ K + I
Sbjct 191 YKGPTLSLPLDKPQVAKMIEAFK--VNKVLHPKYVLMIL----HEARKIFKAMPSVSRIS 244
Query 65 KKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
++ + GD HG+ +D+ ILYK G PS N Y+FNGD
Sbjct 245 TSISNQVTICGDLHGKFDDLCIILYKNGYPSVDNPYIFNGD 285
> ath:AT2G42810 PP5.2; PP5.2 (PROTEIN PHOSPHATASE 5.2); phosphoprotein
phosphatase/ protein binding / protein serine/threonine
phosphatase; K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=538
Score = 51.2 bits (121), Expect = 9e-07, Method: Composition-based stats.
Identities = 33/104 (31%), Positives = 51/104 (49%), Gaps = 7/104 (6%)
Query 3 PGYKGPHLRWP-ITREFALNLIYHFRDHSDVPLPRKYALQLVMAVEEHYKQTMKGAVAEV 61
P Y G + +T +F ++ F++ L ++YA Q+V+ +Q + + V
Sbjct 214 PQYSGARIEGEEVTLDFVKTMMEDFKNQKT--LHKRYAYQIVLQT----RQILLALPSLV 267
Query 62 EIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+I G + + GD HGQ D+L I G PS N YLFNGD
Sbjct 268 DISVPHGKHITVCGDVHGQFYDLLNIFELNGLPSEENPYLFNGD 311
> bbo:BBOV_IV000160 21.m02802; serine/threonine protein phosphatase
5 (EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=545
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 28/72 (38%), Positives = 41/72 (56%), Gaps = 4/72 (5%)
Query 34 LPRKYALQLVMAVEEHYKQTMKGAVAEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGP 93
L RKY +Q+++ V K+ +V ++ + +K + + GD HGQ D+L I G
Sbjct 251 LHRKYVIQIILDVINILKEY--DSVVDLSLTEKDD--ITICGDIHGQFYDLLNIFEINGS 306
Query 94 PSATNVYLFNGD 105
PS TN YLFNGD
Sbjct 307 PSETNGYLFNGD 318
> cpv:cgd2_2960 phosphoprotein phosphatase related
Length=525
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 30/92 (32%), Positives = 45/92 (48%), Gaps = 2/92 (2%)
Query 14 ITREFALNLIYHFRDHSDVPLPRKYALQLVMAVEEHYKQTMKGAVAEVEIPKKPGGRLAL 73
+ RE + + F + + L RKYA +V + + K+ + + I K+ + +
Sbjct 209 LIRESFVTELIDFLKNPENRLHRKYAYMIVYDLIQVLKEVASKPLVRINIGKQE--HITV 266
Query 74 VGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
GD HGQ D+L I G PS N YLFNGD
Sbjct 267 CGDIHGQFFDLLNIFDINGLPSVNNGYLFNGD 298
> tpv:TP02_0944 serine/threonine protein phosphatase; K04460 protein
phosphatase 5 [EC:3.1.3.16]
Length=548
Score = 45.1 bits (105), Expect = 5e-05, Method: Composition-based stats.
Identities = 24/58 (41%), Positives = 30/58 (51%), Gaps = 3/58 (5%)
Query 48 EHYKQTMKGAVAEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+HY Q + V ++ P L + GD HGQ D+L I G PS N YLFNGD
Sbjct 267 KHYSQEYESVV---DVNVYPSDELTVCGDIHGQFYDLLNIFSINGEPSDENSYLFNGD 321
> hsa:5536 PPP5C, FLJ36922, FLJ55954, PP5, PPP5, PPT; protein
phosphatase 5, catalytic subunit (EC:3.1.3.16); K04460 protein
phosphatase 5 [EC:3.1.3.16]
Length=477
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 19/36 (52%), Positives = 24/36 (66%), Gaps = 0/36 (0%)
Query 70 RLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
++ + GDTHGQ D+L I G PS TN Y+FNGD
Sbjct 214 KITVCGDTHGQFYDLLNIFELNGLPSETNPYIFNGD 249
> pfa:MAL13P1.274 PfPP5; serine/threonine protein phosphatase
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=658
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 19/35 (54%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 71 LALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
L + GD HGQ D+L I+ G PS N YLFNGD
Sbjct 398 LTICGDVHGQYYDLLNIMKINGYPSEKNSYLFNGD 432
> tgo:TGME49_112200 serine/threonine protein phosphatase, putative
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=548
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 30/90 (33%), Positives = 45/90 (50%), Gaps = 7/90 (7%)
Query 17 EFALNLIYHFRDHSDVPLPRKYALQLVMAVEEHYKQTMKGAVAEVEIPKKPG-GRLALVG 75
EF L +D S L +++A ++V+ V ++ G + +E + P G + + G
Sbjct 239 EFVGALKEFLKDTSK-RLHKQFAYRMVLDV-----ISLLGKLQTLERIRIPDDGEITVCG 292
Query 76 DTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
D HGQ D+ I G PS N YLFNGD
Sbjct 293 DVHGQYYDLCNIFELNGMPSKENPYLFNGD 322
> sce:YPL179W PPQ1, SAL6; Ppq1p (EC:3.1.3.16); K01090 protein
phosphatase [EC:3.1.3.16]
Length=549
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 21/35 (60%), Positives = 24/35 (68%), Gaps = 1/35 (2%)
Query 71 LALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+ +VGD HGQ ND+L IL G PS TN YLF GD
Sbjct 296 IKVVGDVHGQFNDLLRILKLSGVPSDTN-YLFLGD 329
> ath:AT5G63870 PP7; PP7 (SERINE/THREONINE PHOSPHATASE 7); protein
serine/threonine phosphatase
Length=301
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 24/35 (68%), Gaps = 0/35 (0%)
Query 71 LALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+ +VGD HGQL+D+L++L G P Y+FNGD
Sbjct 79 VVVVGDIHGQLHDLLFLLKDTGFPCQNRCYVFNGD 113
> ath:AT5G10900 calcineurin-like phosphoesterase family protein
Length=600
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 17/36 (47%), Positives = 25/36 (69%), Gaps = 0/36 (0%)
Query 70 RLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
R+ +VGD HGQL+D+L I + G PS ++FNG+
Sbjct 224 RVIVVGDLHGQLHDLLKIFDQSGRPSQNQCFVFNGN 259
> ath:AT1G48120 calcineurin-like phosphoesterase family protein
Length=1338
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 24/37 (64%), Gaps = 0/37 (0%)
Query 69 GRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+ +VGD HGQL+D+L+++ G P Y+FNG+
Sbjct 651 AEVVVVGDLHGQLHDLLYLMQDAGFPDGDRFYVFNGN 687
> cel:T16G12.7 hypothetical protein; K01090 protein phosphatase
[EC:3.1.3.16]
Length=319
Score = 37.4 bits (85), Expect = 0.012, Method: Composition-based stats.
Identities = 22/51 (43%), Positives = 29/51 (56%), Gaps = 7/51 (13%)
Query 55 KGAVAEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+GA+ E+E P K + GD HGQ +DVL + + G P N YLF GD
Sbjct 59 QGAMLELEAPVK------ICGDVHGQYSDVLRLFDRGGFPPLVN-YLFLGD 102
> ath:AT1G64040 TOPP3; TOPP3; protein serine/threonine phosphatase;
K01090 protein phosphatase [EC:3.1.3.16]
Length=322
Score = 35.4 bits (80), Expect = 0.040, Method: Composition-based stats.
Identities = 21/46 (45%), Positives = 26/46 (56%), Gaps = 7/46 (15%)
Query 60 EVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
E+E P K + GDTHGQ +D+L + G P A N YLF GD
Sbjct 51 ELEAPIK------ICGDTHGQFSDLLRLFEYGGYPPAAN-YLFLGD 89
> sce:YML016C PPZ1; Ppz1p (EC:3.1.3.16); K01090 protein phosphatase
[EC:3.1.3.16]
Length=692
Score = 35.4 bits (80), Expect = 0.048, Method: Composition-based stats.
Identities = 17/35 (48%), Positives = 23/35 (65%), Gaps = 1/35 (2%)
Query 71 LALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+ +VGD HGQ D+L + K G P ++N YLF GD
Sbjct 414 VKIVGDVHGQYGDLLRLFTKCGFPPSSN-YLFLGD 447
> cel:R13A5.11 hypothetical protein; K01090 protein phosphatase
[EC:3.1.3.16]
Length=421
Score = 35.4 bits (80), Expect = 0.050, Method: Composition-based stats.
Identities = 16/33 (48%), Positives = 21/33 (63%), Gaps = 1/33 (3%)
Query 73 LVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+VGD HGQ ND++ + G P T VY+F GD
Sbjct 157 VVGDLHGQFNDLINMFILLGRPPET-VYVFTGD 188
> sce:YNR032W PPG1; Ppg1p (EC:3.1.3.16); K01090 protein phosphatase
[EC:3.1.3.16]
Length=368
Score = 34.3 bits (77), Expect = 0.094, Method: Composition-based stats.
Identities = 18/35 (51%), Positives = 22/35 (62%), Gaps = 1/35 (2%)
Query 71 LALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+ +VGD HGQ +D+L I GP TN YLF GD
Sbjct 45 VTVVGDMHGQFHDMLEIFQIGGPVPDTN-YLFLGD 78
> sce:YDR075W PPH3; Catalytic subunit of an evolutionarily conserved
protein phosphatase complex containing Psy2p and the
regulatory subunit Psy4p; required for cisplatin resistance;
involved in activation of Gln3p (EC:3.1.3.16); K01090 protein
phosphatase [EC:3.1.3.16]
Length=308
Score = 33.9 bits (76), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 36/72 (50%), Gaps = 9/72 (12%)
Query 34 LPRKYALQLVMAVEEHYKQTMKGAVAEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGP 93
+P + +L + +E +G V +V+ P + + GD HGQL+D+L + K G
Sbjct 17 IPEETVFRLCLNSQELLMN--EGNVTQVDTP------VTICGDIHGQLHDLLTLFEKSGG 68
Query 94 PSATNVYLFNGD 105
T Y+F GD
Sbjct 69 VEKTR-YIFLGD 79
> cel:F29F11.6 gsp-1; yeast Glc Seven-like Phosphatases family
member (gsp-1); K06269 protein phosphatase 1, catalytic subunit
[EC:3.1.3.16]
Length=329
Score = 33.9 bits (76), Expect = 0.14, Method: Composition-based stats.
Identities = 22/48 (45%), Positives = 28/48 (58%), Gaps = 11/48 (22%)
Query 60 EVEIPKKPGGRLALVGDTHGQLNDVLWILYKFG--PPSATNVYLFNGD 105
E+E P L + GD HGQ ND+L L+++G PP A YLF GD
Sbjct 54 ELEAP------LKICGDIHGQYNDLLR-LFEYGGFPPEAN--YLFLGD 92
> sce:YDL134C PPH21, PPH1; Catalytic subunit of protein phosphatase
2A (PP2A), functionally redundant with Pph22p; methylated
at C terminus; forms alternate complexes with several regulatory
subunits; involved in signal transduction and regulation
of mitosis (EC:3.1.3.16); K04382 protein phosphatase 2
(formerly 2A), catalytic subunit [EC:3.1.3.16]
Length=369
Score = 33.5 bits (75), Expect = 0.18, Method: Composition-based stats.
Identities = 16/35 (45%), Positives = 21/35 (60%), Gaps = 1/35 (2%)
Query 71 LALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+ + GD HGQ +D+L L+K G P YLF GD
Sbjct 112 VTICGDVHGQFHDLLE-LFKIGGPCPDTNYLFMGD 145
> sce:YDL188C PPH22, PPH2; Catalytic subunit of protein phosphatase
2A (PP2A), functionally redundant with Pph21p; methylated
at C terminus; forms alternate complexes with several regulatory
subunits; involved in signal transduction and regulation
of mitosis (EC:3.1.3.16); K04382 protein phosphatase 2
(formerly 2A), catalytic subunit [EC:3.1.3.16]
Length=377
Score = 33.5 bits (75), Expect = 0.18, Method: Composition-based stats.
Identities = 16/35 (45%), Positives = 21/35 (60%), Gaps = 1/35 (2%)
Query 71 LALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+ + GD HGQ +D+L L+K G P YLF GD
Sbjct 120 VTICGDVHGQFHDLLE-LFKIGGPCPDTNYLFMGD 153
> cel:C24H11.2 hypothetical protein; K01090 protein phosphatase
[EC:3.1.3.16]
Length=384
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 20/49 (40%), Positives = 27/49 (55%), Gaps = 7/49 (14%)
Query 57 AVAEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
A+ E++ P + + GDTHGQ ND+L I G + T YLF GD
Sbjct 106 ALLELQAP------INICGDTHGQYNDLLRIFNACGAATKTQ-YLFLGD 147
> sce:YDR436W PPZ2; Ppz2p (EC:3.1.3.16); K01090 protein phosphatase
[EC:3.1.3.16]
Length=710
Score = 33.1 bits (74), Expect = 0.22, Method: Composition-based stats.
Identities = 17/35 (48%), Positives = 21/35 (60%), Gaps = 1/35 (2%)
Query 71 LALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+ +VGD HGQ D+L + K G P N YLF GD
Sbjct 449 VKIVGDVHGQYADLLRLFTKCGFPPMAN-YLFLGD 482
> ath:AT2G39840 TOPP4; TOPP4; protein serine/threonine phosphatase;
K06269 protein phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=321
Score = 33.1 bits (74), Expect = 0.22, Method: Composition-based stats.
Identities = 22/48 (45%), Positives = 29/48 (60%), Gaps = 11/48 (22%)
Query 60 EVEIPKKPGGRLALVGDTHGQLNDVLWILYKFG--PPSATNVYLFNGD 105
E+E P K + GD HGQ +D+L L+++G PPSA YLF GD
Sbjct 64 ELEAPIK------ICGDIHGQYSDLLR-LFEYGGFPPSAN--YLFLGD 102
> cel:Y49E10.3 pph-4.2; Protein PHosphatase family member (pph-4.2)
Length=321
Score = 33.1 bits (74), Expect = 0.22, Method: Composition-based stats.
Identities = 18/47 (38%), Positives = 26/47 (55%), Gaps = 4/47 (8%)
Query 59 AEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
A V++ P + + GD HGQ +D++ + G P TN YLF GD
Sbjct 50 ANVQVIDTP---VTICGDIHGQFHDLMELFRVGGSPPNTN-YLFLGD 92
> cel:C24H11.1 hypothetical protein; K01090 protein phosphatase
[EC:3.1.3.16]
Length=384
Score = 33.1 bits (74), Expect = 0.23, Method: Composition-based stats.
Identities = 20/49 (40%), Positives = 27/49 (55%), Gaps = 7/49 (14%)
Query 57 AVAEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
A+ E++ P + + GDTHGQ ND+L I G + T YLF GD
Sbjct 106 ALLELQAP------INICGDTHGQYNDLLRIFNACGAATKTQ-YLFLGD 147
> dre:393885 MGC56064, Ppp2ca; zgc:56064 (EC:3.1.3.16); K04382
protein phosphatase 2 (formerly 2A), catalytic subunit [EC:3.1.3.16]
Length=309
Score = 33.1 bits (74), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 7/53 (13%)
Query 53 TMKGAVAEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
T + V EV P + + GD HGQ +D++ L++ G S YLF GD
Sbjct 40 TKESNVQEVRCP------VTVCGDVHGQFHDLME-LFRIGGKSPDTNYLFMGD 85
> mmu:19052 Ppp2ca, PP2A, R75353; protein phosphatase 2 (formerly
2A), catalytic subunit, alpha isoform (EC:3.1.3.16); K04382
protein phosphatase 2 (formerly 2A), catalytic subunit [EC:3.1.3.16]
Length=309
Score = 33.1 bits (74), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 7/53 (13%)
Query 53 TMKGAVAEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
T + V EV P + + GD HGQ +D++ L++ G S YLF GD
Sbjct 40 TKESNVQEVRCP------VTVCGDVHGQFHDLME-LFRIGGKSPDTNYLFMGD 85
> hsa:5515 PPP2CA, PP2Ac, PP2CA, PP2Calpha, RP-C; protein phosphatase
2, catalytic subunit, alpha isozyme (EC:3.1.3.16); K04382
protein phosphatase 2 (formerly 2A), catalytic subunit
[EC:3.1.3.16]
Length=309
Score = 33.1 bits (74), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 7/53 (13%)
Query 53 TMKGAVAEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
T + V EV P + + GD HGQ +D++ L++ G S YLF GD
Sbjct 40 TKESNVQEVRCP------VTVCGDVHGQFHDLME-LFRIGGKSPDTNYLFMGD 85
> xla:380045 ppp2ca, MGC53449, pp2ac; protein phosphatase 2, catalytic
subunit, alpha isozyme (EC:3.1.3.16); K04382 protein
phosphatase 2 (formerly 2A), catalytic subunit [EC:3.1.3.16]
Length=309
Score = 32.7 bits (73), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 7/53 (13%)
Query 53 TMKGAVAEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
T + V EV P + + GD HGQ +D++ L++ G S YLF GD
Sbjct 40 TKESNVQEVRCP------VTVCGDVHGQFHDLME-LFRIGGKSPDTNYLFMGD 85
> dre:406582 ppp2cb, fb17h07, fc05c02, fk24a01, wu:fb17h07, wu:fc05c02,
wu:fk24a01, zgc:55808, zgc:77172; protein phosphatase
2 (formerly 2A), catalytic subunit, beta isoform (EC:3.1.3.16);
K04382 protein phosphatase 2 (formerly 2A), catalytic
subunit [EC:3.1.3.16]
Length=309
Score = 32.7 bits (73), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 7/53 (13%)
Query 53 TMKGAVAEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
T + V EV P + + GD HGQ +D++ L++ G S YLF GD
Sbjct 40 TKESNVQEVRCP------VTVCGDVHGQFHDLME-LFRIGGKSPDTNYLFMGD 85
> mmu:19053 Ppp2cb, AI115466, D8Ertd766e, PP2Ac; protein phosphatase
2 (formerly 2A), catalytic subunit, beta isoform (EC:3.1.3.16);
K04382 protein phosphatase 2 (formerly 2A), catalytic
subunit [EC:3.1.3.16]
Length=309
Score = 32.7 bits (73), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 7/53 (13%)
Query 53 TMKGAVAEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
T + V EV P + + GD HGQ +D++ L++ G S YLF GD
Sbjct 40 TKESNVQEVRCP------VTVCGDVHGQFHDLME-LFRIGGKSPDTNYLFMGD 85
> hsa:5516 PPP2CB, PP2Abeta, PP2CB; protein phosphatase 2, catalytic
subunit, beta isozyme (EC:3.1.3.16); K04382 protein phosphatase
2 (formerly 2A), catalytic subunit [EC:3.1.3.16]
Length=309
Score = 32.7 bits (73), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 7/53 (13%)
Query 53 TMKGAVAEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
T + V EV P + + GD HGQ +D++ L++ G S YLF GD
Sbjct 40 TKESNVQEVRCP------VTVCGDVHGQFHDLME-LFRIGGKSPDTNYLFMGD 85
> dre:550585 ppp2ca, zgc:110641; protein phosphatase 2 (formerly
2A), catalytic subunit, alpha isoform (EC:3.1.3.16); K04382
protein phosphatase 2 (formerly 2A), catalytic subunit [EC:3.1.3.16]
Length=309
Score = 32.7 bits (73), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 7/53 (13%)
Query 53 TMKGAVAEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+ + V EV P + + GD HGQ +D++ L+K G S YLF GD
Sbjct 40 SKESNVQEVRCP------VTVCGDVHGQFHDLME-LFKIGGKSPDTNYLFMGD 85
> cel:C47A4.3 hypothetical protein; K01090 protein phosphatase
[EC:3.1.3.16]
Length=316
Score = 32.3 bits (72), Expect = 0.34, Method: Composition-based stats.
Identities = 19/48 (39%), Positives = 25/48 (52%), Gaps = 7/48 (14%)
Query 58 VAEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVYLFNGD 105
+ EV P K + GD HGQ D+L + ++ G P N YLF GD
Sbjct 49 MVEVNAPIK------VCGDIHGQFPDLLRLFHRGGWPPTAN-YLFLGD 89
> ath:AT4G11240 TOPP7; TOPP7; protein serine/threonine phosphatase
Length=322
Score = 32.3 bits (72), Expect = 0.34, Method: Composition-based stats.
Identities = 24/65 (36%), Positives = 33/65 (50%), Gaps = 9/65 (13%)
Query 41 QLVMAVEEHYKQTMKGAVAEVEIPKKPGGRLALVGDTHGQLNDVLWILYKFGPPSATNVY 100
QL +A +E + + + E+E P K + GD HGQ D+L + G P A N Y
Sbjct 34 QLCLASKEVFLS--QPNLLELEAPIK------ICGDVHGQFPDLLRLFEYGGYPPAAN-Y 84
Query 101 LFNGD 105
LF GD
Sbjct 85 LFLGD 89
Lambda K H
0.321 0.142 0.456
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2017521068
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40