bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0997_orf1
Length=177
Score E
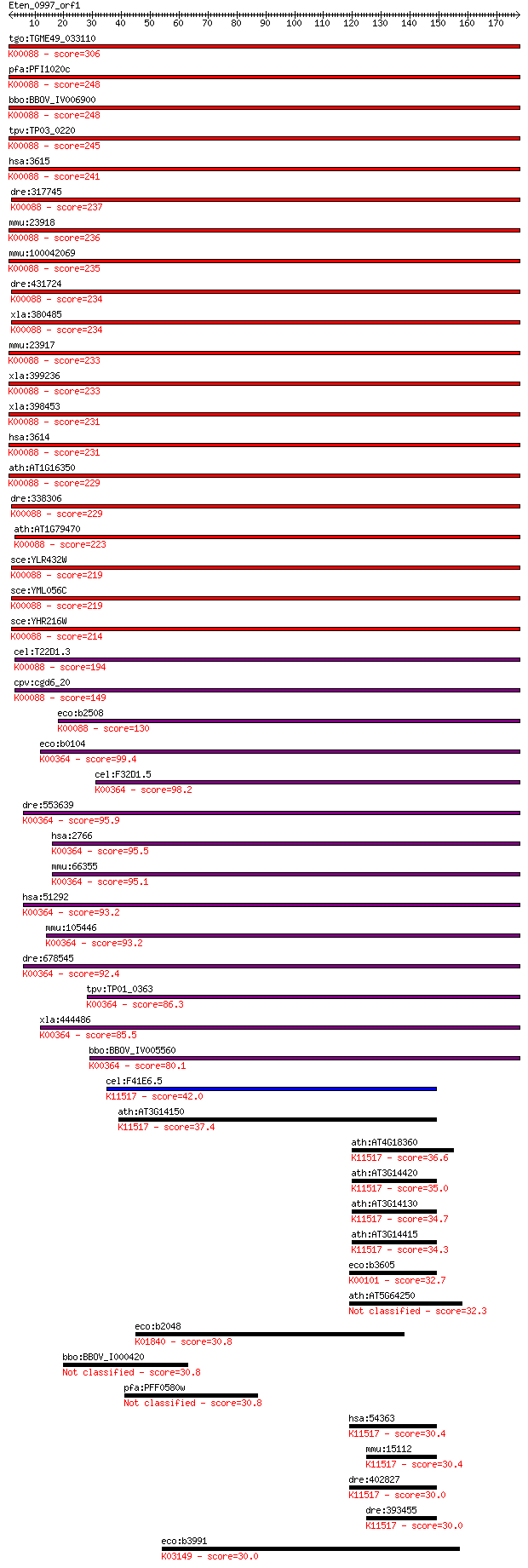
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_033110 inosine-5'-monophosphate dehydrogenase, puta... 306 3e-83
pfa:PFI1020c inosine-5'-monophosphate dehydrogenase (EC:1.1.1.... 248 7e-66
bbo:BBOV_IV006900 23.m05893; inosine-5'-monophosphate dehydrog... 248 9e-66
tpv:TP03_0220 inosine-5'-monophosphate dehydrogenase (EC:1.1.1... 245 6e-65
hsa:3615 IMPDH2, IMPD2, IMPDH-II; IMP (inosine 5'-monophosphat... 241 8e-64
dre:317745 impdh2, cb635, wu:fb64g02, wu:fc43f09; IMP (inosine... 237 1e-62
mmu:23918 Impdh2, IMPD; inosine 5'-phosphate dehydrogenase 2 (... 236 3e-62
mmu:100042069 Gm15210, OTTMUSG00000019498; predicted gene 1521... 235 5e-62
dre:431724 impdh1a, zgc:91911; inosine 5'-phosphate dehydrogen... 234 8e-62
xla:380485 impdh1, MGC52763, imp, imp1, imp2; IMP (inosine 5'-... 234 2e-61
mmu:23917 Impdh1, B930086D20Rik; inosine 5'-phosphate dehydrog... 233 2e-61
xla:399236 impdh2, MGC53611; inosine 5'-phosphate dehydrogenas... 233 4e-61
xla:398453 impdh2, MGC53627, imp2; IMP (inosine 5'-monophospha... 231 9e-61
hsa:3614 IMPDH1, DKFZp781N0678, IMPD, IMPD1, LCA11, RP10, sWSS... 231 1e-60
ath:AT1G16350 inosine-5'-monophosphate dehydrogenase, putative... 229 3e-60
dre:338306 impdh1b, IMPDH1, id:ibd5035, si:dkey-31f5.7, wu:fa0... 229 3e-60
ath:AT1G79470 inosine-5'-monophosphate dehydrogenase (EC:1.1.1... 223 4e-58
sce:YLR432W IMD3; Imd3p (EC:1.1.1.205); K00088 IMP dehydrogena... 219 4e-57
sce:YML056C IMD4; Imd4p (EC:1.1.1.205); K00088 IMP dehydrogena... 219 4e-57
sce:YHR216W IMD2, PUR5; Imd2p (EC:1.1.1.205); K00088 IMP dehyd... 214 9e-56
cel:T22D1.3 hypothetical protein; K00088 IMP dehydrogenase [EC... 194 1e-49
cpv:cgd6_20 inosine-5-monophosphate dehydrogenase (EC:1.1.1.20... 149 5e-36
eco:b2508 guaB, ECK2504, guaR, JW5401; IMP dehydrogenase (EC:1... 130 2e-30
eco:b0104 guaC, ECK0104, JW0101; GMP reductase (EC:1.7.1.7); K... 99.4 6e-21
cel:F32D1.5 hypothetical protein; K00364 GMP reductase [EC:1.7... 98.2 1e-20
dre:553639 gmpr, MGC110617, zgc:110617; guanosine monophosphat... 95.9 7e-20
hsa:2766 GMPR, GMPR1; guanosine monophosphate reductase (EC:1.... 95.5 8e-20
mmu:66355 Gmpr, 2310004P21Rik, AV028449; guanosine monophospha... 95.1 1e-19
hsa:51292 GMPR2, MGC15084, MGC830; guanosine monophosphate red... 93.2 4e-19
mmu:105446 Gmpr2, 1810008P16Rik, 5730544D12Rik, AA959850; guan... 93.2 4e-19
dre:678545 MGC136869; zgc:136869 (EC:1.7.1.7); K00364 GMP redu... 92.4 7e-19
tpv:TP01_0363 guanosine monophosphate reductase; K00364 GMP re... 86.3 6e-17
xla:444486 gmpr2, MGC81876; guanosine monophosphate reductase ... 85.5 8e-17
bbo:BBOV_IV005560 23.m05800; GMP reductase (EC:1.7.1.7); K0036... 80.1 4e-15
cel:F41E6.5 hypothetical protein; K11517 (S)-2-hydroxy-acid ox... 42.0 0.001
ath:AT3G14150 (S)-2-hydroxy-acid oxidase, peroxisomal, putativ... 37.4 0.029
ath:AT4G18360 (S)-2-hydroxy-acid oxidase, peroxisomal, putativ... 36.6 0.040
ath:AT3G14420 (S)-2-hydroxy-acid oxidase, peroxisomal, putativ... 35.0 0.14
ath:AT3G14130 (S)-2-hydroxy-acid oxidase, peroxisomal, putativ... 34.7 0.15
ath:AT3G14415 (S)-2-hydroxy-acid oxidase, peroxisomal, putativ... 34.3 0.25
eco:b3605 lldD, ECK3595, JW3580, lct, lctD; L-lactate dehydrog... 32.7 0.73
ath:AT5G64250 2-nitropropane dioxygenase family / NPD family 32.3 0.96
eco:b2048 cpsG, ECK2042, JW2033, manB, rfbK; phosphomannomutas... 30.8 2.5
bbo:BBOV_I000420 16.m00758; hypothetical protein 30.8 2.7
pfa:PFF0580w lsm12, putative 30.8 2.8
hsa:54363 HAO1, GOX, GOX1, HAOX1, MGC142225, MGC142227; hydrox... 30.4 3.1
mmu:15112 Hao1, GOX, Gox1, Hao-1, MGC141211; hydroxyacid oxida... 30.4 3.5
dre:402827 hao1; hydroxyacid oxidase (glycolate oxidase) 1 (EC... 30.0 4.1
dre:393455 hao2, MGC63690, zgc:63690; hydroxyacid oxidase 2 (l... 30.0 4.3
eco:b3991 thiG, ECK3982, JW5549, thiB; thiamin biosynthesis Th... 30.0 4.6
> tgo:TGME49_033110 inosine-5'-monophosphate dehydrogenase, putative
(EC:3.4.23.29 1.1.1.205); K00088 IMP dehydrogenase [EC:1.1.1.205]
Length=551
Score = 306 bits (783), Expect = 3e-83, Method: Compositional matrix adjust.
Identities = 146/177 (82%), Positives = 163/177 (92%), Gaps = 0/177 (0%)
Query 1 NKQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQII 60
NKQLLVGAA+ST+ D+ERAKAL + GADVLVVDSSQGDS++QVD+VK+LK+A+P QII
Sbjct 225 NKQLLVGAAVSTKPHDIERAKALQEAGADVLVVDSSQGDSIYQVDLVKRLKAAFPELQII 284
Query 61 GGNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANI 120
GGNVVTARQAKSLIDAGVD LRIGMGSGSICTTQVVCAVGRAQATAVYHV KYARE ++
Sbjct 285 GGNVVTARQAKSLIDAGVDGLRIGMGSGSICTTQVVCAVGRAQATAVYHVCKYAREHGDL 344
Query 121 PCIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
PCIADGGIQNSGHV+KALALGA+ VM+GS+LA TEEAPG YYFHNG RVK+YRGMGS
Sbjct 345 PCIADGGIQNSGHVMKALALGANAVMMGSMLAGTEEAPGEYYFHNGVRVKTYRGMGS 401
> pfa:PFI1020c inosine-5'-monophosphate dehydrogenase (EC:1.1.1.205);
K00088 IMP dehydrogenase [EC:1.1.1.205]
Length=510
Score = 248 bits (634), Expect = 7e-66, Method: Compositional matrix adjust.
Identities = 120/177 (67%), Positives = 147/177 (83%), Gaps = 1/177 (0%)
Query 1 NKQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQII 60
NKQL+VGA++STR DLERA L++ DV+ +DSSQG+S++Q+D +K++KSA+P+ II
Sbjct 224 NKQLIVGASISTREHDLERANQLIKNMIDVICIDSSQGNSIYQIDTIKKIKSAHPDIPII 283
Query 61 GGNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANI 120
GGNVVT++QAK+LIDAG D LRIGMGSGSICTTQ VCAVGRAQ TAVYHVSKYA NI
Sbjct 284 GGNVVTSQQAKNLIDAGADVLRIGMGSGSICTTQDVCAVGRAQGTAVYHVSKYAH-TRNI 342
Query 121 PCIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
IADGGI+NSG++VKAL+LGA VM+G+LLAATEE+ YYF N R+K YRGMGS
Sbjct 343 KTIADGGIKNSGNIVKALSLGADFVMLGNLLAATEESCSEYYFENNVRLKIYRGMGS 399
> bbo:BBOV_IV006900 23.m05893; inosine-5'-monophosphate dehydrogenase
(EC:1.1.1.205); K00088 IMP dehydrogenase [EC:1.1.1.205]
Length=505
Score = 248 bits (632), Expect = 9e-66, Method: Compositional matrix adjust.
Identities = 119/178 (66%), Positives = 146/178 (82%), Gaps = 1/178 (0%)
Query 1 NKQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQII 60
N QLLVG A+ST+ +E+AK L+ GADVLV+DSSQG+SV+Q+D++KQL+ +YPN QII
Sbjct 225 NMQLLVGVAISTQPGSIEKAKKLMDAGADVLVIDSSQGNSVYQIDLIKQLRQSYPNVQII 284
Query 61 GGNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVAN- 119
GGNVVT QAK+LIDAGVDALR+GMGSGSIC+TQ V VGR QATAVYHV+KYA E N
Sbjct 285 GGNVVTGSQAKNLIDAGVDALRVGMGSGSICSTQGVVGVGRPQATAVYHVAKYANEYGNG 344
Query 120 IPCIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
P IADGGI++SG ++KALALGAS M+G +A T E+PG +++HNG RVK YRGMGS
Sbjct 345 CPIIADGGIRSSGDIMKALALGASCCMLGGAIAGTNESPGDFFYHNGIRVKQYRGMGS 402
> tpv:TP03_0220 inosine-5'-monophosphate dehydrogenase (EC:1.1.1.205);
K00088 IMP dehydrogenase [EC:1.1.1.205]
Length=503
Score = 245 bits (625), Expect = 6e-65, Method: Compositional matrix adjust.
Identities = 117/178 (65%), Positives = 148/178 (83%), Gaps = 1/178 (0%)
Query 1 NKQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQII 60
NKQLLVGAA+STR L+ AK L+ D+LVVDSSQG+SVFQ+D++KQLKS YP+ Q++
Sbjct 225 NKQLLVGAAISTRGNGLDTAKKLIDAKVDILVVDSSQGNSVFQIDLIKQLKSVYPDFQVM 284
Query 61 GGNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVAN- 119
GNVVTA+QAK+L++AG D++++GMG GSICTTQ +C VGR QA+AVY+VS+YA E N
Sbjct 285 AGNVVTAQQAKNLLEAGCDSIKVGMGIGSICTTQNICGVGRGQASAVYYVSRYAFEHWNG 344
Query 120 IPCIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
IP IADGGI++SG +VKAL+LGAS VM GSL A ++E PG YYF+NG R+KSYRGMGS
Sbjct 345 IPIIADGGIKSSGDIVKALSLGASCVMGGSLFAGSKETPGEYYFNNGVRMKSYRGMGS 402
> hsa:3615 IMPDH2, IMPD2, IMPDH-II; IMP (inosine 5'-monophosphate)
dehydrogenase 2 (EC:1.1.1.205); K00088 IMP dehydrogenase
[EC:1.1.1.205]
Length=514
Score = 241 bits (615), Expect = 8e-64, Method: Compositional matrix adjust.
Identities = 120/177 (67%), Positives = 140/177 (79%), Gaps = 1/177 (0%)
Query 1 NKQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQII 60
KQLL GAA+ T D R L Q G DV+V+DSSQG+S+FQ++++K +K YPN Q+I
Sbjct 241 KKQLLCGAAIGTHEDDKYRLDLLAQAGVDVVVLDSSQGNSIFQINMIKYIKDKYPNLQVI 300
Query 61 GGNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANI 120
GGNVVTA QAK+LIDAGVDALR+GMGSGSIC TQ V A GR QATAVY VS+YAR +
Sbjct 301 GGNVVTAAQAKNLIDAGVDALRVGMGSGSICITQEVLACGRPQATAVYKVSEYARRFG-V 359
Query 121 PCIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
P IADGGIQN GH+ KALALGASTVM+GSLLAAT EAPG Y+F +G R+K YRGMGS
Sbjct 360 PVIADGGIQNVGHIAKALALGASTVMMGSLLAATTEAPGEYFFSDGIRLKKYRGMGS 416
> dre:317745 impdh2, cb635, wu:fb64g02, wu:fc43f09; IMP (inosine
monophosphate) dehydrogenase 2 (EC:1.1.1.205); K00088 IMP
dehydrogenase [EC:1.1.1.205]
Length=514
Score = 237 bits (605), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 118/176 (67%), Positives = 138/176 (78%), Gaps = 1/176 (0%)
Query 2 KQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQIIG 61
KQLL GAA+ T D R L Q G DV+V+DSSQG+S+FQ++++K +K YPN Q+IG
Sbjct 242 KQLLCGAAIGTHNDDKYRLDLLAQAGVDVVVLDSSQGNSIFQINMIKYIKEKYPNVQVIG 301
Query 62 GNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIP 121
GNVVTA QAK+LIDAG DALR+GMGSGSIC TQ V A GR QATAVY VS+YAR +P
Sbjct 302 GNVVTAAQAKNLIDAGADALRVGMGSGSICITQEVLACGRPQATAVYKVSEYARRFG-VP 360
Query 122 CIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
IADGGIQ GH+ KALALGASTVM+GSLLAAT EAPG Y+F +G R+K YRGMGS
Sbjct 361 VIADGGIQTVGHIAKALALGASTVMMGSLLAATSEAPGEYFFSDGIRLKKYRGMGS 416
> mmu:23918 Impdh2, IMPD; inosine 5'-phosphate dehydrogenase 2
(EC:1.1.1.205); K00088 IMP dehydrogenase [EC:1.1.1.205]
Length=514
Score = 236 bits (602), Expect = 3e-62, Method: Compositional matrix adjust.
Identities = 118/177 (66%), Positives = 139/177 (78%), Gaps = 1/177 (0%)
Query 1 NKQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQII 60
KQLL GAA+ T D R L G DV+V+DSSQG+S+FQ++++K +K YP+ Q+I
Sbjct 241 KKQLLCGAAIGTHEDDKYRLDLLALAGVDVVVLDSSQGNSIFQINMIKYIKEKYPSLQVI 300
Query 61 GGNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANI 120
GGNVVTA QAK+LIDAGVDALR+GMGSGSIC TQ V A GR QATAVY VS+YAR +
Sbjct 301 GGNVVTAAQAKNLIDAGVDALRVGMGSGSICITQEVLACGRPQATAVYKVSEYARRFG-V 359
Query 121 PCIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
P IADGGIQN GH+ KALALGASTVM+GSLLAAT EAPG Y+F +G R+K YRGMGS
Sbjct 360 PVIADGGIQNVGHIAKALALGASTVMMGSLLAATTEAPGEYFFSDGIRLKKYRGMGS 416
> mmu:100042069 Gm15210, OTTMUSG00000019498; predicted gene 15210;
K00088 IMP dehydrogenase [EC:1.1.1.205]
Length=544
Score = 235 bits (600), Expect = 5e-62, Method: Compositional matrix adjust.
Identities = 117/177 (66%), Positives = 138/177 (77%), Gaps = 1/177 (0%)
Query 1 NKQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQII 60
KQLL GAA+ T D R L G DV+V+DSSQG+S+FQ++++K +K YP+ Q+I
Sbjct 271 KKQLLCGAAIGTHEDDKYRLDLLALAGVDVVVLDSSQGNSIFQINMIKYIKEKYPSLQVI 330
Query 61 GGNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANI 120
GGNVVTA QAK+LIDAGVDALR+GMGSGSIC TQ V A GR QATAVY VS+YAR +
Sbjct 331 GGNVVTAAQAKNLIDAGVDALRVGMGSGSICITQEVLACGRPQATAVYKVSEYARRFG-V 389
Query 121 PCIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
P IADGGIQN GH+ KALA GASTVM+GSLLAAT EAPG Y+F +G R+K YRGMGS
Sbjct 390 PVIADGGIQNVGHIAKALAFGASTVMMGSLLAATTEAPGEYFFSDGIRLKKYRGMGS 446
> dre:431724 impdh1a, zgc:91911; inosine 5'-phosphate dehydrogenase
1a (EC:1.1.1.205); K00088 IMP dehydrogenase [EC:1.1.1.205]
Length=544
Score = 234 bits (598), Expect = 8e-62, Method: Compositional matrix adjust.
Identities = 116/176 (65%), Positives = 137/176 (77%), Gaps = 1/176 (0%)
Query 2 KQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQIIG 61
KQLL GAA+ TR D R L+Q G DV+V+DSSQG+SVFQ+ ++ +K YP Q++G
Sbjct 242 KQLLCGAAIGTREDDKYRLDLLMQAGVDVIVLDSSQGNSVFQISMINYIKQKYPELQVVG 301
Query 62 GNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIP 121
GNVVTA QAK+LIDAGVDALR+GMG GSIC TQ V A GR Q T+VY V++YAR +P
Sbjct 302 GNVVTAAQAKNLIDAGVDALRVGMGCGSICITQEVMACGRPQGTSVYKVAEYARRFG-VP 360
Query 122 CIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
IADGGIQ GHVVKALALGASTVM+GSLLAAT EAPG Y+F +G R+K YRGMGS
Sbjct 361 VIADGGIQTVGHVVKALALGASTVMMGSLLAATTEAPGEYFFSDGVRLKKYRGMGS 416
> xla:380485 impdh1, MGC52763, imp, imp1, imp2; IMP (inosine 5'-monophosphate)
dehydrogenase 1 (EC:1.1.1.205); K00088 IMP
dehydrogenase [EC:1.1.1.205]
Length=514
Score = 234 bits (596), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 115/176 (65%), Positives = 138/176 (78%), Gaps = 1/176 (0%)
Query 2 KQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQIIG 61
KQLL GAA+ TR D R L+Q G DV+V+DSSQG+SV+Q++++ +K YP Q++G
Sbjct 242 KQLLCGAAIGTREDDKYRLDLLMQAGVDVVVLDSSQGNSVYQINMIHYIKQKYPELQVVG 301
Query 62 GNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIP 121
GNVVTA QAK+LIDAGVDALR+GMG GSIC TQ V A GR Q TAVY V++YAR +P
Sbjct 302 GNVVTAAQAKNLIDAGVDALRVGMGCGSICITQEVMACGRPQGTAVYKVAEYARRFG-VP 360
Query 122 CIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
+ADGGIQ GHVVKALALGASTVM+GSLLAAT EAPG Y+F +G R+K YRGMGS
Sbjct 361 VVADGGIQTVGHVVKALALGASTVMMGSLLAATTEAPGEYFFSDGVRLKKYRGMGS 416
> mmu:23917 Impdh1, B930086D20Rik; inosine 5'-phosphate dehydrogenase
1 (EC:1.1.1.205); K00088 IMP dehydrogenase [EC:1.1.1.205]
Length=514
Score = 233 bits (594), Expect = 2e-61, Method: Compositional matrix adjust.
Identities = 118/177 (66%), Positives = 138/177 (77%), Gaps = 1/177 (0%)
Query 1 NKQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQII 60
+KQLL GAA+ TR D R L Q GADV+V+DSSQG+SV+Q+ +V +K YP+ Q+I
Sbjct 241 HKQLLCGAAVGTREDDKYRLDLLTQAGADVIVLDSSQGNSVYQIAMVHYIKQKYPHLQVI 300
Query 61 GGNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANI 120
GGNVVTA QAK+LIDAGVD LR+GMG GSIC TQ V A GR Q TAVY V++YAR +
Sbjct 301 GGNVVTAAQAKNLIDAGVDGLRVGMGCGSICITQEVMACGRPQGTAVYKVAEYARRFG-V 359
Query 121 PCIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
P IADGGIQ GHVVKALALGASTVM+GSLLAAT EAPG Y+F +G R+K YRGMGS
Sbjct 360 PVIADGGIQTVGHVVKALALGASTVMMGSLLAATTEAPGEYFFSDGVRLKKYRGMGS 416
> xla:399236 impdh2, MGC53611; inosine 5'-phosphate dehydrogenase
2 (EC:1.1.1.205); K00088 IMP dehydrogenase [EC:1.1.1.205]
Length=514
Score = 233 bits (593), Expect = 4e-61, Method: Compositional matrix adjust.
Identities = 115/177 (64%), Positives = 137/177 (77%), Gaps = 1/177 (0%)
Query 1 NKQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQII 60
KQLL GAA+ T D R L+Q G D +V+DSSQG+S+FQ++++K +K YP+ Q+I
Sbjct 241 KKQLLCGAAIGTHEDDKYRLDLLVQAGVDAVVLDSSQGNSIFQINMIKYIKEKYPDLQVI 300
Query 61 GGNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANI 120
GNVVTA QAK+LIDAG DALR+GMGSGSIC TQ V A GR QATAVY VS+YAR +
Sbjct 301 AGNVVTAAQAKNLIDAGADALRVGMGSGSICITQEVLACGRPQATAVYKVSEYARRFG-V 359
Query 121 PCIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
P IADGGIQ GH+ KALALGASTVM+GSLLAAT EAPG Y+F +G R+K YRGMGS
Sbjct 360 PVIADGGIQTVGHIAKALALGASTVMMGSLLAATTEAPGEYFFSDGIRLKKYRGMGS 416
> xla:398453 impdh2, MGC53627, imp2; IMP (inosine 5'-monophosphate)
dehydrogenase 2; K00088 IMP dehydrogenase [EC:1.1.1.205]
Length=514
Score = 231 bits (589), Expect = 9e-61, Method: Compositional matrix adjust.
Identities = 115/177 (64%), Positives = 136/177 (76%), Gaps = 1/177 (0%)
Query 1 NKQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQII 60
KQLL GAA T D R L+Q G D +V+DSSQG+S+FQ++++K +K YP+ Q+I
Sbjct 241 KKQLLCGAATGTHEDDKYRLDLLVQAGVDAVVLDSSQGNSIFQINMIKYIKEKYPDLQVI 300
Query 61 GGNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANI 120
GNVVTA QAK+LIDAG DALR+GMGSGSIC TQ V A GR QATAVY VS+YAR +
Sbjct 301 AGNVVTAAQAKNLIDAGADALRVGMGSGSICITQEVLACGRPQATAVYKVSEYARRFG-V 359
Query 121 PCIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
P IADGGIQ GH+ KALALGASTVM+GSLLAAT EAPG Y+F +G R+K YRGMGS
Sbjct 360 PVIADGGIQTVGHIAKALALGASTVMMGSLLAATTEAPGEYFFSDGIRLKKYRGMGS 416
> hsa:3614 IMPDH1, DKFZp781N0678, IMPD, IMPD1, LCA11, RP10, sWSS2608;
IMP (inosine 5'-monophosphate) dehydrogenase 1 (EC:1.1.1.205);
K00088 IMP dehydrogenase [EC:1.1.1.205]
Length=599
Score = 231 bits (588), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 117/177 (66%), Positives = 136/177 (76%), Gaps = 1/177 (0%)
Query 1 NKQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQII 60
KQLL GAA+ TR D R L Q G DV+V+DSSQG+SV+Q+ +V +K YP+ Q+I
Sbjct 326 QKQLLCGAAVGTREDDKYRLDLLTQAGVDVIVLDSSQGNSVYQIAMVHYIKQKYPHLQVI 385
Query 61 GGNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANI 120
GGNVVTA QAK+LIDAGVD LR+GMG GSIC TQ V A GR Q TAVY V++YAR +
Sbjct 386 GGNVVTAAQAKNLIDAGVDGLRVGMGCGSICITQEVMACGRPQGTAVYKVAEYARRFG-V 444
Query 121 PCIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
P IADGGIQ GHVVKALALGASTVM+GSLLAAT EAPG Y+F +G R+K YRGMGS
Sbjct 445 PIIADGGIQTVGHVVKALALGASTVMMGSLLAATTEAPGEYFFSDGVRLKKYRGMGS 501
> ath:AT1G16350 inosine-5'-monophosphate dehydrogenase, putative
(EC:1.1.1.205); K00088 IMP dehydrogenase [EC:1.1.1.205]
Length=502
Score = 229 bits (585), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 111/177 (62%), Positives = 142/177 (80%), Gaps = 1/177 (0%)
Query 1 NKQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQII 60
+K+ +VGAA+ TR +D ER + L++ GA+V+V+DSSQG+S++Q++++K +K+ YP ++
Sbjct 231 DKKWMVGAAIGTRESDKERLEHLVKAGANVVVLDSSQGNSIYQLEMIKYVKNTYPELDVV 290
Query 61 GGNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANI 120
GGNVVT QA++LI AGVD LR+GMGSGSICTTQ VCAVGR QATAVY VS A + +
Sbjct 291 GGNVVTMYQAENLIKAGVDGLRVGMGSGSICTTQEVCAVGRGQATAVYKVSTLAAQ-HGV 349
Query 121 PCIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
P IADGGI NSGH+VKAL LGASTVM+GS LA + EAPGAY + NG RVK YRGMGS
Sbjct 350 PVIADGGISNSGHIVKALVLGASTVMMGSFLAGSTEAPGAYEYRNGRRVKKYRGMGS 406
> dre:338306 impdh1b, IMPDH1, id:ibd5035, si:dkey-31f5.7, wu:fa09h11,
wu:fa99c03, zgc:113446; inosine 5'-phosphate dehydrogenase
1b (EC:1.1.1.205); K00088 IMP dehydrogenase [EC:1.1.1.205]
Length=514
Score = 229 bits (585), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 113/176 (64%), Positives = 137/176 (77%), Gaps = 1/176 (0%)
Query 2 KQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQIIG 61
KQLL GAA+ TR D R L Q G D++V+DSSQG+SV+Q++++ +K YP Q++G
Sbjct 242 KQLLCGAAIGTREDDKYRLDLLTQSGVDMVVLDSSQGNSVYQINMIHYIKQKYPELQVVG 301
Query 62 GNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIP 121
GNVVTA QAK+LIDAGVDALR+GMG GSIC TQ V A GR Q T+VY V++YAR +P
Sbjct 302 GNVVTAAQAKNLIDAGVDALRVGMGCGSICITQEVMACGRPQGTSVYKVAEYARRFG-VP 360
Query 122 CIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
IADGGIQ GHVVKAL+LGASTVM+GSLLAAT EAPG Y+F +G R+K YRGMGS
Sbjct 361 VIADGGIQTVGHVVKALSLGASTVMMGSLLAATTEAPGEYFFSDGVRLKKYRGMGS 416
> ath:AT1G79470 inosine-5'-monophosphate dehydrogenase (EC:1.1.1.205);
K00088 IMP dehydrogenase [EC:1.1.1.205]
Length=503
Score = 223 bits (567), Expect = 4e-58, Method: Compositional matrix adjust.
Identities = 108/175 (61%), Positives = 136/175 (77%), Gaps = 1/175 (0%)
Query 3 QLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQIIGG 62
+ +VGAA+ TR +D ER + L+ VG + +V+DSSQG+S++Q++++K +K YP +IGG
Sbjct 234 EWMVGAAIGTRESDKERLEHLVNVGVNAVVLDSSQGNSIYQLEMIKYVKKTYPELDVIGG 293
Query 63 NVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIPC 122
NVVT QA++LI AGVD LR+GMGSGSICTTQ VCAVGR QATAVY V A + + IP
Sbjct 294 NVVTMYQAQNLIQAGVDGLRVGMGSGSICTTQEVCAVGRGQATAVYKVCSIAAQ-SGIPV 352
Query 123 IADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
IADGGI NSGH+VKAL LGASTVM+GS LA + EAPG Y + NG R+K YRGMGS
Sbjct 353 IADGGISNSGHIVKALVLGASTVMMGSFLAGSTEAPGGYEYTNGKRIKKYRGMGS 407
> sce:YLR432W IMD3; Imd3p (EC:1.1.1.205); K00088 IMP dehydrogenase
[EC:1.1.1.205]
Length=523
Score = 219 bits (558), Expect = 4e-57, Method: Compositional matrix adjust.
Identities = 102/176 (57%), Positives = 136/176 (77%), Gaps = 1/176 (0%)
Query 2 KQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQIIG 61
KQLL GAA+ T AD ER + L++ G DV+++DSSQG+S+FQ++++K +K +P+ +II
Sbjct 246 KQLLCGAAIGTIDADKERLRLLVEAGLDVVILDSSQGNSIFQLNMIKWIKETFPDLEIIA 305
Query 62 GNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIP 121
GNV T QA +LI AG D LRIGMGSGSIC TQ V A GR Q TAVY+V ++A + IP
Sbjct 306 GNVATREQAANLIAAGADGLRIGMGSGSICITQEVMACGRPQGTAVYNVCEFANQFG-IP 364
Query 122 CIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
C+ADGG+QN GH+ KALALG+STVM+G +LA T E+PG Y++ +G R+K+YRGMGS
Sbjct 365 CMADGGVQNIGHITKALALGSSTVMMGGMLAGTTESPGEYFYQDGKRLKAYRGMGS 420
> sce:YML056C IMD4; Imd4p (EC:1.1.1.205); K00088 IMP dehydrogenase
[EC:1.1.1.205]
Length=524
Score = 219 bits (558), Expect = 4e-57, Method: Compositional matrix adjust.
Identities = 102/176 (57%), Positives = 136/176 (77%), Gaps = 1/176 (0%)
Query 2 KQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQIIG 61
KQLL GAA+ T AD ER + L++ G DV+++DSSQG+SVFQ++++K +K +P+ +II
Sbjct 247 KQLLCGAAIGTIEADKERLRLLVEAGLDVVILDSSQGNSVFQLNMIKWIKETFPDLEIIA 306
Query 62 GNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIP 121
GNV T QA +LI AG D LRIGMGSGSIC TQ V A GR Q TAVY+V ++A + +P
Sbjct 307 GNVATREQAANLIAAGADGLRIGMGSGSICITQEVMACGRPQGTAVYNVCQFANQFG-VP 365
Query 122 CIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
C+ADGG+QN GH+ KALALG+STVM+G +LA T E+PG Y++ +G R+K+YRGMGS
Sbjct 366 CMADGGVQNIGHITKALALGSSTVMMGGMLAGTTESPGEYFYKDGKRLKAYRGMGS 421
> sce:YHR216W IMD2, PUR5; Imd2p (EC:1.1.1.205); K00088 IMP dehydrogenase
[EC:1.1.1.205]
Length=523
Score = 214 bits (546), Expect = 9e-56, Method: Compositional matrix adjust.
Identities = 98/176 (55%), Positives = 137/176 (77%), Gaps = 1/176 (0%)
Query 2 KQLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQIIG 61
KQLL GA++ T AD ER + L++ G DV+++DSSQG+S+F+++++K +K ++P ++I
Sbjct 246 KQLLCGASIGTMDADKERLRLLVKAGLDVVILDSSQGNSIFELNMLKWVKESFPGLEVIA 305
Query 62 GNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIP 121
GNVVT QA +LI AG D LRIGMG+GSIC TQ V A GR Q TAVY+V ++A + +P
Sbjct 306 GNVVTREQAANLIAAGADGLRIGMGTGSICITQEVMACGRPQGTAVYNVCEFANQFG-VP 364
Query 122 CIADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
C+ADGG+QN GH+ KALALG+STVM+G +LA T E+PG Y++ +G R+K+YRGMGS
Sbjct 365 CMADGGVQNIGHITKALALGSSTVMMGGMLAGTTESPGEYFYQDGKRLKAYRGMGS 420
> cel:T22D1.3 hypothetical protein; K00088 IMP dehydrogenase [EC:1.1.1.205]
Length=534
Score = 194 bits (493), Expect = 1e-49, Method: Compositional matrix adjust.
Identities = 99/176 (56%), Positives = 128/176 (72%), Gaps = 2/176 (1%)
Query 3 QLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQIIGG 62
QLL GAA++TR +++ G DVL++DSS G S +Q+ +++ +K +P+ Q+I G
Sbjct 261 QLLCGAAVNTRGESQYTVDRVVEAGVDVLIIDSSNGSSTYQISMLRYIKEKHPHVQVIAG 320
Query 63 NVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIPC 122
NVVT QAK LID G D LRIGMGSGSIC TQ V AVGRAQ TAVY V++YA + IP
Sbjct 321 NVVTRAQAKLLIDQGADGLRIGMGSGSICITQDVMAVGRAQGTAVYDVARYANQ-RGIPI 379
Query 123 IADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYF-HNGARVKSYRGMGS 177
+ADGGI++ G++ KA++LGAS VM+G LLAAT EAPG Y++ G RVK YRGMGS
Sbjct 380 VADGGIRDVGYITKAISLGASAVMMGGLLAATTEAPGEYFWGPGGVRVKKYRGMGS 435
> cpv:cgd6_20 inosine-5-monophosphate dehydrogenase (EC:1.1.1.205);
K00088 IMP dehydrogenase [EC:1.1.1.205]
Length=402
Score = 149 bits (376), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 84/175 (48%), Positives = 115/175 (65%), Gaps = 4/175 (2%)
Query 3 QLLVGAALSTRAADLERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQIIGG 62
+L VGAA+ ++ERAK L++ G DV+V+DS+ G S+ + +K++KS N +I G
Sbjct 136 RLRVGAAIGVN--EIERAKLLVEAGVDVIVLDSAHGHSLNIIRTLKEIKSKM-NIDVIVG 192
Query 63 NVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIPC 122
NVVT K LI+ G D +++G+G GSICTT++V VG Q TA+ S A + IP
Sbjct 193 NVVTEEATKELIENGADGIKVGIGPGSICTTRIVAGVGVPQITAIEKCSSVASKFG-IPI 251
Query 123 IADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
IADGGI+ SG + KALA+GAS+VM+GS+LA TEE+PG K YRGMGS
Sbjct 252 IADGGIRYSGDIGKALAVGASSVMIGSILAGTEESPGEKELIGDTVYKYYRGMGS 306
> eco:b2508 guaB, ECK2504, guaR, JW5401; IMP dehydrogenase (EC:1.1.1.205);
K00088 IMP dehydrogenase [EC:1.1.1.205]
Length=488
Score = 130 bits (327), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 83/160 (51%), Positives = 109/160 (68%), Gaps = 1/160 (0%)
Query 18 ERAKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQIIGGNVVTARQAKSLIDAG 77
ER AL+ G DVL++DSS G S + +++ ++ YP+ QIIGGNV TA A++L +AG
Sbjct 232 ERVDALVAAGVDVLLIDSSHGHSEGVLQRIRETRAKYPDLQIIGGNVATAAGARALAEAG 291
Query 78 VDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIPCIADGGIQNSGHVVKA 137
A+++G+G GSICTT++V VG Q TAV + A E IP IADGGI+ SG + KA
Sbjct 292 CSAVKVGIGPGSICTTRIVTGVGVPQITAVADAVE-ALEGTGIPVIADGGIRFSGDIAKA 350
Query 138 LALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
+A GAS VMVGS+LA TEE+PG + G KSYRGMGS
Sbjct 351 IAAGASAVMVGSMLAGTEESPGEIELYQGRSYKSYRGMGS 390
> eco:b0104 guaC, ECK0104, JW0101; GMP reductase (EC:1.7.1.7);
K00364 GMP reductase [EC:1.7.1.7]
Length=347
Score = 99.4 bits (246), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 59/168 (35%), Positives = 88/168 (52%), Gaps = 3/168 (1%)
Query 12 TRAADLERAKALLQVGA--DVLVVDSSQGDSVFQVDIVKQLKSAYPNTQIIGGNVVTARQ 69
T AD E+ K +L + + + +D + G S V V + + A+P I GNVVT
Sbjct 105 TSDADFEKTKQILDLNPALNFVCIDVANGYSEHFVQFVAKAREAWPTKTICAGNVVTGEM 164
Query 70 AKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIPCIADGGIQ 129
+ LI +G D +++G+G GS+CTT+V VG Q +AV + A + + ++DGG
Sbjct 165 CEELILSGADIVKVGIGPGSVCTTRVKTGVGYPQLSAVIECADAAHGLGGM-IVSDGGCT 223
Query 130 NSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
G V KA GA VM+G +LA EE+ G NG + + GM S
Sbjct 224 TPGDVAKAFGGGADFVMLGGMLAGHEESGGRIVEENGEKFMLFYGMSS 271
> cel:F32D1.5 hypothetical protein; K00364 GMP reductase [EC:1.7.1.7]
Length=358
Score = 98.2 bits (243), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 54/147 (36%), Positives = 83/147 (56%), Gaps = 1/147 (0%)
Query 31 LVVDSSQGDSVFQVDIVKQLKSAYPNTQIIGGNVVTARQAKSLIDAGVDALRIGMGSGSI 90
+ +D + G S V+ +++++ AYP I+ GNVVT + LI +G D +++G+G GS+
Sbjct 127 ICLDVANGYSESFVEFIRRVREAYPKHTIMAGNVVTGEMVEELILSGADIVKVGIGPGSV 186
Query 91 CTTQVVCAVGRAQATAVYHVSKYAREVANIPCIADGGIQNSGHVVKALALGASTVMVGSL 150
CTT+ VG Q +AV + A + N ++DGG N G V KA GA VM+G L
Sbjct 187 CTTRKKAGVGYPQLSAVLECADAAHGL-NGHVMSDGGCSNPGDVAKAFGAGADFVMIGGL 245
Query 151 LAATEEAPGAYYFHNGARVKSYRGMGS 177
A +++ G HNG + K + GM S
Sbjct 246 FAGHDQSGGDLIEHNGKKFKLFYGMSS 272
> dre:553639 gmpr, MGC110617, zgc:110617; guanosine monophosphate
reductase (EC:1.7.1.7); K00364 GMP reductase [EC:1.7.1.7]
Length=345
Score = 95.9 bits (237), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 59/174 (33%), Positives = 95/174 (54%), Gaps = 3/174 (1%)
Query 6 VGAALSTRAADLERAKALLQVGADV--LVVDSSQGDSVFQVDIVKQLKSAYPNTQIIGGN 63
V A+ + AADLE+ ++L+ + + +D + G S V+ VK+++ +P I+ GN
Sbjct 99 VAASSGSGAADLEKLCSILEAIPLIQYICLDVANGYSEHFVEFVKRVRERFPKHTIMAGN 158
Query 64 VVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIPCI 123
VVT + LI +G D +++G+G GS+CTT++ VG Q +AV + A + I
Sbjct 159 VVTGEMVEELILSGADIIKVGIGPGSVCTTRIKTGVGYPQLSAVIECADSAHGLKG-HII 217
Query 124 ADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
+DGG G V KA GA VM+G +LA ++ G +G + K + GM S
Sbjct 218 SDGGCSCPGDVAKAFGAGADFVMLGGMLAGHDQCAGEVIEKDGKKFKLFYGMSS 271
> hsa:2766 GMPR, GMPR1; guanosine monophosphate reductase (EC:1.7.1.7);
K00364 GMP reductase [EC:1.7.1.7]
Length=345
Score = 95.5 bits (236), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 56/164 (34%), Positives = 88/164 (53%), Gaps = 3/164 (1%)
Query 16 DLERAKALLQVGADV--LVVDSSQGDSVFQVDIVKQLKSAYPNTQIIGGNVVTARQAKSL 73
DLE+ ++L+ V + +D + G S V+ VK +++ +P I+ GNVVT + L
Sbjct 109 DLEKMTSILEAVPQVKFICLDVANGYSEHFVEFVKLVRAKFPEHTIMAGNVVTGEMVEEL 168
Query 74 IDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIPCIADGGIQNSGH 133
I +G D +++G+G GS+CTT+ VG Q +AV + A + I+DGG G
Sbjct 169 ILSGADIIKVGVGPGSVCTTRTKTGVGYPQLSAVIECADSAHGLKG-HIISDGGCTCPGD 227
Query 134 VVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
V KA GA VM+G + + E G + NG ++K + GM S
Sbjct 228 VAKAFGAGADFVMLGGMFSGHTECAGEVFERNGRKLKLFYGMSS 271
> mmu:66355 Gmpr, 2310004P21Rik, AV028449; guanosine monophosphate
reductase (EC:1.7.1.7); K00364 GMP reductase [EC:1.7.1.7]
Length=345
Score = 95.1 bits (235), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 58/164 (35%), Positives = 86/164 (52%), Gaps = 3/164 (1%)
Query 16 DLERAKALLQVGADV--LVVDSSQGDSVFQVDIVKQLKSAYPNTQIIGGNVVTARQAKSL 73
DLER +L+ V + +D + G S V+ VK ++S +P I+ GNVVT + L
Sbjct 109 DLERMSRILEAVPQVKFICLDVANGYSEHFVEFVKLVRSKFPEHTIMAGNVVTGEMVEEL 168
Query 74 IDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIPCIADGGIQNSGH 133
I +G D +++G+G GS+CTT+ VG Q +AV + A + I+DGG G
Sbjct 169 ILSGADIIKVGVGPGSVCTTRTKTGVGYPQLSAVIECADSAHGLKG-HIISDGGCTCPGD 227
Query 134 VVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
V KA GA VM+G + + E G NG ++K + GM S
Sbjct 228 VAKAFGAGADFVMLGGMFSGHTECAGEVIERNGQKLKLFYGMSS 271
> hsa:51292 GMPR2, MGC15084, MGC830; guanosine monophosphate reductase
2 (EC:1.7.1.7); K00364 GMP reductase [EC:1.7.1.7]
Length=348
Score = 93.2 bits (230), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 58/174 (33%), Positives = 92/174 (52%), Gaps = 3/174 (1%)
Query 6 VGAALSTRAADLERAKALLQVGADV--LVVDSSQGDSVFQVDIVKQLKSAYPNTQIIGGN 63
+ A+ T ++D E+ + +L+ V + +D + G S V+ VK ++ +P I+ GN
Sbjct 99 LAASSGTGSSDFEQLEQILEAIPQVKYICLDVANGYSEHFVEFVKDVRKRFPQHTIMAGN 158
Query 64 VVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIPCI 123
VVT + LI +G D +++G+G GS+CTT+ VG Q +AV + A + I
Sbjct 159 VVTGEMVEELILSGADIIKVGIGPGSVCTTRKKTGVGYPQLSAVMECADAAHGLKG-HII 217
Query 124 ADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
+DGG G V KA GA VM+G +LA E+ G +G + K + GM S
Sbjct 218 SDGGCSCPGDVAKAFGAGADFVMLGGMLAGHSESGGELIERDGKKYKLFYGMSS 271
> mmu:105446 Gmpr2, 1810008P16Rik, 5730544D12Rik, AA959850; guanosine
monophosphate reductase 2 (EC:1.7.1.7); K00364 GMP reductase
[EC:1.7.1.7]
Length=348
Score = 93.2 bits (230), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 57/166 (34%), Positives = 88/166 (53%), Gaps = 3/166 (1%)
Query 14 AADLERAKALLQVGADV--LVVDSSQGDSVFQVDIVKQLKSAYPNTQIIGGNVVTARQAK 71
+AD E+ + +L+ V + +D + G S V+ VK ++ +P I+ GNVVT +
Sbjct 107 SADFEQLEQILEAIPQVKYICLDVANGYSEHFVEFVKDVRKRFPQHTIMAGNVVTGEMVE 166
Query 72 SLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIPCIADGGIQNS 131
LI +G D +++G+G GS+CTT+ VG Q +AV + A + I+DGG
Sbjct 167 ELILSGADIIKVGIGPGSVCTTRKKTGVGYPQLSAVMECADAAHGLKG-HIISDGGCSCP 225
Query 132 GHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
G V KA GA VM+G +LA E+ G +G + K + GM S
Sbjct 226 GDVAKAFGAGADFVMLGGMLAGHSESGGELIERDGKKYKLFYGMSS 271
> dre:678545 MGC136869; zgc:136869 (EC:1.7.1.7); K00364 GMP reductase
[EC:1.7.1.7]
Length=348
Score = 92.4 bits (228), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 60/174 (34%), Positives = 88/174 (50%), Gaps = 3/174 (1%)
Query 6 VGAALSTRAADLERAKALLQVGADV--LVVDSSQGDSVFQVDIVKQLKSAYPNTQIIGGN 63
V + T D E+ A++ + + VD + G S V+ VK ++ +P I+ GN
Sbjct 99 VAVSTGTSEGDFEKLGAIVAAVPQIRYICVDVANGYSEHFVNFVKDVRQKFPTHTIMAGN 158
Query 64 VVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIPCI 123
VVT + LI AG D +++G+G GS+CTT+ VG Q +AV + A + I
Sbjct 159 VVTGEMVEELILAGADIIKVGIGPGSVCTTRKKTGVGYPQLSAVIECADAAHGLGG-HII 217
Query 124 ADGGIQNSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
+DGG G V KA GA VM+G +LA E+ G NG + K + GM S
Sbjct 218 SDGGCTCPGDVSKAFGAGADFVMLGGMLAGHCESGGEVIEKNGKKYKLFYGMSS 271
> tpv:TP01_0363 guanosine monophosphate reductase; K00364 GMP
reductase [EC:1.7.1.7]
Length=326
Score = 86.3 bits (212), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 51/153 (33%), Positives = 79/153 (51%), Gaps = 6/153 (3%)
Query 28 ADVLVVDSSQGDSVFQVDIVKQLKSAYPN-TQIIGGNVVTARQAKSLIDAGVDALRIGMG 86
+ + +D S G + +++ +++ + N T +I GNV T + K + D G DA+++G+G
Sbjct 111 PEFITIDISHGHNPRVKGMIEHIRANFGNETFVIAGNVTTPQGIKDMEDWGADAIKVGLG 170
Query 87 SGSICTTQVVCAVGR--AQATAVYHVSKYAREVANIPCIADGGIQNSGHVVKALALGAST 144
G CTT G Q +AV +KYA I DGG+ SG + KA+ +GA
Sbjct 171 PGHACTTSPRTGFGSRGWQLSAVAECAKYATRAV---VICDGGVSKSGDIAKAIHMGADW 227
Query 145 VMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
+M G L + T EAPG +G + K Y G S
Sbjct 228 IMSGYLFSGTIEAPGEVVVKDGVKYKPYYGSSS 260
> xla:444486 gmpr2, MGC81876; guanosine monophosphate reductase
2 (EC:1.7.1.7); K00364 GMP reductase [EC:1.7.1.7]
Length=346
Score = 85.5 bits (210), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 57/168 (33%), Positives = 86/168 (51%), Gaps = 3/168 (1%)
Query 12 TRAADLERAKALLQVGADV--LVVDSSQGDSVFQVDIVKQLKSAYPNTQIIGGNVVTARQ 69
T +D + + +L V + +D + G S V VK +++ +P I+ GNVVT
Sbjct 105 TGQSDFQHLEKVLSAVPQVRYICLDVANGYSEQFVQHVKDVRARFPKHTIMAGNVVTGEM 164
Query 70 AKSLIDAGVDALRIGMGSGSICTTQVVCAVGRAQATAVYHVSKYAREVANIPCIADGGIQ 129
+ LI +G D +++G+G GS+CTT+ VG Q +AV + A + N I+DGG
Sbjct 165 VEELILSGADIIKVGIGPGSVCTTRKKTGVGYPQLSAVIECADAAHGL-NGHIISDGGCS 223
Query 130 NSGHVVKALALGASTVMVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
G V KA GA VM+G +L+ E G NG + K + GM S
Sbjct 224 CPGDVSKAFGAGADFVMLGGMLSGHTECGGEVIEKNGKKYKLFYGMSS 271
> bbo:BBOV_IV005560 23.m05800; GMP reductase (EC:1.7.1.7); K00364
GMP reductase [EC:1.7.1.7]
Length=327
Score = 80.1 bits (196), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 52/152 (34%), Positives = 79/152 (51%), Gaps = 6/152 (3%)
Query 29 DVLVVDSSQGDSVFQVDIVKQLKSAY-PNTQIIGGNVVTARQAKSLIDAGVDALRIGMGS 87
D + +D + G S+ ++ +K A+ P T II GNV TA L + G DA+++G+G
Sbjct 111 DYVTIDVAHGHSLAMQRMISYIKKAFGPKTFIIAGNVATAEGVVDLENWGADAIKVGLGP 170
Query 88 GSICTTQVVCAVG--RAQATAVYHVSKYAREVANIPCIADGGIQNSGHVVKALALGASTV 145
G +C+T + G Q AV ++ A++ IADGG + SG +VKAL +GA V
Sbjct 171 GYVCSTSIRTGFGTRNWQLAAVRECARVAKKAV---IIADGGCRLSGDIVKALYMGADWV 227
Query 146 MVGSLLAATEEAPGAYYFHNGARVKSYRGMGS 177
M G A ++P +G +K Y G S
Sbjct 228 MSGYFYAGFIKSPSETEIVDGVPMKVYYGNAS 259
> cel:F41E6.5 hypothetical protein; K11517 (S)-2-hydroxy-acid
oxidase [EC:1.1.3.15]
Length=371
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 35/115 (30%), Positives = 56/115 (48%), Gaps = 11/115 (9%)
Query 35 SSQGDSVFQVDIVKQLKSAYPNTQIIGGNVVTARQAKSLIDAGVDALRIGMGSGSICTTQ 94
SSQ D + +K +++ N +I V+ A ++AGVD + + G Q
Sbjct 213 SSQIDPSLDWNTLKWIRTK-TNLPVIVKGVMRGDDALLALEAGVDGIIVSNHGGR----Q 267
Query 95 VVCAVGRAQATAVYHVSKYAREVAN-IPCIADGGIQNSGHVVKALALGASTVMVG 148
+ C V ++ + + R V N IP DGG++N ++KA+ALGA V VG
Sbjct 268 MDCTVATIES-----LPEVLRAVDNRIPVWMDGGVRNGRDILKAVALGARGVFVG 317
> ath:AT3G14150 (S)-2-hydroxy-acid oxidase, peroxisomal, putative
/ glycolate oxidase, putative / short chain alpha-hydroxy
acid oxidase, putative (EC:1.1.3.15); K11517 (S)-2-hydroxy-acid
oxidase [EC:1.1.3.15]
Length=363
Score = 37.4 bits (85), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 31/110 (28%), Positives = 50/110 (45%), Gaps = 9/110 (8%)
Query 39 DSVFQVDIVKQLKSAYPNTQIIGGNVVTARQAKSLIDAGVDALRIGMGSGSICTTQVVCA 98
D+ F ++ L+S ++ G ++T A ++AGVD + + G
Sbjct 206 DASFSWKDIEWLRSITELPILVKG-ILTREDALKAVEAGVDGIIVSNHGGR------QLD 258
Query 99 VGRAQATAVYHVSKYAREVANIPCIADGGIQNSGHVVKALALGASTVMVG 148
A T + V + R IP + DGG++ V KALALGA V++G
Sbjct 259 YSPATITVLEEVVQVVR--GRIPVLLDGGVRRGTDVFKALALGAQAVLIG 306
> ath:AT4G18360 (S)-2-hydroxy-acid oxidase, peroxisomal, putative
/ glycolate oxidase, putative / short chain alpha-hydroxy
acid oxidase, putative; K11517 (S)-2-hydroxy-acid oxidase
[EC:1.1.3.15]
Length=314
Score = 36.6 bits (83), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 18/35 (51%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 120 IPCIADGGIQNSGHVVKALALGASTVMVGSLLAAT 154
IP DGG++ V KALALGAS V V S + T
Sbjct 280 IPVFLDGGVRRGTDVFKALALGASGVFVSSFIIYT 314
> ath:AT3G14420 (S)-2-hydroxy-acid oxidase, peroxisomal, putative
/ glycolate oxidase, putative / short chain alpha-hydroxy
acid oxidase, putative; K11517 (S)-2-hydroxy-acid oxidase
[EC:1.1.3.15]
Length=348
Score = 35.0 bits (79), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 15/29 (51%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 120 IPCIADGGIQNSGHVVKALALGASTVMVG 148
IP DGG++ V KALALGAS + +G
Sbjct 261 IPVFLDGGVRRGTDVFKALALGASGIFIG 289
> ath:AT3G14130 (S)-2-hydroxy-acid oxidase, peroxisomal, putative
/ glycolate oxidase, putative / short chain alpha-hydroxy
acid oxidase, putative (EC:1.1.3.15); K11517 (S)-2-hydroxy-acid
oxidase [EC:1.1.3.15]
Length=363
Score = 34.7 bits (78), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 15/29 (51%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 120 IPCIADGGIQNSGHVVKALALGASTVMVG 148
IP + DGG++ V KALALGA V++G
Sbjct 278 IPVLLDGGVRRGTDVFKALALGAQAVLIG 306
> ath:AT3G14415 (S)-2-hydroxy-acid oxidase, peroxisomal, putative
/ glycolate oxidase, putative / short chain alpha-hydroxy
acid oxidase, putative (EC:1.1.3.15); K11517 (S)-2-hydroxy-acid
oxidase [EC:1.1.3.15]
Length=367
Score = 34.3 bits (77), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 120 IPCIADGGIQNSGHVVKALALGASTVMVG 148
+P DGG++ V KALALGAS + +G
Sbjct 280 VPVFLDGGVRRGTDVFKALALGASGIFIG 308
> eco:b3605 lldD, ECK3595, JW3580, lct, lctD; L-lactate dehydrogenase,
FMN-linked (EC:1.1.2.3); K00101 L-lactate dehydrogenase
(cytochrome) [EC:1.1.2.3]
Length=396
Score = 32.7 bits (73), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 119 NIPCIADGGIQNSGHVVKALALGASTVMVG 148
+I +AD GI+N VV+ +ALGA TV++G
Sbjct 300 DIAILADSGIRNGLDVVRMIALGADTVLLG 329
> ath:AT5G64250 2-nitropropane dioxygenase family / NPD family
Length=333
Score = 32.3 bits (72), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 24/39 (61%), Gaps = 0/39 (0%)
Query 119 NIPCIADGGIQNSGHVVKALALGASTVMVGSLLAATEEA 157
+IP IA GGI + V AL+LGA V +G+ AT E+
Sbjct 164 DIPVIAAGGIVDGRGYVAALSLGAQGVCLGTRFVATHES 202
> eco:b2048 cpsG, ECK2042, JW2033, manB, rfbK; phosphomannomutase
(EC:5.4.2.8); K01840 phosphomannomutase [EC:5.4.2.8]
Length=456
Score = 30.8 bits (68), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 50/111 (45%), Gaps = 19/111 (17%)
Query 45 DIVKQLKSAY-----PNTQIIGGNVVTARQ------AKSLIDAGVDALRIGM-GSGSICT 92
DI ++ AY P T ++GG+V + AK L DAGVD L IGM G+ I
Sbjct 23 DIAWRIGRAYGEFLKPKTIVLGGDVRLTSETLKLALAKGLQDAGVDVLDIGMSGTEEIYF 82
Query 93 TQVVCAV-GRAQATAVYHVSKY-----AREVANIPCIADGGIQNSGHVVKA 137
V G + TA ++ Y RE A P D G+++ + +A
Sbjct 83 ATFHLGVDGGIEVTASHNPMDYNGMKLVREGAR-PISGDTGLRDVQRLAEA 132
> bbo:BBOV_I000420 16.m00758; hypothetical protein
Length=1736
Score = 30.8 bits (68), Expect = 2.7, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 26/43 (60%), Gaps = 3/43 (6%)
Query 20 AKALLQVGADVLVVDSSQGDSVFQVDIVKQLKSAYPNTQIIGG 62
+KALL+ +L + +SQG+++ DI K LK Y N +GG
Sbjct 995 SKALLEEKIRILKIYTSQGNTM---DIYKMLKKHYMNNPFMGG 1034
> pfa:PFF0580w lsm12, putative
Length=176
Score = 30.8 bits (68), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 41 VFQVDIVKQLKSAYPNTQIIGGNVVTARQAKSLIDAGVDALRIGMG 86
+ V+I ++K+ N +I N+V + K+L+D LRIG+G
Sbjct 61 ILDVEIKPKVKNTNDNIPMINKNIVQKIEKKALMDFEKAKLRIGIG 106
> hsa:54363 HAO1, GOX, GOX1, HAOX1, MGC142225, MGC142227; hydroxyacid
oxidase (glycolate oxidase) 1 (EC:1.1.3.15); K11517
(S)-2-hydroxy-acid oxidase [EC:1.1.3.15]
Length=370
Score = 30.4 bits (67), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 119 NIPCIADGGIQNSGHVVKALALGASTVMVG 148
+ DGG++ V+KALALGA V VG
Sbjct 285 KVEVFLDGGVRKGTDVLKALALGAKAVFVG 314
> mmu:15112 Hao1, GOX, Gox1, Hao-1, MGC141211; hydroxyacid oxidase
1, liver (EC:1.1.3.15); K11517 (S)-2-hydroxy-acid oxidase
[EC:1.1.3.15]
Length=370
Score = 30.4 bits (67), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 14/24 (58%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 125 DGGIQNSGHVVKALALGASTVMVG 148
DGG++ V+KALALGA V VG
Sbjct 291 DGGVRKGTDVLKALALGAKAVFVG 314
> dre:402827 hao1; hydroxyacid oxidase (glycolate oxidase) 1 (EC:1.1.3.15);
K11517 (S)-2-hydroxy-acid oxidase [EC:1.1.3.15]
Length=369
Score = 30.0 bits (66), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 119 NIPCIADGGIQNSGHVVKALALGASTVMVG 148
+ DGG++ V+KALALGA V +G
Sbjct 285 QVEVFMDGGVRMGSDVLKALALGAKAVFIG 314
> dre:393455 hao2, MGC63690, zgc:63690; hydroxyacid oxidase 2
(long chain) (EC:1.1.3.15); K11517 (S)-2-hydroxy-acid oxidase
[EC:1.1.3.15]
Length=357
Score = 30.0 bits (66), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 13/24 (54%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 125 DGGIQNSGHVVKALALGASTVMVG 148
DGGI+ V+KA+ALGA V +G
Sbjct 283 DGGIRTGNDVLKAIALGARCVFIG 306
> eco:b3991 thiG, ECK3982, JW5549, thiB; thiamin biosynthesis
ThiGH complex subunit; K03149 thiamine biosynthesis ThiG
Length=256
Score = 30.0 bits (66), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 25/114 (21%), Positives = 50/114 (43%), Gaps = 11/114 (9%)
Query 54 YPNTQ-IIGGNVVTARQAKSLIDAGVDALRIGMGSGSICT--TQVVCA--------VGRA 102
+P+ + ++ + T + A++L+ G L +C +V CA +G
Sbjct 99 HPDARWLLPDPIETLKAAETLVQQGFVVLPYCGADPVLCKRLEEVGCAAVMPLGAPIGSN 158
Query 103 QATAVYHVSKYAREVANIPCIADGGIQNSGHVVKALALGASTVMVGSLLAATEE 156
Q + + + A +P + D GI H +AL +GA V+V + +A ++
Sbjct 159 QGLETRAMLEIIIQQATVPVVVDAGIGVPSHAAQALEMGADAVLVNTAIAVADD 212
Lambda K H
0.317 0.130 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4665550176
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40