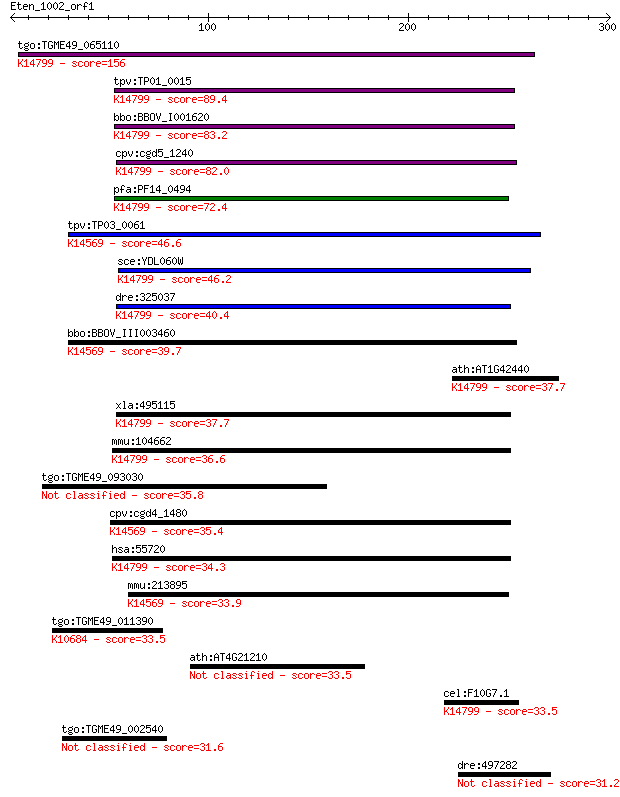
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1002_orf1
Length=300
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_065110 hypothetical protein ; K14799 pre-rRNA-proce... 156 9e-38
tpv:TP01_0015 hypothetical protein; K14799 pre-rRNA-processing... 89.4 2e-17
bbo:BBOV_I001620 19.m02052; protein of unknown function (DUF66... 83.2 1e-15
cpv:cgd5_1240 Tsr1p GTpase, multitransmembrane region protein ... 82.0 2e-15
pfa:PF14_0494 ribosome biogenesis protein tsr1, putative; K147... 72.4 2e-12
tpv:TP03_0061 hypothetical protein; K14569 ribosome biogenesis... 46.6 1e-04
sce:YDL060W TSR1; Protein required for processing of 20S pre-r... 46.2 1e-04
dre:325037 tsr1, MGC123319, fc52b09, wu:fc52b09, zgc:123319; T... 40.4 0.007
bbo:BBOV_III003460 17.m07327; hypothetical protein; K14569 rib... 39.7 0.015
ath:AT1G42440 hypothetical protein; K14799 pre-rRNA-processing... 37.7 0.049
xla:495115 tsr1; TSR1, 20S rRNA accumulation, homolog; K14799 ... 37.7 0.058
mmu:104662 Tsr1, AU040765, AW550801, mKIAA1401; TSR1, 20S rRNA... 36.6 0.10
tgo:TGME49_093030 hypothetical protein 35.8 0.18
cpv:cgd4_1480 BMS1 like GTpase involved in ribosome biogenesis... 35.4 0.24
hsa:55720 TSR1, FLJ10534, KIAA1401, MGC131829; TSR1, 20S rRNA ... 34.3 0.55
mmu:213895 Bms1, AA408648, AU020092, AU043373, BB007109, BC030... 33.9 0.67
tgo:TGME49_011390 hypothetical protein ; K10684 ubiquitin-like... 33.5 1.0
ath:AT4G21210 ATRP1; ATRP1 (PPDK REGULATORY PROTEIN); phosphop... 33.5 1.1
cel:F10G7.1 tag-151; Temporarily Assigned Gene name family mem... 33.5 1.1
tgo:TGME49_002540 3',5'-cyclic nucleotide phosphodiesterase, p... 31.6 3.8
dre:497282 smchd1, kiaa0650l, kiaao650l; structural maintenanc... 31.2 5.5
> tgo:TGME49_065110 hypothetical protein ; K14799 pre-rRNA-processing
protein TSR1
Length=1007
Score = 156 bits (394), Expect = 9e-38, Method: Compositional matrix adjust.
Identities = 113/346 (32%), Positives = 154/346 (44%), Gaps = 116/346 (33%)
Query 5 YTFSLPAYARNPLIPKRYQQHVLLIPAPRISETAGKETMLEEGEPPERPSELLRYMDLCN 64
+ F LP YAR +P R QQ +LL+PAP P L +DLC
Sbjct 175 FLFPLPNYARCASLPLRSQQRILLLPAPP-------------------PRNTLPLLDLCK 215
Query 65 CCDVLVCLFGGSCTYERSAFSRRGYKILQALKLQGLPPSVIGVGCVDPSVLE-------- 116
C D+L+C+F G CTY + AF GY+ L ALK+QG+PP V+GVG P L+
Sbjct 216 CADLLLCVFSGECTYTKPAFDEDGYRFLTALKMQGVPP-VVGVGS--PVALQAHLGTCLS 272
Query 117 ------PADAPK-----------------------------------------GKSSASE 129
P D P G+ A+E
Sbjct 273 GNGAALPCDFPSSSLTGVSKMEMAAEAMNDVDVSSMSMDNESIQTPAALGGKAGRKQAAE 332
Query 130 SLKFMRRFFESELGAERKFFSIGSDADWHNVLRAVGAAASLTQGDTNTKTVAKGAEAAAS 189
+L+F +R+F SE G+++KF ++ ADW V+R +G A+ + ++ AEAA +
Sbjct 333 TLRFQQRYFSSEFGSDKKFCAVSLPADWQKVVRMIGGASV----PLDVARSSQAAEAAGT 388
Query 190 -------RRRGHLLALSWHIAW-----------------------HHVS---EDPAAQPE 216
R RG+LLAL W I +VS ED A+ E
Sbjct 389 HGDLQWRRGRGYLLALDWEILTGAQRQLAGAASPPGSELPKSERRENVSGAGEDDEAE-E 447
Query 217 PCLEACGLVRGAGLTCNLPVHITGVGDFVLSRIETMEPAYEAKRRE 262
L A G VRG G+TC LP+HITGVGDF+ RI+ + + KRRE
Sbjct 448 TYLHAYGFVRGVGMTCRLPIHITGVGDFIAERIDILHDD-DWKRRE 492
> tpv:TP01_0015 hypothetical protein; K14799 pre-rRNA-processing
protein TSR1
Length=740
Score = 89.4 bits (220), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 62/200 (31%), Positives = 100/200 (50%), Gaps = 36/200 (18%)
Query 53 PSELLRYMDLCNCCDVLVCLFGGSCTYERSAFSRRGYKILQALKLQGLPPSVIGVGCVDP 112
P L + + D+++CLF G+ + + AF GY+IL +L+LQG+P S +GV
Sbjct 131 PRNLTDILYSASASDIILCLFRGA-SQDSPAFDELGYRILSSLRLQGIP-SPVGVNF--- 185
Query 113 SVLEPADAPKGKSSASESLKFMRRFFESELGAERKFFSIGSDADWHNVLRAVGAAASLTQ 172
A G S +L +RR+F SE G ++KF S+ S+ D VL +G ++
Sbjct 186 -----ESALPGDRQPSSTL--VRRYFHSEFGLDKKFTSVFSEKDLRTVLSLIGCVST--- 235
Query 173 GDTNTKTVAKGAEAAASRRRGHLLALSWHIAWHHVSEDPAAQPEPCLEACGLVRGAGLTC 232
++ + R RG++L+L+ H D + E LE G V+G G T
Sbjct 236 -----------SDLSWRRDRGYMLSLN-----HTYDSD---KKELVLE--GYVKGLGFTV 274
Query 233 NLPVHITGVGDFVLSRIETM 252
PVH+T GDF++ +I+ +
Sbjct 275 RHPVHLTSFGDFIIKKIDLL 294
> bbo:BBOV_I001620 19.m02052; protein of unknown function (DUF663)
and AARP2CN (NUC121) domain containing protein; K14799
pre-rRNA-processing protein TSR1
Length=732
Score = 83.2 bits (204), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 61/200 (30%), Positives = 95/200 (47%), Gaps = 36/200 (18%)
Query 53 PSELLRYMDLCNCCDVLVCLFGGSCTYERSAFSRRGYKILQALKLQGLPPSVIGVGCVDP 112
P L + + D+L+CLF GS E AF GYK L ++ QG+P + +D
Sbjct 130 PRSLSDILYAASVADILICLFRGSSQTE-PAFDDFGYKALSVIRQQGVPTPI----GIDL 184
Query 113 SVLEPADAPKGKSSASESLKFMRRFFESELGAERKFFSIGSDADWHNVLRAVGAAASLTQ 172
+ P D S ++R+F ELG +++F +I + + L A+G
Sbjct 185 ELGLPGDR-------FASYNLVKRYFHDELGGDKRFVAILAPDNIKAALSAIGC------ 231
Query 173 GDTNTKTVAKGAEAAASRRRGHLLALSWHIAWHHVSEDPAAQPEPCLEACGLVRGAGLTC 232
V+ G+ + S R G++L+L + S DP+ Q L G RG G +
Sbjct 232 -------VSIGSLSWRSGR-GYMLSLGY-------SYDPSLQQ---LRVTGYARGVGFSV 273
Query 233 NLPVHITGVGDFVLSRIETM 252
+ PVH+TGVGDF+L RI+ +
Sbjct 274 HHPVHVTGVGDFILERIDML 293
> cpv:cgd5_1240 Tsr1p GTpase, multitransmembrane region protein
; K14799 pre-rRNA-processing protein TSR1
Length=750
Score = 82.0 bits (201), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 53/200 (26%), Positives = 93/200 (46%), Gaps = 28/200 (14%)
Query 54 SELLRYMDLCNCCDVLVCLFGGSCTYERSAFSRRGYKILQALKLQGLPPSVIGVGCVDPS 113
+++ +DL C D ++ LFG + ++ AF GYK+L+++KLQG ++
Sbjct 138 NKIFDTLDLIKCSDFVMALFGFNDEFQ--AFDEIGYKLLKSIKLQGQTDTI--------G 187
Query 114 VLEPADAPKGKSSASESLKFMRRFFESELGAERKFFSIGSDADWHNVLRAVGAAASLTQG 173
+ ++ + + K ++ FE E KFFS+ + D N+L ++
Sbjct 188 LFFHNESHNFGNKILDCEKVFKKSFEMEFEKNMKFFSLQKENDIRNLLSSIPNMGY---- 243
Query 174 DTNTKTVAKGAEAAASRRRGHLLALSWHIAWHHVSEDPAAQPEPCLEACGLVRGAGLTCN 233
++ RG++L+ S I ++ S + Q L G VRG GL+ N
Sbjct 244 ----------SKICLREGRGYMLSESSSIVVNNYSNEEKCQ----LVIRGFVRGVGLSMN 289
Query 234 LPVHITGVGDFVLSRIETME 253
PVHIT +GDFV+ I ++
Sbjct 290 FPVHITNIGDFVIDEIRPID 309
> pfa:PF14_0494 ribosome biogenesis protein tsr1, putative; K14799
pre-rRNA-processing protein TSR1
Length=1029
Score = 72.4 bits (176), Expect = 2e-12, Method: Composition-based stats.
Identities = 57/197 (28%), Positives = 88/197 (44%), Gaps = 36/197 (18%)
Query 53 PSELLRYMDLCNCCDVLVCLFGGSCTYERSAFSRRGYKILQALKLQGLPPSVIGVGCVDP 112
P ++ +D C DV++CLF + E S F GYK+L LK+QG+P S+IG+G
Sbjct 142 PRDIYGIIDGTKCADVVLCLFKDG-SIENSCFDELGYKLLSVLKIQGVP-SIIGIGY--- 196
Query 113 SVLEPADAPKGKSSASESLKFMRRFFESELGAERKFFSIGSDADWHNVLRAVGAAASLTQ 172
S S KF+ R+F SE + K F I + + S Q
Sbjct 197 ---------NTNMSNKGSHKFVMRYFNSEFTMDDKIFFISENKN------------SDFQ 235
Query 173 GDTNTKTVAKGAEAAASRRRGHLLALSWHIAWHHVSEDPAAQPEPCLEACGLVRGAGLTC 232
N T K + RG+++A S A++ ++ C+ G ++G G
Sbjct 236 KLYNEITNMKIKNVSYREGRGYMMADS--CAYNSEND--------CIYIKGFIKGVGFNY 285
Query 233 NLPVHITGVGDFVLSRI 249
+ P+HIT VGD+ + I
Sbjct 286 HNPIHITDVGDYYIDNI 302
> tpv:TP03_0061 hypothetical protein; K14569 ribosome biogenesis
protein BMS1
Length=601
Score = 46.6 bits (109), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 51/238 (21%), Positives = 94/238 (39%), Gaps = 37/238 (15%)
Query 30 PAPRISETAGKETMLEEGEPPERPSELLRYMDLCNCCDVLVCLFGGSCTYERSAFSRRGY 89
P +S + + T++E G + +L +D C D+ + + GS YE F
Sbjct 111 PITLVSSKSRRITLIECG------NSMLDMIDCCKIADIAIVMVDGSYGYEMETF----- 159
Query 90 KILQALKLQGLPPSVIGVGCVDPSVLEPADAPKGKSSASESLKFMRRFFESEL-GAERKF 148
+ + +++ GL P VIG V+ D+ K + + K +++ F SEL + F
Sbjct 160 EFVNMMQVHGL-PRVIG-------VVTHLDSFKDNKTMRRTKKILKKRFWSELYDGAKMF 211
Query 149 FSIGSDADWHNVLRAVGAAASLTQGDTNTKTVAKGAEAAASRRRGHLLALSWHIAWHHVS 208
+ G D + + A + ++ + R H +S H +
Sbjct 212 YFTGLQYDRYKKNEVLNLA----------RYISSQKPPTINWRLSHPYTVSLR---HEII 258
Query 209 EDPAAQPEPC-LEACGLVRGAGLTCNLPVHITGVGDFVLSRIETMEPAYEAKRREPLL 265
E + + C + G V G L +HI G GDF + + M+ E + ++ L
Sbjct 259 E---TKDDSCKVFFYGYVYGGKLNTTNGIHIPGAGDFTIDSVTNMQDPCEIQSKDRTL 313
> sce:YDL060W TSR1; Protein required for processing of 20S pre-rRNA
in the cytoplasm, associates with pre-40S ribosomal particles;
K14799 pre-rRNA-processing protein TSR1
Length=788
Score = 46.2 bits (108), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 54/210 (25%), Positives = 93/210 (44%), Gaps = 37/210 (17%)
Query 55 ELLRYMDLCNCCDVL-VCLFGGSCTYERSAFSRRGYKILQALKLQGLPPSVIGVGCVDPS 113
++ ++++ +C V +FG S E G +I++AL+LQG+ S IGV +
Sbjct 141 DMTNFLNILDCAKVADFVVFGLSGVQEVD--EEFGEQIIRALELQGIA-SYIGVISNLSA 197
Query 114 VLEPADAPKGKSSASESLK-FMRRFFESELGAERKFFSIGSDADWHNVLRAV--GAAASL 170
V E K + +SL+ + + FF SE + +++ ++D NVLR + S+
Sbjct 198 VHE---KEKFQLDVKQSLESYFKHFFPSE----ERVYNLEKNSDALNVLRTLCQRLPRSI 250
Query 171 TQGDTNTKTVAKGAEAAASRRRGHLLALSWHIAWHHVSEDPAAQPEPCLEACGLVRGAGL 230
D RG+++A + + S D L G VRG G
Sbjct 251 NWRDN----------------RGYVVAD--FVDFVETSPDSGD-----LVIEGTVRGIGF 287
Query 231 TCNLPVHITGVGDFVLSRIETMEPAYEAKR 260
N VHI GDF L++IE + + + ++
Sbjct 288 NANRLVHIPDFGDFQLNKIEKISESSQKRK 317
> dre:325037 tsr1, MGC123319, fc52b09, wu:fc52b09, zgc:123319;
TSR1, 20S rRNA accumulation, homolog (yeast); K14799 pre-rRNA-processing
protein TSR1
Length=822
Score = 40.4 bits (93), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 52/198 (26%), Positives = 75/198 (37%), Gaps = 34/198 (17%)
Query 54 SELLRYMDLCNCCDVLVCLFGGSCTYERSAFSRRGYKILQALKLQGLPP-SVIGVGCVDP 112
+L +D+ D LV + + + G L L QGLP +++ G D
Sbjct 152 DDLHSLLDVAKIADSLVFVLDSN-----EGWDSYGEYCLSCLFAQGLPSHALVCQGVADL 206
Query 113 SVLEPADAPKGKSSASESLKFMRRFFESELGAERKFFSIGSDADWHNVLRAVGAAASLTQ 172
+V +ES + + R ES E + F + D D +LR + A
Sbjct 207 AV----------KKRTESRRALSRLVESRF-PEARLFPVDCDQDAVLLLRHLSAQ----- 250
Query 173 GDTNTKTVAKGAEAAASRRRGHLLALSWHIAWHHVSEDPAAQPEPCLEACGLVRGAGLTC 232
K RR HLLA A + + + L G +RG L
Sbjct 251 ---------KQRRLGFRSRRSHLLA---QRATYVPNNSQSGTGLGTLCVSGYIRGCPLQV 298
Query 233 NLPVHITGVGDFVLSRIE 250
N VHITG GDF LS+I+
Sbjct 299 NRLVHITGHGDFQLSQID 316
> bbo:BBOV_III003460 17.m07327; hypothetical protein; K14569 ribosome
biogenesis protein BMS1
Length=924
Score = 39.7 bits (91), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 52/232 (22%), Positives = 92/232 (39%), Gaps = 45/232 (19%)
Query 30 PAPRISETAGKETMLEEGEPPERPSELLRYMDLCNCCDVLVCLFGGSCTYERSAFSRRGY 89
P +S + + T++E G S ++ +D C D+++ + GS YE
Sbjct 120 PITLVSSSTRRLTLIECG------SSVVDMLDCCKIADLVLVMIDGSVGYEMETL----- 168
Query 90 KILQALKLQGLPPSVIGVGCVDPSVLEPADAPKGKSSASESLKFMRRFFESEL--GAERK 147
+ L ++ G P C +V+ + S+ + K ++ F SE+ GA+
Sbjct 169 EFLNIMQTHGFPRV-----C---AVVNHLEGFPDNSTLRKVKKRLKNRFWSEIYDGAKMV 220
Query 148 FFS-----IGSDADWHNVLRAVGAAASLTQGDTNTKTVAKGAEAAASRRRGHLLALSW-H 201
+ S D N+ RA+ +Q TN S R+ H +S H
Sbjct 221 YLSGLKYGRYKSNDILNLARAIA-----SQKPTNI-----------SWRQTHFYCVSLRH 264
Query 202 IAWHHVSEDPAAQPEPCLEACGLVRGAGLTCNLPVHITGVGDFVLSRIETME 253
V++D + + G V G+ + N +HI G+GDF+ I M+
Sbjct 265 ELLSSVTDDSGSSVKAAF--YGYVYGSRIVNNQAIHIPGIGDFIPDSITNMQ 314
> ath:AT1G42440 hypothetical protein; K14799 pre-rRNA-processing
protein TSR1
Length=793
Score = 37.7 bits (86), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 30/56 (53%), Gaps = 3/56 (5%)
Query 222 CGLVRGAGLTCNLPVHITGVGDFVLSRIETME---PAYEAKRREPLLVNGEAXAEV 274
G +R L+ N VH++GVGDF S+IE ++ P E K + + ++ EV
Sbjct 275 SGYLRARKLSVNQLVHVSGVGDFQFSKIEVLKDPFPLNERKNQNSMELDDSHDEEV 330
> xla:495115 tsr1; TSR1, 20S rRNA accumulation, homolog; K14799
pre-rRNA-processing protein TSR1
Length=815
Score = 37.7 bits (86), Expect = 0.058, Method: Compositional matrix adjust.
Identities = 50/204 (24%), Positives = 76/204 (37%), Gaps = 48/204 (23%)
Query 54 SELLRYMDLCNCCDVLVCLFGGSCTYERSAFSRRGYKILQALKLQGLPPSVIGV-GCVDP 112
+L +DL D L+ L + + G L L QGLP V+ V G
Sbjct 146 DDLCSLLDLAKVADTLLFLLDP-----QEGWDSYGDYCLSCLFAQGLPSYVLAVQGMNYI 200
Query 113 SVLEPADAPKGKSSASESLKFMRRFFESELGAERKFFSIGSDADWHNVLRAVGAAASLTQ 172
+ + AD K S E+ RF ++ K F + ++ + ++R +
Sbjct 201 PIKKRADIKKQLSKVIEN-----RFTDA------KLFQLDTEQEAAVLIRQI-------- 241
Query 173 GDTNTKTVAKGAEAAASRRRGHLLALSWHIAWHHVSEDPAAQPEP------CLEACGLVR 226
+ K A RR ++LA + QP L+ G VR
Sbjct 242 ------STQKQRHLAFRSRRSYMLA-----------QRADFQPTDESGLVGTLKLSGYVR 284
Query 227 GAGLTCNLPVHITGVGDFVLSRIE 250
G L N VHI G GDF +S+I+
Sbjct 285 GQELNVNRLVHIVGHGDFHMSQID 308
> mmu:104662 Tsr1, AU040765, AW550801, mKIAA1401; TSR1, 20S rRNA
accumulation, homolog (yeast); K14799 pre-rRNA-processing
protein TSR1
Length=803
Score = 36.6 bits (83), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 49/201 (24%), Positives = 77/201 (38%), Gaps = 38/201 (18%)
Query 52 RPSELLRYMDLCNCCDVLVCLFGGSCTYERSAFSRRGYKILQALKLQGLPPSVIGVGCVD 111
RP +L +D+ D ++ L ++ + G L L QGLP + V +
Sbjct 139 RPGDLHTLLDMAKVADTILFLLDPLEGWDST-----GDYCLSCLFAQGLPTYTLAVQGLS 193
Query 112 PSVLEPADAPKGKSSASESLKFM--RRFFESELGAERKFFSIGSDADWHNVLRAVGAAAS 169
PK + A + L M +RF E K + + + +LR +
Sbjct 194 ------GFPPKKQIDARKKLSKMVEKRF------PEDKLLLLDTQQESGMLLRQLANQ-- 239
Query 170 LTQGDTNTKTVAKGAEAAASRRRGHLLALSWHIAWHHVSEDPAAQPEPCLEACGLVRGAG 229
K A RR +L A H+A SE+ + L+ G VRG
Sbjct 240 ------------KQRHLAFRDRRAYLFA---HVADFVPSEE--SDLVGTLKISGYVRGRT 282
Query 230 LTCNLPVHITGVGDFVLSRIE 250
L N +HI G GDF +++I+
Sbjct 283 LNVNSLLHIVGHGDFQMNQID 303
> tgo:TGME49_093030 hypothetical protein
Length=2318
Score = 35.8 bits (81), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 43/157 (27%), Positives = 61/157 (38%), Gaps = 26/157 (16%)
Query 17 LIPKRYQQH--VLLIPAP---RIS----ETAGKETMLEEGEPPERPSELLRYMDLCNCCD 67
L P R+Q +L I AP R+S T ++++L EPP RPS +R + C C
Sbjct 107 LTPTRHQDSSCLLFIDAPSPSRVSLLRAPTGSEQSVLYPNEPPYRPS-TVRSLSFCECVS 165
Query 68 VLV---CLFGGSCTYERSAFSRRGYKILQALKLQGLPPSVIGVGCVDPSVLEPADAPKGK 124
L C E +++G + PS G P+ L A P
Sbjct 166 PLTLPPSSPSSHCDAEEKLLAQKGQ----------VSPSGDSTGACSPASLSSASRPVSP 215
Query 125 SSASESLKFMRRFFES---ELGAERKFFSIGSDADWH 158
SAS R E ELG E++ S + + H
Sbjct 216 GSASPRFPQTRLRIEKKSEELGLEKQRPSPSNSSPEH 252
> cpv:cgd4_1480 BMS1 like GTpase involved in ribosome biogenesis
; K14569 ribosome biogenesis protein BMS1
Length=1051
Score = 35.4 bits (80), Expect = 0.24, Method: Composition-based stats.
Identities = 45/208 (21%), Positives = 88/208 (42%), Gaps = 30/208 (14%)
Query 51 ERPSELLRYMDLCNCCDVLVCLFGGSCTYERSAFSRRGYKILQALKLQGLPPSVIGVGCV 110
E P+++ +D+ D+++ L S ++E F + L L++ G P V+GV
Sbjct 93 ECPNDMHGMIDVAKVADLVLLLIDASFSFEMETF-----EFLNILQVHGFP-RVLGV--- 143
Query 111 DPSVLEPADAPKGKSSASESLKFMRRFFESELGAERKFFSIGSDADWHNVLRAVGAAASL 170
L D + + ++ K ++ F +E+ K F + + HN +
Sbjct 144 ----LTHLDKIEDNKTVRKTKKKLKNRFWTEIYNGAKLFYL---SGIHN------GFYNK 190
Query 171 TQGDTNTKTVAKGAEAAASRRRGHLLALSWHIA------WHHVSEDPA--AQPEPCLEAC 222
T+ ++ +A S R H +S I +H ++ + PE +
Sbjct 191 TEIRNLSRFIAVQKFENLSWRSSHPYIVSLRIEEINDDDYHQNNKQSSESKNPETSVYFY 250
Query 223 GLVRGAGLTCNLPVHITGVGDFVLSRIE 250
G V+G + N VHI G+GD++++ I+
Sbjct 251 GFVKGGIMRKNQSVHIPGLGDYLINDID 278
> hsa:55720 TSR1, FLJ10534, KIAA1401, MGC131829; TSR1, 20S rRNA
accumulation, homolog (S. cerevisiae); K14799 pre-rRNA-processing
protein TSR1
Length=804
Score = 34.3 bits (77), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 48/202 (23%), Positives = 73/202 (36%), Gaps = 40/202 (19%)
Query 52 RPSELLRYMDLCNCCDVLVCLFGGSCTYERSAFSRRGYKILQALKLQGLPPSVIGV-GCV 110
RP +L +D+ D ++ L ++ + G L L QGLP + V G
Sbjct 139 RPGDLHVVLDMAKVADTILFLLDPLEGWDST-----GDYCLSCLFAQGLPTYTLAVQGIS 193
Query 111 DPSVLEPADAPKGKSSASESLKFMRRFFESELGAERKFFSIGSDADWHNVLRAVGAAASL 170
+ + D K S A E +RF K + + + +LR +
Sbjct 194 GLPLKKQIDTRKKLSKAVE-----KRF------PHDKLLLLDTQQEAGMLLRQLANQ--- 239
Query 171 TQGDTNTKTVAKGAEAAASRRRGHLLALSWHIAWHHVSEDPAAQPE--PCLEACGLVRGA 228
K A RR +L A H V P+ + L+ G VRG
Sbjct 240 -----------KQQHLAFRDRRAYLFA-------HAVDFVPSEENNLVGTLKISGYVRGQ 281
Query 229 GLTCNLPVHITGVGDFVLSRIE 250
L N +HI G GDF + +I+
Sbjct 282 TLNVNRLLHIVGYGDFQMKQID 303
> mmu:213895 Bms1, AA408648, AU020092, AU043373, BB007109, BC030906,
Bms1l, KIAA0187, mKIAA0187; BMS1 homolog, ribosome assembly
protein (yeast); K14569 ribosome biogenesis protein BMS1
Length=1284
Score = 33.9 bits (76), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 45/209 (21%), Positives = 76/209 (36%), Gaps = 58/209 (27%)
Query 60 MDLCNCCDVLVCLFGGSCTYERSAFSRRGYKILQALKLQGLPPSVIGVGCVDPSVLEPAD 119
+DL D+++ L S +E F + L ++ G P ++GV L D
Sbjct 141 IDLAKVADLVLMLIDASFGFEMETF-----EFLNICQVHGFP-KIMGV-------LTHLD 187
Query 120 APKGKSSASESLKFMRRFFESELGAERKFFSIG-------SDADWHNVLRAVGAAASLTQ 172
+ K ++ K ++ F +E+ K F + + + HN+ R +
Sbjct 188 SFKHNKQLKKTKKRLKHRFWTEVYPGAKLFYLSGMVHGEYQNQEIHNLGRFI-------- 239
Query 173 GDTNTKTVAKGAEAAASRRRGHLLALSWHIAWHHVSED---PAAQPEPC---------LE 220
TV K L+W + ++ D PE +
Sbjct 240 ------TVMK------------FRPLTWQTSHPYILADRMEDLTNPEAIRTNVKCDRKVS 281
Query 221 ACGLVRGAGLTCNLPVHITGVGDFVLSRI 249
G +RGA L N +H+ GVGDFV S +
Sbjct 282 LYGYLRGAYLKNNSQIHMPGVGDFVASDV 310
> tgo:TGME49_011390 hypothetical protein ; K10684 ubiquitin-like
1-activating enzyme E1 A [EC:6.3.2.19]
Length=405
Score = 33.5 bits (75), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 16/57 (28%), Positives = 27/57 (47%), Gaps = 2/57 (3%)
Query 22 YQQHVLLIPAPR--ISETAGKETMLEEGEPPERPSELLRYMDLCNCCDVLVCLFGGS 76
+ H +PAP+ S+ AG + GE RP ++R + + D+L C G +
Sbjct 202 FHHHTFRVPAPKAQTSQAAGGKATATNGEEESRPHHVMREISFPSLDDMLHCSLGNA 258
> ath:AT4G21210 ATRP1; ATRP1 (PPDK REGULATORY PROTEIN); phosphoprotein
phosphatase/ protein kinase
Length=376
Score = 33.5 bits (75), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 37/91 (40%), Gaps = 19/91 (20%)
Query 91 ILQALKLQGLPPSVIGVGCVDPSVLEPADAPKGKSSASESLKFMRRFFESELGAERKFFS 150
+L A+KLQG PP + S L P P G S S SE G+ERK
Sbjct 3 LLSAMKLQGRPPPI-------SSNLNPNSKPAGSDSVS--------LNASEPGSERKPRK 47
Query 151 IGSDADWHNVLRAVGAAASL----TQGDTNT 177
S + N R + + A L T G NT
Sbjct 48 FSSQLNRWNRARTLRSGAKLDSTITNGSNNT 78
> cel:F10G7.1 tag-151; Temporarily Assigned Gene name family member
(tag-151); K14799 pre-rRNA-processing protein TSR1
Length=785
Score = 33.5 bits (75), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Query 218 CLEACGLVRGAGLTCNLPVHITGVGDFVLSRIE-TMEP 254
L A G +RG N VH+ G GDF +S+IE T++P
Sbjct 270 TLVAQGYLRGPEWNANNLVHLPGFGDFQISKIESTVDP 307
> tgo:TGME49_002540 3',5'-cyclic nucleotide phosphodiesterase,
putative (EC:3.1.4.17 1.6.99.5)
Length=1670
Score = 31.6 bits (70), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 27 LLIPAPRISETAGKETMLEEGEPPERPSELLRYMDLCNCCDVLVCLFGGSCT 78
++ A R + G+ET++ G E E R D +C VC FGGSC+
Sbjct 201 VIAAAERGRKREGEETIVLRGNQGECWQEAQRTADSPDCSSSSVCCFGGSCS 252
> dre:497282 smchd1, kiaa0650l, kiaao650l; structural maintenance
of chromosomes flexible hinge domain containing 1
Length=326
Score = 31.2 bits (69), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 22/46 (47%), Gaps = 5/46 (10%)
Query 225 VRGAGLTCNLPVHITGVGDFVLSRIETMEPAYEAKRREPLLVNGEA 270
V G GL + V +TG GD V+ E E KRR L NGE
Sbjct 241 VAGEGLDGQVVVTVTGTGDIVIPLFENKE-----KRRSYSLTNGET 281
Lambda K H
0.320 0.136 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 11965793168
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40