bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
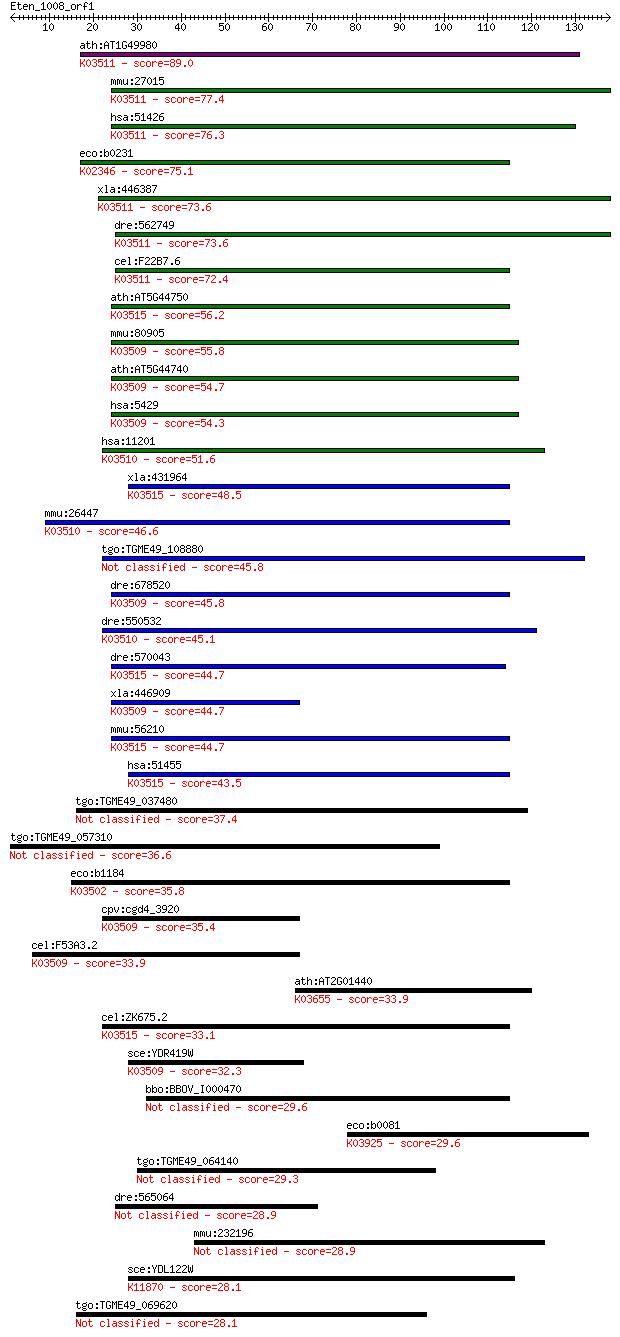
Query= Eten_1008_orf1
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
ath:AT1G49980 DNA-directed DNA polymerase/ damaged DNA binding... 89.0 4e-18
mmu:27015 Polk, Dinb1; polymerase (DNA directed), kappa (EC:2.... 77.4 1e-14
hsa:51426 POLK, DINB1, DINP, POLQ; polymerase (DNA directed) k... 76.3 3e-14
eco:b0231 dinB, dinP, ECK0232, JW0221; DNA polymerase IV (EC:2... 75.1 7e-14
xla:446387 polk, MGC83722; polymerase (DNA directed) kappa (EC... 73.6 2e-13
dre:562749 polk, si:ch211-254o18.3; polymerase (DNA directed) ... 73.6 2e-13
cel:F22B7.6 polk-1; POLK (DNA polymerase kappa) homolog family... 72.4 4e-13
ath:AT5G44750 REV1; REV1; DNA-directed DNA polymerase; K03515 ... 56.2 3e-08
mmu:80905 Polh, RAD30A, XPV; polymerase (DNA directed), eta (R... 55.8 4e-08
ath:AT5G44740 POLH; POLH (Y-FAMILT DNA POLYMERASE H); DNA-dire... 54.7 8e-08
hsa:5429 POLH, FLJ16395, FLJ21978, RAD30, RAD30A, XP-V, XPV; p... 54.3 1e-07
hsa:11201 POLI, RAD30B, RAD3OB; polymerase (DNA directed) iota... 51.6 7e-07
xla:431964 rev1, MGC83743; REV1 homolog; K03515 DNA repair pro... 48.5 7e-06
mmu:26447 Poli, Rad30b; polymerase (DNA directed), iota (EC:2.... 46.6 3e-05
tgo:TGME49_108880 DNA polymerase, putative (EC:2.7.7.7 3.2.1.91) 45.8 4e-05
dre:678520 MGC136881, fc50a12, wu:fc50a12, zgc:152642; zgc:136... 45.8 4e-05
dre:550532 wu:fc27h11; zgc:110185 (EC:2.7.7.7); K03510 DNA pol... 45.1 6e-05
dre:570043 si:dkey-57a22.7; K03515 DNA repair protein REV1 [EC... 44.7 8e-05
xla:446909 polh, MGC81702; polymerase (DNA directed), eta (EC:... 44.7 8e-05
mmu:56210 Rev1, 1110027I23Rik, AU022044, MGC66950, Rev1l; REV1... 44.7 1e-04
hsa:51455 REV1, FLJ21523, MGC163283, MGC26225, REV1L; REV1 hom... 43.5 2e-04
tgo:TGME49_037480 ImpB/MucB/SamB family domain-containing prot... 37.4 0.014
tgo:TGME49_057310 nuclear movement domain-containing protein 36.6 0.021
eco:b1184 umuC, ECK1172, JW1173, uvm; DNA polymerase V, subuni... 35.8 0.036
cpv:cgd4_3920 DinB/family X-type DNA polymerase ; K03509 DNA p... 35.4 0.048
cel:F53A3.2 polh-1; POLH (DNA polymerase eta) homolog family m... 33.9 0.14
ath:AT2G01440 ATP binding / ATP-dependent DNA helicase/ ATP-de... 33.9 0.17
cel:ZK675.2 rev-1; REV1 (translesion DNA polymerase) homolog f... 33.1 0.26
sce:YDR419W RAD30, DBH1; DNA polymerase eta, involved in tranl... 32.3 0.45
bbo:BBOV_I000470 16.m00783; ImpB/MucB/SamB family protein 29.6 2.9
eco:b0081 mraZ, ECK0082, JW0079, yabB; conserved protein, MraZ... 29.6 3.2
tgo:TGME49_064140 hypothetical protein 29.3 4.2
dre:565064 serpina10a, si:dkey-85n7.5; serpin peptidase inhibi... 28.9 5.0
mmu:232196 MGC25529; expressed sequence C87436 28.9 5.4
sce:YDL122W UBP1; Ubiquitin-specific protease that removes ubi... 28.1 8.5
tgo:TGME49_069620 hypothetical protein 28.1 9.3
> ath:AT1G49980 DNA-directed DNA polymerase/ damaged DNA binding;
K03511 DNA polymerase kappa subunit [EC:2.7.7.7]
Length=671
Score = 89.0 bits (219), Expect = 4e-18, Method: Composition-based stats.
Identities = 50/118 (42%), Positives = 73/118 (61%), Gaps = 11/118 (9%)
Query 17 CRRRVTA---LVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATT 73
CR R + + E+R++V S TGL+CSAG A+R++AK+ SD+NKPNGQ +++ +T
Sbjct 210 CRERGLSGGEIAEELRSSVYSETGLTCSAGVAANRLLAKVCSDINKPNGQFVLQNDRST- 268
Query 74 AAATAATAAFLDSLNVRKIPGVGKYREKML-NALGIHTCGELLQHAPLLLYLLPKITA 130
F+ L VRKI G+GK E +L +ALGI TC E++Q LL L + +A
Sbjct 269 ------VMTFVSFLPVRKIGGIGKVTEHILKDALGIKTCEEMVQKGSLLYALFSQSSA 320
> mmu:27015 Polk, Dinb1; polymerase (DNA directed), kappa (EC:2.7.7.7);
K03511 DNA polymerase kappa subunit [EC:2.7.7.7]
Length=852
Score = 77.4 bits (189), Expect = 1e-14, Method: Composition-based stats.
Identities = 44/114 (38%), Positives = 63/114 (55%), Gaps = 7/114 (6%)
Query 24 LVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAF 83
+V EIR + T L+ SAG + ++AK+ SD NKPNGQ + + +A F
Sbjct 292 VVKEIRFRIEQKTTLTASAGIAPNTMLAKVCSDKNKPNGQYQI-------LPSRSAVMDF 344
Query 84 LDSLNVRKIPGVGKYREKMLNALGIHTCGELLQHAPLLLYLLPKITAVHLIRMA 137
+ L +RK+ G+GK EKML ALGI TC EL Q LL L + + + + +A
Sbjct 345 IKDLPIRKVSGIGKVTEKMLMALGIVTCTELYQQRALLSLLFSETSWHYFLHIA 398
> hsa:51426 POLK, DINB1, DINP, POLQ; polymerase (DNA directed)
kappa (EC:2.7.7.7); K03511 DNA polymerase kappa subunit [EC:2.7.7.7]
Length=870
Score = 76.3 bits (186), Expect = 3e-14, Method: Composition-based stats.
Identities = 43/106 (40%), Positives = 57/106 (53%), Gaps = 7/106 (6%)
Query 24 LVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAF 83
+V EIR + T L+ SAG + ++AK+ SD NKPNGQ + A F
Sbjct 293 VVKEIRFRIEQKTTLTASAGIAPNTMLAKVCSDKNKPNGQYQILPN-------RQAVMDF 345
Query 84 LDSLNVRKIPGVGKYREKMLNALGIHTCGELLQHAPLLLYLLPKIT 129
+ L +RK+ G+GK EKML ALGI TC EL Q LL L + +
Sbjct 346 IKDLPIRKVSGIGKVTEKMLKALGIITCTELYQQRALLSLLFSETS 391
> eco:b0231 dinB, dinP, ECK0232, JW0221; DNA polymerase IV (EC:2.7.7.7);
K02346 DNA polymerase IV [EC:2.7.7.7]
Length=351
Score = 75.1 bits (183), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 40/98 (40%), Positives = 54/98 (55%), Gaps = 9/98 (9%)
Query 17 CRRRVTALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAA 76
C T + EIR + + L+ SAG + +AKIASDMNKPNGQ ++
Sbjct 115 CHGSATLIAQEIRQTIFNELQLTASAGVAPVKFLAKIASDMNKPNGQFVI---------T 165
Query 77 TAATAAFLDSLNVRKIPGVGKYREKMLNALGIHTCGEL 114
A AFL +L + KIPGVGK L A+G+ TCG++
Sbjct 166 PAEVPAFLQTLPLAKIPGVGKVSAAKLEAMGLRTCGDV 203
> xla:446387 polk, MGC83722; polymerase (DNA directed) kappa (EC:2.7.7.7);
K03511 DNA polymerase kappa subunit [EC:2.7.7.7]
Length=862
Score = 73.6 bits (179), Expect = 2e-13, Method: Composition-based stats.
Identities = 42/117 (35%), Positives = 62/117 (52%), Gaps = 7/117 (5%)
Query 21 VTALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAAT 80
V V E+R + T L+ SAG + ++AK+ SD NKPNGQ + A A
Sbjct 288 VEEAVKEMRFRIEQKTTLTASAGIAPNMMLAKVCSDKNKPNGQYRI-------ANDRQAI 340
Query 81 AAFLDSLNVRKIPGVGKYREKMLNALGIHTCGELLQHAPLLLYLLPKITAVHLIRMA 137
F+ L +RK+PG+GK EKML AL + TC +L Q LL L + + + + ++
Sbjct 341 KDFIKDLPIRKVPGIGKVTEKMLKALEVVTCTDLYQQRALLSLLFSETSWQNFVHIS 397
> dre:562749 polk, si:ch211-254o18.3; polymerase (DNA directed)
kappa; K03511 DNA polymerase kappa subunit [EC:2.7.7.7]
Length=888
Score = 73.6 bits (179), Expect = 2e-13, Method: Composition-based stats.
Identities = 43/113 (38%), Positives = 59/113 (52%), Gaps = 7/113 (6%)
Query 25 VSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAFL 84
V E+R + T L+ SAG + ++AK+ SD NKPNGQ + F+
Sbjct 258 VREMRFRIEQKTSLTASAGIAPNMMLAKVCSDKNKPNGQYRI-------PPERQVVMDFM 310
Query 85 DSLNVRKIPGVGKYREKMLNALGIHTCGELLQHAPLLLYLLPKITAVHLIRMA 137
L VRK+ GVGK EKML ALGI TC +L Q LL L + + H + ++
Sbjct 311 KDLPVRKVSGVGKVTEKMLAALGIVTCSQLGQQMALLSLLFSETSWHHFLHIS 363
> cel:F22B7.6 polk-1; POLK (DNA polymerase kappa) homolog family
member (polk-1); K03511 DNA polymerase kappa subunit [EC:2.7.7.7]
Length=596
Score = 72.4 bits (176), Expect = 4e-13, Method: Composition-based stats.
Identities = 38/90 (42%), Positives = 53/90 (58%), Gaps = 7/90 (7%)
Query 25 VSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAFL 84
V EIR V TGL+CSAG ++ ++AKI SD+NKPNGQ ++E A FL
Sbjct 256 VREIRFRVEQLTGLTCSAGIASNFMLAKICSDLNKPNGQYVLEND-------KNAIMEFL 308
Query 85 DSLNVRKIPGVGKYREKMLNALGIHTCGEL 114
L +RK+ G+G+ E L A+ I T G++
Sbjct 309 KDLPIRKVGGIGRVCEAQLKAMDIQTVGDM 338
> ath:AT5G44750 REV1; REV1; DNA-directed DNA polymerase; K03515
DNA repair protein REV1 [EC:2.7.7.-]
Length=1105
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 33/91 (36%), Positives = 46/91 (50%), Gaps = 9/91 (9%)
Query 24 LVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAF 83
L S IR + TG S SAG G + ++A++A+ + KP GQ + A F
Sbjct 493 LASTIRNEILETTGCSASAGIGGTMLMARLATRVAKPAGQLYISA---------EKVEEF 543
Query 84 LDSLNVRKIPGVGKYREKMLNALGIHTCGEL 114
LD L V +PGVG ++ L I TCG+L
Sbjct 544 LDQLPVGTLPGVGSVLKEKLVKQNIQTCGQL 574
> mmu:80905 Polh, RAD30A, XPV; polymerase (DNA directed), eta
(RAD 30 related) (EC:2.7.7.7); K03509 DNA polymerase eta subunit
[EC:2.7.7.7]
Length=694
Score = 55.8 bits (133), Expect = 4e-08, Method: Composition-based stats.
Identities = 32/94 (34%), Positives = 50/94 (53%), Gaps = 10/94 (10%)
Query 24 LVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAF 83
+V E+RAA+ S TG CSAG ++++AK+A +NKPN Q +V + +
Sbjct 195 IVEEMRAAIESKTGFQCSAGISHNKVLAKLACGLNKPNRQTLV---------SHGSVPQL 245
Query 84 LDSLNVRKIPGV-GKYREKMLNALGIHTCGELLQ 116
+ +RKI + GK ++ LGI G+L Q
Sbjct 246 FSQMPIRKIRSLGGKLGASVIEVLGIEYMGDLTQ 279
> ath:AT5G44740 POLH; POLH (Y-FAMILT DNA POLYMERASE H); DNA-directed
DNA polymerase; K03509 DNA polymerase eta subunit [EC:2.7.7.7]
Length=672
Score = 54.7 bits (130), Expect = 8e-08, Method: Composition-based stats.
Identities = 34/94 (36%), Positives = 51/94 (54%), Gaps = 10/94 (10%)
Query 24 LVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAF 83
+V+E+R V T +CSAG ++++AK+AS MNKP Q +V AA
Sbjct 191 IVAELRKQVLKETEFTCSAGIAHNKMLAKLASGMNKPAQQTVV---------PYAAVQEL 241
Query 84 LDSLNVRKIPGV-GKYREKMLNALGIHTCGELLQ 116
L SL ++K+ + GK + LG+ T G+LLQ
Sbjct 242 LSSLPIKKMKQLGGKLGTSLQTDLGVDTVGDLLQ 275
> hsa:5429 POLH, FLJ16395, FLJ21978, RAD30, RAD30A, XP-V, XPV;
polymerase (DNA directed), eta (EC:2.7.7.7); K03509 DNA polymerase
eta subunit [EC:2.7.7.7]
Length=713
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 32/94 (34%), Positives = 49/94 (52%), Gaps = 10/94 (10%)
Query 24 LVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAF 83
+V E+RAA+ TG CSAG ++++AK+A +NKPN Q +V + +
Sbjct 196 IVEEMRAAIERETGFQCSAGISHNKVLAKLACGLNKPNRQTLV---------SHGSVPQL 246
Query 84 LDSLNVRKIPGV-GKYREKMLNALGIHTCGELLQ 116
+ +RKI + GK ++ LGI GEL Q
Sbjct 247 FSQMPIRKIRSLGGKLGASVIEILGIEYMGELTQ 280
> hsa:11201 POLI, RAD30B, RAD3OB; polymerase (DNA directed) iota
(EC:2.7.7.7); K03510 DNA polymerase iota subunit [EC:2.7.7.7]
Length=740
Score = 51.6 bits (122), Expect = 7e-07, Method: Composition-based stats.
Identities = 31/102 (30%), Positives = 56/102 (54%), Gaps = 10/102 (9%)
Query 22 TALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATA 81
+ + +E+R A+ + GL+ AG +++++AK+ S + KPN Q T +
Sbjct 202 SQIAAEMREAMYNQLGLTGCAGVASNKLLAKLVSGVFKPNQQ---------TVLLPESCQ 252
Query 82 AFLDSLN-VRKIPGVGKYREKMLNALGIHTCGELLQHAPLLL 122
+ SLN +++IPG+G K L ALGI++ +L +P +L
Sbjct 253 HLIHSLNHIKEIPGIGYKTAKCLEALGINSVRDLQTFSPKIL 294
> xla:431964 rev1, MGC83743; REV1 homolog; K03515 DNA repair protein
REV1 [EC:2.7.7.-]
Length=1230
Score = 48.5 bits (114), Expect = 7e-06, Method: Composition-based stats.
Identities = 27/87 (31%), Positives = 46/87 (52%), Gaps = 9/87 (10%)
Query 28 IRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAFLDSL 87
IR + TG + S G G++ ++A++A+ KP+GQ ++ F+
Sbjct 582 IRTEIKEKTGCAASIGIGSNILLARMATRRAKPDGQYHLKPEEVDD---------FIRGQ 632
Query 88 NVRKIPGVGKYREKMLNALGIHTCGEL 114
V +PGVG+ + L++LG+ TCGEL
Sbjct 633 LVNNLPGVGRTMDCKLSSLGVKTCGEL 659
> mmu:26447 Poli, Rad30b; polymerase (DNA directed), iota (EC:2.7.7.7);
K03510 DNA polymerase iota subunit [EC:2.7.7.7]
Length=737
Score = 46.6 bits (109), Expect = 3e-05, Method: Composition-based stats.
Identities = 32/110 (29%), Positives = 57/110 (51%), Gaps = 13/110 (11%)
Query 9 QKVSVRRECRRRV---TALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKI 65
Q V++ RR+ + + +E+R A+ + GL+ AG ++++AK+ S + KPN Q
Sbjct 181 QSVNLHNIMHRRLVVGSQIAAEMREAMYNQLGLTGCAGVAPNKLLAKLVSGVFKPNQQ-- 238
Query 66 VEATGATTAAATAATAAFLDSLN-VRKIPGVGKYREKMLNALGIHTCGEL 114
T + + SLN +++IPG+G K L LGI++ +L
Sbjct 239 -------TVLLPESCQHLIHSLNHIKEIPGIGYKTAKRLEVLGINSVHDL 281
> tgo:TGME49_108880 DNA polymerase, putative (EC:2.7.7.7 3.2.1.91)
Length=1613
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 35/110 (31%), Positives = 55/110 (50%), Gaps = 15/110 (13%)
Query 22 TALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATA 81
T L S IR + + GL+ +AG ++ +AK+A NKP+ Q ++
Sbjct 620 THLASAIRRYIGTHMGLTSTAGISTNKSLAKLAGAFNKPSRQTLL---------LPQHRR 670
Query 82 AFLDSLNVRKIPGVGKYREKMLNALGIHTCGELLQHAPLLLYLLPKITAV 131
FL L ++KIPGVG ++L+ G+ TC A LL LP++ A+
Sbjct 671 GFLAPLALQKIPGVGSALVRLLSRAGLRTC------ADLLAVSLPRLAAI 714
> dre:678520 MGC136881, fc50a12, wu:fc50a12, zgc:152642; zgc:136881
(EC:2.7.7.7); K03509 DNA polymerase eta subunit [EC:2.7.7.7]
Length=743
Score = 45.8 bits (107), Expect = 4e-05, Method: Composition-based stats.
Identities = 28/92 (30%), Positives = 45/92 (48%), Gaps = 10/92 (10%)
Query 24 LVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAF 83
+V ++RAAV TG CSAG ++++AK+A +NKPN Q T ++
Sbjct 195 IVEQMRAAVEEHTGFRCSAGISHNKVLAKLACGLNKPNRQ---------TVLPLSSVPQL 245
Query 84 LDSLNVRKIPGV-GKYREKMLNALGIHTCGEL 114
+L + KI + GK + L + G+L
Sbjct 246 FSTLPISKIRNLGGKLGSSITETLSVENMGDL 277
> dre:550532 wu:fc27h11; zgc:110185 (EC:2.7.7.7); K03510 DNA polymerase
iota subunit [EC:2.7.7.7]
Length=710
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 35/101 (34%), Positives = 54/101 (53%), Gaps = 13/101 (12%)
Query 22 TALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATA 81
+ + +E+R A+ S GL+ AG +++++AK+ S KPN Q T + A
Sbjct 182 SVIAAEMRQAIFSTLGLTGCAGVASNKLLAKLVSGSFKPNQQ---------TTLLPHSNA 232
Query 82 AFLDSLN-VRKIPGVG-KYREKMLNALGIHTCGELLQHAPL 120
+ SL + K+PG+G K REK L ALG+ + LQ PL
Sbjct 233 ELMSSLTGLIKVPGIGYKTREK-LKALGLVNVRD-LQLYPL 271
> dre:570043 si:dkey-57a22.7; K03515 DNA repair protein REV1 [EC:2.7.7.-]
Length=1268
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 25/90 (27%), Positives = 42/90 (46%), Gaps = 9/90 (10%)
Query 24 LVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAF 83
L IR + TG S G ++ ++A++A+ KP GQ ++ + F
Sbjct 629 LARSIREDIKEKTGCCASVGMSSNILLARMATRKAKPKGQYLLR---------SEEVDDF 679
Query 84 LDSLNVRKIPGVGKYREKMLNALGIHTCGE 113
+ V +PGVG+ L +LG+ TCG+
Sbjct 680 IRDQPVSSLPGVGRSMSSKLTSLGVSTCGD 709
> xla:446909 polh, MGC81702; polymerase (DNA directed), eta (EC:2.7.7.7);
K03509 DNA polymerase eta subunit [EC:2.7.7.7]
Length=684
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 20/43 (46%), Positives = 29/43 (67%), Gaps = 0/43 (0%)
Query 24 LVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIV 66
+V E+RAAV T CSAG ++++AK+A +NKPN Q I+
Sbjct 195 IVEEMRAAVEEETTFQCSAGIAHNKVLAKLACGLNKPNRQTIL 237
> mmu:56210 Rev1, 1110027I23Rik, AU022044, MGC66950, Rev1l; REV1
homolog (S. cerevisiae); K03515 DNA repair protein REV1 [EC:2.7.7.-]
Length=1249
Score = 44.7 bits (104), Expect = 1e-04, Method: Composition-based stats.
Identities = 26/91 (28%), Positives = 45/91 (49%), Gaps = 9/91 (9%)
Query 24 LVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAF 83
+ +R + T + S G G++ ++A++A+ KP+GQ ++ F
Sbjct 588 FAAALRIEIKDKTKCAASVGIGSNILLARMATKKAKPDGQYHLQPDEVDD---------F 638
Query 84 LDSLNVRKIPGVGKYREKMLNALGIHTCGEL 114
+ V +PGVG+ E L +LGI TCG+L
Sbjct 639 IRGQLVTNLPGVGRSMESKLASLGIKTCGDL 669
> hsa:51455 REV1, FLJ21523, MGC163283, MGC26225, REV1L; REV1 homolog
(S. cerevisiae); K03515 DNA repair protein REV1 [EC:2.7.7.-]
Length=1250
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 43/87 (49%), Gaps = 9/87 (10%)
Query 28 IRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAFLDSL 87
+R + T + S G G++ ++A++A+ KP+GQ ++ F+
Sbjct 593 VRMEIKDQTKCAASVGIGSNILLARMATRKAKPDGQYHLKPEEVDD---------FIRGQ 643
Query 88 NVRKIPGVGKYREKMLNALGIHTCGEL 114
V +PGVG E L +LGI TCG+L
Sbjct 644 LVTNLPGVGHSMESKLASLGIKTCGDL 670
> tgo:TGME49_037480 ImpB/MucB/SamB family domain-containing protein
(EC:2.7.7.7)
Length=2474
Score = 37.4 bits (85), Expect = 0.014, Method: Composition-based stats.
Identities = 28/111 (25%), Positives = 49/111 (44%), Gaps = 8/111 (7%)
Query 16 ECRRRVTALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAA 75
E + + E+R + TG S GAG + + AK+A KP ++ + +
Sbjct 903 EAEKAILDACVELRQRIFERTGCWVSIGAGPNLLAAKLAGLQAKPKASSSSSSSSSLSRD 962
Query 76 ATAATA-------AFLDSLNVRKIPGVGKYR-EKMLNALGIHTCGELLQHA 118
AF+D + +RK+PGVG EK+ A + CG++++
Sbjct 963 GVCVIRPRESFLWAFMDQVPLRKLPGVGSMLGEKVEKAGRLRLCGQVMKEG 1013
> tgo:TGME49_057310 nuclear movement domain-containing protein
Length=355
Score = 36.6 bits (83), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 32/101 (31%), Positives = 44/101 (43%), Gaps = 9/101 (8%)
Query 1 LLVLLLLQQKVSVRR--ECRRRVTALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMN 58
L LL++KV+ RR + + V SE R + TG AGA A A+D
Sbjct 205 LWFWELLEKKVNWRRLAQLAKDVNGQ-SEERTPATDGTG----AGADACPTPESRAADQT 259
Query 59 KPNGQKIVEATGATTAAATAATAAFLDSLNVRKIPGV-GKY 98
+ Q + E G A FL+ +N+RK P V G Y
Sbjct 260 EKEAQNLTETEGEAAGEGQEEVAVFLE-INLRKKPEVAGTY 299
> eco:b1184 umuC, ECK1172, JW1173, uvm; DNA polymerase V, subunit
C; K03502 DNA polymerase V
Length=422
Score = 35.8 bits (81), Expect = 0.036, Method: Composition-based stats.
Identities = 28/100 (28%), Positives = 46/100 (46%), Gaps = 4/100 (4%)
Query 15 RECRRRVTALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTA 74
R CR +T EIRA V T L+ G ++ +AK+A+ K + + G
Sbjct 111 RNCRD-LTDFGREIRATVLQRTHLTVGVGIAQTKTLAKLANHAAKKWQR---QTGGVVDL 166
Query 75 AATAATAAFLDSLNVRKIPGVGKYREKMLNALGIHTCGEL 114
+ + +L V + G+G+ K L+A+GI T +L
Sbjct 167 SNLERQRKLMSALPVDDVWGIGRRISKKLDAMGIKTVLDL 206
> cpv:cgd4_3920 DinB/family X-type DNA polymerase ; K03509 DNA
polymerase eta subunit [EC:2.7.7.7]
Length=358
Score = 35.4 bits (80), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 22 TALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIV 66
+ L+ R + T +CS+G ++I +K+ S + KPNGQ ++
Sbjct 271 SILIYRARKRLQEETTYTCSSGIFINKIYSKMISSLKKPNGQTVL 315
> cel:F53A3.2 polh-1; POLH (DNA polymerase eta) homolog family
member (polh-1); K03509 DNA polymerase eta subunit [EC:2.7.7.7]
Length=584
Score = 33.9 bits (76), Expect = 0.14, Method: Composition-based stats.
Identities = 17/66 (25%), Positives = 33/66 (50%), Gaps = 5/66 (7%)
Query 6 LLQQKVSVRRECRRRVTALVS-----EIRAAVSSATGLSCSAGAGASRIVAKIASDMNKP 60
+L + + R C + L++ +IR + T CSAG G ++++AK+ +KP
Sbjct 172 VLLEYIENARNCTENLLLLIAAVTVEQIRQQIHEETQFFCSAGVGNNKMMAKLVCARHKP 231
Query 61 NGQKIV 66
Q ++
Sbjct 232 RQQTLI 237
> ath:AT2G01440 ATP binding / ATP-dependent DNA helicase/ ATP-dependent
helicase/ helicase/ nucleic acid binding; K03655 ATP-dependent
DNA helicase RecG [EC:3.6.4.12]
Length=973
Score = 33.9 bits (76), Expect = 0.17, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 26/54 (48%), Gaps = 5/54 (9%)
Query 66 VEATGATTAAATAATAAFLDSLNVRKIPGVGKYREKMLNALGIHTCGELLQHAP 119
VEAT AA A +DS+ PG+ K L++ G HT +LL H P
Sbjct 203 VEATSDDVFAAQRFLATSIDSM-----PGLSKRHSNQLDSCGFHTMKKLLHHFP 251
> cel:ZK675.2 rev-1; REV1 (translesion DNA polymerase) homolog
family member (rev-1); K03515 DNA repair protein REV1 [EC:2.7.7.-]
Length=1027
Score = 33.1 bits (74), Expect = 0.26, Method: Composition-based stats.
Identities = 27/98 (27%), Positives = 41/98 (41%), Gaps = 17/98 (17%)
Query 22 TALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATA 81
T L IR + T S G G++ ++A++A+ KP+G V A
Sbjct 562 TILAEHIRKVIREKTQCPASVGIGSTSLLARLATRHAKPDGVFWVNAHKKN--------- 612
Query 82 AFLDSLNVRKIPGVGKYREKMLNAL-----GIHTCGEL 114
F+ V+ +PG G +M+N L I C EL
Sbjct 613 EFISEEKVKDLPGFGY---EMMNRLTSFFGDITKCREL 647
> sce:YDR419W RAD30, DBH1; DNA polymerase eta, involved in tranlesion
synthesis during post-replication repair; catalyzes
the synthesis of DNA opposite cyclobutane pyrimidine dimers
and other lesions; mutations in human pol eta are responsible
for XPV (EC:2.7.7.7); K03509 DNA polymerase eta subunit [EC:2.7.7.7]
Length=632
Score = 32.3 bits (72), Expect = 0.45, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 24/40 (60%), Gaps = 0/40 (0%)
Query 28 IRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVE 67
IR ++ G + S G +++ V K+AS+ KP+ Q IV+
Sbjct 248 IRDSIKDILGYTTSCGLSSTKNVCKLASNYKKPDAQTIVK 287
> bbo:BBOV_I000470 16.m00783; ImpB/MucB/SamB family protein
Length=1246
Score = 29.6 bits (65), Expect = 2.9, Method: Composition-based stats.
Identities = 24/88 (27%), Positives = 43/88 (48%), Gaps = 6/88 (6%)
Query 32 VSSATGLSCSAGAGASRIVAKIASDMNKP--NGQKIVEATGATTAAATAA---TAAFLDS 86
+ + T S G G + ++AK+AS K +G + V TG + A AF+ S
Sbjct 562 IETETECPLSIGIGDNMMLAKLASQRCKALKHGSEEV-PTGYVLSGNVCAIQDVEAFIAS 620
Query 87 LNVRKIPGVGKYREKMLNALGIHTCGEL 114
+++ ++PGVG +L + G C ++
Sbjct 621 VSLHELPGVGDRTLDILKSCGYVYCSDI 648
> eco:b0081 mraZ, ECK0082, JW0079, yabB; conserved protein, MraZ
family; K03925 MraZ protein
Length=152
Score = 29.6 bits (65), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 28/56 (50%), Gaps = 3/56 (5%)
Query 78 AATAAFLDSLNVRKIPGVGKYREKML-NALGIHTCGELLQHAPLLLYLLPKITAVH 132
AT LDS +P +YRE++L NA G C + H LLLY LP+ +
Sbjct 4 GATLVNLDSKGRLSVPT--RYREQLLENAAGQMVCTIDIYHPCLLLYPLPEWEIIE 57
> tgo:TGME49_064140 hypothetical protein
Length=4532
Score = 29.3 bits (64), Expect = 4.2, Method: Composition-based stats.
Identities = 19/68 (27%), Positives = 33/68 (48%), Gaps = 0/68 (0%)
Query 30 AAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAFLDSLNV 89
+ VSSA+ LS A +G + + + N P ++++ +G AAT +F D +V
Sbjct 3185 SVVSSASRLSTRAQSGDTELHLEEDKGSNSPESRRLITHSGIERTAATEKKLSFSDEGSV 3244
Query 90 RKIPGVGK 97
G+ K
Sbjct 3245 GTGEGLTK 3252
> dre:565064 serpina10a, si:dkey-85n7.5; serpin peptidase inhibitor,
clade A (alpha-1 antiproteinase, antitrypsin), member
10a
Length=391
Score = 28.9 bits (63), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 26/46 (56%), Gaps = 1/46 (2%)
Query 25 VSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVEATG 70
V EI ++ TG+S S G S + K+A D+++ G + EA+G
Sbjct 306 VKEIFGNTANLTGISSSEGLKLSEVEQKVAVDVDESGG-SLAEASG 350
> mmu:232196 MGC25529; expressed sequence C87436
Length=574
Score = 28.9 bits (63), Expect = 5.4, Method: Composition-based stats.
Identities = 24/80 (30%), Positives = 37/80 (46%), Gaps = 5/80 (6%)
Query 43 GAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAFLDSLNVRKIPGVGKYREKM 102
G A V+ S +KP +K E +GA ++ A + N+RK G R +
Sbjct 285 GCHAESSVSACESAASKPRKRKKDEVSGAQVNSSLMPQDAV--NSNLRK---SGLKRPVV 339
Query 103 LNALGIHTCGELLQHAPLLL 122
++L H CG+LL A + L
Sbjct 340 TSSLKRHVCGQLLDEAQVTL 359
> sce:YDL122W UBP1; Ubiquitin-specific protease that removes ubiquitin
from ubiquitinated proteins; cleaves at the C terminus
of ubiquitin fusions irrespective of their size; capable
of cleaving polyubiquitin chains (EC:3.1.2.15); K11870 ubiquitin
carboxyl-terminal hydrolase 1 [EC:3.1.2.15]
Length=809
Score = 28.1 bits (61), Expect = 8.5, Method: Composition-based stats.
Identities = 25/91 (27%), Positives = 44/91 (48%), Gaps = 6/91 (6%)
Query 28 IRAAVSSATGLSC-SAGAGASRIVAKIASDMNKPNGQKIVEATGATTAAATAATAAFLDS 86
IR +V S L+ + G++ ++++ SD +KP +I+E A TAA +
Sbjct 346 IRYSVFSGLSLNLPNENIGSTLKLSQLLSDWSKP---EIIEGVECNRCALTAAHSHLFGQ 402
Query 87 LNVRKIPGVGKYREKMLNALG--IHTCGELL 115
L + G EK++NA+ +H E+L
Sbjct 403 LKEFEKKPEGSIPEKLINAVKDRVHQIEEVL 433
> tgo:TGME49_069620 hypothetical protein
Length=554
Score = 28.1 bits (61), Expect = 9.3, Method: Composition-based stats.
Identities = 27/90 (30%), Positives = 40/90 (44%), Gaps = 12/90 (13%)
Query 16 ECRRRVTALVSEIRAAVSSATGLSCSAGAGASRIVAKIASDMNKPNGQKIVE---ATGAT 72
E RR + ++ RA VS++ G+ C G+G A A + P G+ G
Sbjct 355 EQRRHIVPDATDTRA-VSASMGIECPQGSGLPPSGAHAAVSAH-PGGEYAANGCVGVGHE 412
Query 73 TAAATAATAA-------FLDSLNVRKIPGV 95
+A+ TAAT A F +VR+ P V
Sbjct 413 SASGTAATVAEARGRLSFESGADVREKPSV 442
Lambda K H
0.321 0.132 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2428006156
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40