bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1025_orf1
Length=61
Score E
Sequences producing significant alignments: (Bits) Value
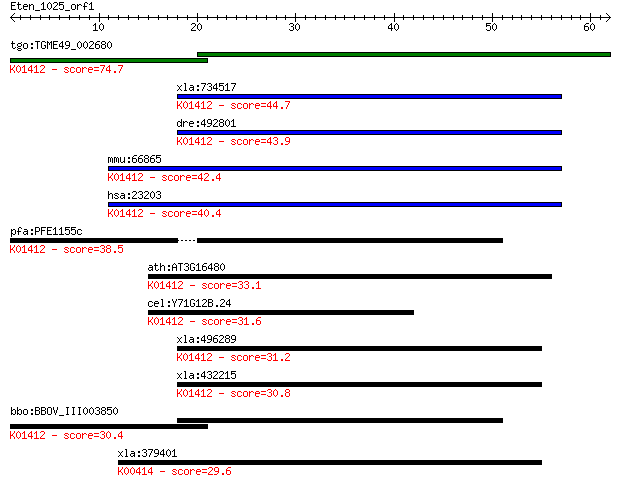
tgo:TGME49_002680 mitochondrial-processing peptidase alpha sub... 74.7 6e-14
xla:734517 pmpca, MGC114896; peptidase (mitochondrial processi... 44.7 7e-05
dre:492801 pmpca, wu:fi19e06, wu:fj83d11, zgc:101647; peptidas... 43.9 1e-04
mmu:66865 Pmpca, 1200002L24Rik, 4933435E07Rik, Alpha-MPP, P-55... 42.4 4e-04
hsa:23203 PMPCA, Alpha-MPP, FLJ26258, INPP5E, KIAA0123, MGC104... 40.4 0.001
pfa:PFE1155c mitochondrial processing peptidase alpha subunit,... 38.5 0.005
ath:AT3G16480 MPPalpha (mitochondrial processing peptidase alp... 33.1 0.24
cel:Y71G12B.24 mppa-1; Mitochondrial Processing Peptidase Alph... 31.6 0.71
xla:496289 hypothetical LOC496289; K01412 mitochondrial proces... 31.2 0.76
xla:432215 pmpcb, MGC78954; peptidase (mitochondrial processin... 30.8 1.2
bbo:BBOV_III003850 17.m07356; mitochondrial processing peptida... 30.4 1.5
xla:379401 uqcrc1, MGC53748; ubiquinol-cytochrome c reductase ... 29.6 2.2
> tgo:TGME49_002680 mitochondrial-processing peptidase alpha subunit,
putative (EC:3.4.24.64); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=563
Score = 74.7 bits (182), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 32/42 (76%), Positives = 38/42 (90%), Gaps = 0/42 (0%)
Query 20 DIRRVAEKMFSKPPTVVAYGDICSVPHYEEIRAALRAAGVGK 61
DI+RV + M+ KPPTVVAYGD+ +VPHYEE+RAALRAAGVGK
Sbjct 522 DIKRVVDAMYKKPPTVVAYGDVSTVPHYEEVRAALRAAGVGK 563
Score = 31.6 bits (70), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 13/20 (65%), Positives = 15/20 (75%), Gaps = 0/20 (0%)
Query 1 MLHQTAWHNNTLWLKLFATD 20
+LH TAWHNNTL KL T+
Sbjct 269 LLHTTAWHNNTLGHKLHCTE 288
> xla:734517 pmpca, MGC114896; peptidase (mitochondrial processing)
alpha (EC:3.4.24.64); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=518
Score = 44.7 bits (104), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 26/39 (66%), Gaps = 0/39 (0%)
Query 18 ATDIRRVAEKMFSKPPTVVAYGDICSVPHYEEIRAALRA 56
A+DI+RVA KM P V A GD+ +P YE I+AAL +
Sbjct 467 ASDIKRVATKMLRNKPAVAALGDLTELPDYEHIQAALSS 505
> dre:492801 pmpca, wu:fi19e06, wu:fj83d11, zgc:101647; peptidase
(mitochondrial processing) alpha (EC:3.4.24.64); K01412
mitochondrial processing peptidase [EC:3.4.24.64]
Length=517
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 19/39 (48%), Positives = 26/39 (66%), Gaps = 0/39 (0%)
Query 18 ATDIRRVAEKMFSKPPTVVAYGDICSVPHYEEIRAALRA 56
A+DI+RV KM P V A GD+ +P YE+I+AAL +
Sbjct 466 ASDIKRVTMKMLRSKPAVAALGDLTELPSYEDIQAALSS 504
> mmu:66865 Pmpca, 1200002L24Rik, 4933435E07Rik, Alpha-MPP, P-55;
peptidase (mitochondrial processing) alpha (EC:3.4.24.64);
K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=524
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 21/46 (45%), Positives = 27/46 (58%), Gaps = 0/46 (0%)
Query 11 TLWLKLFATDIRRVAEKMFSKPPTVVAYGDICSVPHYEEIRAALRA 56
TL + DI+RVA KM P V A GD+ +P YE I+AAL +
Sbjct 466 TLIRNVKPEDIKRVASKMLRGKPAVAALGDLTDLPTYEHIQAALSS 511
> hsa:23203 PMPCA, Alpha-MPP, FLJ26258, INPP5E, KIAA0123, MGC104197;
peptidase (mitochondrial processing) alpha (EC:3.4.24.64);
K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=525
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 11 TLWLKLFATDIRRVAEKMFSKPPTVVAYGDICSVPHYEEIRAALRA 56
TL + D++RVA KM P V A GD+ +P YE I+ AL +
Sbjct 467 TLIRNVKPEDVKRVASKMLRGKPAVAALGDLTDLPTYEHIQTALSS 512
> pfa:PFE1155c mitochondrial processing peptidase alpha subunit,
putative; K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=534
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 16/31 (51%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 20 DIRRVAEKMFSKPPTVVAYGDICSVPHYEEI 50
DI+RV PTVV YG+I PHY+EI
Sbjct 497 DIQRVVHNFLKTKPTVVVYGNINYSPHYDEI 527
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 11/17 (64%), Positives = 14/17 (82%), Gaps = 0/17 (0%)
Query 1 MLHQTAWHNNTLWLKLF 17
+LH TAW+NNTL KL+
Sbjct 243 LLHNTAWYNNTLGNKLY 259
> ath:AT3G16480 MPPalpha (mitochondrial processing peptidase alpha
subunit); catalytic/ metal ion binding / metalloendopeptidase/
zinc ion binding; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=499
Score = 33.1 bits (74), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 13/41 (31%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 15 KLFATDIRRVAEKMFSKPPTVVAYGDICSVPHYEEIRAALR 55
+L DI K+ +KP T+ +GD+ +VP Y+ + R
Sbjct 459 QLTLKDIADFTSKVITKPLTMATFGDVLNVPSYDSVSKRFR 499
> cel:Y71G12B.24 mppa-1; Mitochondrial Processing Peptidase Alpha
family member (mppa-1); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=477
Score = 31.6 bits (70), Expect = 0.71, Method: Composition-based stats.
Identities = 12/27 (44%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 15 KLFATDIRRVAEKMFSKPPTVVAYGDI 41
K+ +DI RV E++ + P++V YGDI
Sbjct 428 KVTNSDIIRVTERLLASKPSLVGYGDI 454
> xla:496289 hypothetical LOC496289; K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=479
Score = 31.2 bits (69), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Query 18 ATDIRRVAEK-MFSKPPTVVAYGDICSVPHYEEIRAAL 54
A IR V K +++K P V A G I +P+Y+ IR+ +
Sbjct 437 AETIREVCTKYIYNKSPAVAAVGPIGELPNYDRIRSGM 474
> xla:432215 pmpcb, MGC78954; peptidase (mitochondrial processing)
beta (EC:3.4.24.64); K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=479
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 22/38 (57%), Gaps = 1/38 (2%)
Query 18 ATDIRRVAEK-MFSKPPTVVAYGDICSVPHYEEIRAAL 54
A IR V K +++K P V A G I +P Y+ IR+ +
Sbjct 437 AETIREVCTKYIYNKSPAVAAVGPIGQLPDYDRIRSGM 474
> bbo:BBOV_III003850 17.m07356; mitochondrial processing peptidase
alpha subunit; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=496
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 21/34 (61%), Gaps = 1/34 (2%)
Query 18 ATDIRRVAEKMFS-KPPTVVAYGDICSVPHYEEI 50
A DI+R + M P+VVA G++ +PH EE+
Sbjct 457 ALDIQRCVQSMLKGSKPSVVALGNLAFMPHPEEL 490
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 10/20 (50%), Positives = 14/20 (70%), Gaps = 0/20 (0%)
Query 1 MLHQTAWHNNTLWLKLFATD 20
+LH AWHNNTL L + ++
Sbjct 206 LLHSVAWHNNTLGLPNYCSE 225
> xla:379401 uqcrc1, MGC53748; ubiquinol-cytochrome c reductase
core protein I (EC:1.10.2.2); K00414 ubiquinol-cytochrome
c reductase core subunit 1 [EC:1.10.2.2]
Length=478
Score = 29.6 bits (65), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 25/47 (53%), Gaps = 4/47 (8%)
Query 12 LWLKLFATDIRRVAE----KMFSKPPTVVAYGDICSVPHYEEIRAAL 54
L ++ A D ++V+E ++ K P V G I +P Y IR+A+
Sbjct 427 LNARIDAVDAKKVSEICSKYLYDKCPAVAGVGPIEQIPDYNRIRSAM 473
Lambda K H
0.323 0.134 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2064920148
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40