bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1027_orf1
Length=226
Score E
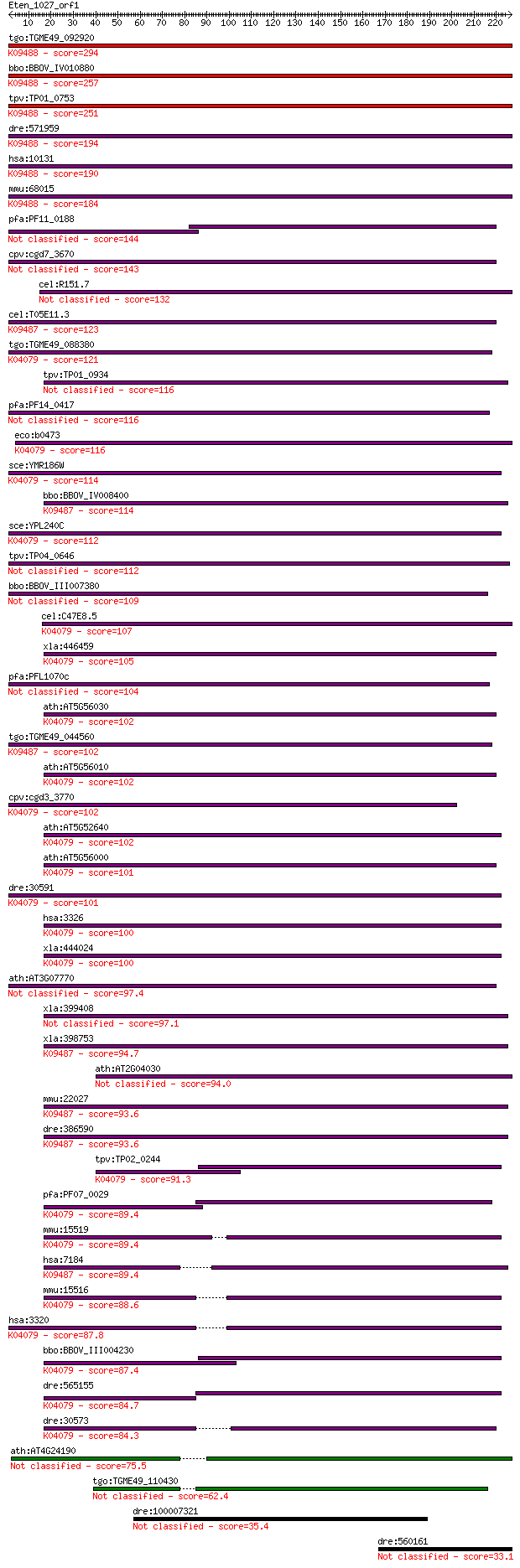
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_092920 heat shock protein 90, putative ; K09488 TNF... 294 1e-79
bbo:BBOV_IV010880 23.m05763; heat shock protein 75; K09488 TNF... 257 3e-68
tpv:TP01_0753 heat shock protein 75; K09488 TNF receptor-assoc... 251 2e-66
dre:571959 trap1, fc85a11, wu:fc85a11; TNF receptor-associated... 194 2e-49
hsa:10131 TRAP1, HSP75, HSP90L; TNF receptor-associated protei... 190 3e-48
mmu:68015 Trap1, 2410002K23Rik, HSP75; TNF receptor-associated... 184 2e-46
pfa:PF11_0188 heat shock protein 90, putative 144 2e-34
cpv:cgd7_3670 heat shock protein 90 (Hsp90), signal peptide pl... 143 4e-34
cel:R151.7 hypothetical protein 132 9e-31
cel:T05E11.3 hypothetical protein; K09487 heat shock protein 9... 123 5e-28
tgo:TGME49_088380 heat shock protein 90 ; K04079 molecular cha... 121 2e-27
tpv:TP01_0934 heat shock protein 90 116 6e-26
pfa:PF14_0417 HSP90 116 7e-26
eco:b0473 htpG, ECK0467, JW0462; molecular chaperone HSP90 fam... 116 8e-26
sce:YMR186W HSC82, HSP90; Cytoplasmic chaperone of the Hsp90 f... 114 2e-25
bbo:BBOV_IV008400 23.m06066; heat shock protein 90; K09487 hea... 114 4e-25
sce:YPL240C HSP82, HSP90; Hsp82p; K04079 molecular chaperone HtpG 112 8e-25
tpv:TP04_0646 heat shock protein 90 112 1e-24
bbo:BBOV_III007380 17.m07646; heat shock protein 90 109 1e-23
cel:C47E8.5 daf-21; abnormal DAuer Formation family member (da... 107 5e-23
xla:446459 hsp90ab1, hsp90b, hsp90beta; heat shock protein 90k... 105 2e-22
pfa:PFL1070c endoplasmin homolog precursor, putative 104 2e-22
ath:AT5G56030 HSP81-2 (HEAT SHOCK PROTEIN 81-2); ATP binding; ... 102 8e-22
tgo:TGME49_044560 heat shock protein 90, putative (EC:2.7.13.3... 102 9e-22
ath:AT5G56010 HSP81-3; ATP binding / unfolded protein binding;... 102 1e-21
cpv:cgd3_3770 Hsp90 ; K04079 molecular chaperone HtpG 102 1e-21
ath:AT5G52640 ATHSP90.1 (HEAT SHOCK PROTEIN 90.1); ATP binding... 102 1e-21
ath:AT5G56000 heat shock protein 81-4 (HSP81-4); K04079 molecu... 101 2e-21
dre:30591 hsp90a.1, fb17b01, hsp90, hsp90a, hsp90alpha, wu:fb1... 101 2e-21
hsa:3326 HSP90AB1, D6S182, FLJ26984, HSP90-BETA, HSP90B, HSPC2... 100 3e-21
xla:444024 hsp90aa1.1, MGC82579, hsp86, hsp89, hsp90, hsp90a, ... 100 5e-21
ath:AT3G07770 ATP binding 97.4 4e-20
xla:399408 hsp90b1, ecgp, gp96, grp94, tra1; heat shock protei... 97.1 4e-20
xla:398753 hypothetical protein MGC68448; K09487 heat shock pr... 94.7 2e-19
ath:AT2G04030 CR88; CR88; ATP binding 94.0 4e-19
mmu:22027 Hsp90b1, ERp99, GRP94, TA-3, Targ2, Tra-1, Tra1, end... 93.6 5e-19
dre:386590 hsp90b1, GP96, GRP94, fb61d09, tra-1, tra1, wu:fb61... 93.6 6e-19
tpv:TP02_0244 heat shock protein 90; K04079 molecular chaperon... 91.3 2e-18
pfa:PF07_0029 heat shock protein 86; K04079 molecular chaperon... 89.4 9e-18
mmu:15519 Hsp90aa1, 86kDa, 89kDa, AL024080, AL024147, Hsp86-1,... 89.4 1e-17
hsa:7184 HSP90B1, ECGP, GP96, GRP94, TRA1; heat shock protein ... 89.4 1e-17
mmu:15516 Hsp90ab1, 90kDa, AL022974, C81438, Hsp84, Hsp84-1, H... 88.6 2e-17
hsa:3320 HSP90AA1, FLJ31884, HSP86, HSP89A, HSP90A, HSP90N, HS... 87.8 3e-17
bbo:BBOV_III004230 17.m07381; hsp90 protein; K04079 molecular ... 87.4 4e-17
dre:565155 hsp90a.2, cb820, hsp90a2; heat shock protein 90-alp... 84.7 2e-16
dre:30573 hsp90ab1, hsp90b, hsp90beta, wu:fa29f01, wu:fa91e11,... 84.3 3e-16
ath:AT4G24190 SHD; SHD (SHEPHERD); ATP binding / unfolded prot... 75.5 1e-13
tgo:TGME49_110430 heat shock protein 90, putative (EC:3.2.1.3 ... 62.4 1e-09
dre:100007321 fe05a04; wu:fe05a04 35.4 0.17
dre:560161 nucleotide-binding oligomerization domain containin... 33.1 0.86
> tgo:TGME49_092920 heat shock protein 90, putative ; K09488 TNF
receptor-associated protein 1
Length=861
Score = 294 bits (753), Expect = 1e-79, Method: Compositional matrix adjust.
Identities = 142/231 (61%), Positives = 181/231 (78%), Gaps = 6/231 (2%)
Query 1 RVEVFTRSRE-GGKVLRWCSDGFGTFSLSSAPADSLSSAS---GTKIVCHLKQDCLEFAN 56
RV+V+TR+ E G K W SDG G F++ + S A GTKIVCHLK+DCLEF+N
Sbjct 291 RVDVYTRAHEEGAKAYLWSSDGAGEFNVKELSEEEASEAGLKRGTKIVCHLKKDCLEFSN 350
Query 57 PHRVKECAKKFSSFVNFPVYM-EENGKEVQISSQEALWLKPSPTPEEHKQFFRHLTNQSW 115
H VKECA KFSSFVNFP+Y+ EE+GK +I+SQ+ALWL+ S T +EH+Q FR+L+N SW
Sbjct 351 IHHVKECATKFSSFVNFPIYVKEEDGKNTKITSQQALWLQTSATEDEHRQCFRYLSNTSW 410
Query 116 GEPFYSIMFSADAPLCIRSVLYFPADPPNRLFQTGPLESGVSLLARRVLVKKSATDLLPK 175
G+P YSIMF DAPL I++V Y P DPP+RLFQ E GVSL +RRVLVKKSATD++PK
Sbjct 411 GDPMYSIMFRTDAPLSIKAVFYIPEDPPSRLFQPAN-EVGVSLHSRRVLVKKSATDIIPK 469
Query 176 WLWFLRGIVDCEDLPLNVSREHMQDSVLQRKLSTVIVKRILKFLNDQVKSD 226
WL F++G++DC+D+PLNVSRE+MQ+SVL KLS ++V+RILKFL+DQ KS+
Sbjct 470 WLGFVKGVIDCDDIPLNVSRENMQNSVLIEKLSQILVRRILKFLDDQSKSN 520
> bbo:BBOV_IV010880 23.m05763; heat shock protein 75; K09488 TNF
receptor-associated protein 1
Length=623
Score = 257 bits (656), Expect = 3e-68, Method: Compositional matrix adjust.
Identities = 118/229 (51%), Positives = 165/229 (72%), Gaps = 8/229 (3%)
Query 1 RVEVFTRS---REGGKVLRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANP 57
+VEVFTRS +G K W SDG G+F++ GTKIVCHL+ DC+ FAN
Sbjct 152 KVEVFTRSYDPEKGEKGYHWVSDGTGSFTIREVQ----ELPRGTKIVCHLRDDCVVFANT 207
Query 58 HRVKECAKKFSSFVNFPVYMEENGKEVQISSQEALWLKPSPTPEEHKQFFRHLTNQSWGE 117
VK+ A+KFS+FVNFP+Y++E E +I++Q+ LW++ + + EEH +FFR+L N SWGE
Sbjct 208 ANVKKVAEKFSAFVNFPLYIQEKDAETEITTQKPLWIEKNASDEEHTKFFRYLNNTSWGE 267
Query 118 PFYSIMFSADAPLCIRSVLYFPADPPNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWL 177
P Y I+F +D PL I+S+LY P D P++LF E GVSL +R++L++KSAT ++PKWL
Sbjct 268 PMYKILFHSDVPLSIKSLLYIPEDAPSKLFHNTN-EVGVSLHSRKILIQKSATAIIPKWL 326
Query 178 WFLRGIVDCEDLPLNVSREHMQDSVLQRKLSTVIVKRILKFLNDQVKSD 226
+F++G++DCED+PLNVSRE +QDS L +KL +VKRILKF ++Q K D
Sbjct 327 FFIKGVIDCEDIPLNVSRELVQDSQLVKKLGNTVVKRILKFFHEQSKKD 375
> tpv:TP01_0753 heat shock protein 75; K09488 TNF receptor-associated
protein 1
Length=724
Score = 251 bits (640), Expect = 2e-66, Method: Compositional matrix adjust.
Identities = 120/231 (51%), Positives = 168/231 (72%), Gaps = 10/231 (4%)
Query 1 RVEVFTRS---REGGKVLRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANP 57
RVEVFTRS + K W SDG G+F+L D+L GTKI+C+LK D L F N
Sbjct 213 RVEVFTRSYDPEKDPKGYHWVSDGTGSFTLKEV--DNL--PRGTKIICYLKDDSLLFCNS 268
Query 58 HRVKECAKKFSSFVNFPVYMEENGKEVQISSQEALWLKPSPTPEEHKQFFRHLTNQSWGE 117
+ VK+ A+KFSSF+NFP++++E K+V+I++Q+ LW++ +PEEH +F+R L N SWGE
Sbjct 269 NNVKKVAEKFSSFINFPLFLQEKDKDVEITTQKPLWVEKKSSPEEHTKFYRFLCNTSWGE 328
Query 118 PFYSIMFSADAPLCIRSVLYFPADPPNRLFQTGPLESGVSLLAR--RVLVKKSATDLLPK 175
P Y++ F +D+PL I+S+ Y P D PNR+FQ E GVSL +R +VL+KKSA +++PK
Sbjct 329 PMYTLNFHSDSPLSIKSLFYIPEDAPNRMFQASN-ELGVSLYSRYLKVLIKKSAENIIPK 387
Query 176 WLWFLRGIVDCEDLPLNVSREHMQDSVLQRKLSTVIVKRILKFLNDQVKSD 226
WL+F++G++DCED+PLNVSRE+MQD+ L KLS +V ++LKFL Q KSD
Sbjct 388 WLFFVKGVIDCEDMPLNVSRENMQDNQLMAKLSNTVVTKLLKFLQQQSKSD 438
> dre:571959 trap1, fc85a11, wu:fc85a11; TNF receptor-associated
protein 1; K09488 TNF receptor-associated protein 1
Length=719
Score = 194 bits (492), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 99/229 (43%), Positives = 155/229 (67%), Gaps = 12/229 (5%)
Query 1 RVEVFTRSREG-GKVLRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHR 59
+VEV+++S E +W SDG G F ++ A GTKIV HLK DC EF++ R
Sbjct 229 KVEVYSQSAEADAPGYKWSSDGSGVFEVAEASG----VRQGTKIVLHLKDDCKEFSSEDR 284
Query 60 VKECAKKFSSFVNFPVYMEENGKEVQISSQEALWL-KPSPTPE-EHKQFFRHLTNQSWGE 117
VKE K+S+FV+FP+++ NG+ ++++ +ALW+ +P E +H++F+R++ Q++ +
Sbjct 285 VKEVVTKYSNFVSFPIFL--NGR--RLNTLQALWMMEPKDISEWQHEEFYRYVA-QAYDK 339
Query 118 PFYSIMFSADAPLCIRSVLYFPADPPNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWL 177
P Y++ + ADAPL IRS+ Y P P+ + + S V+L +R++L++ ATD+LPKWL
Sbjct 340 PRYTLHYRADAPLNIRSIFYVPEMKPSMFDVSREMGSSVALYSRKILIQTKATDILPKWL 399
Query 178 WFLRGIVDCEDLPLNVSREHMQDSVLQRKLSTVIVKRILKFLNDQVKSD 226
FLRG+VD ED+PLN+SRE +Q+S L RKL V+ +R+++FL DQ K D
Sbjct 400 RFLRGVVDSEDIPLNLSRELLQESALIRKLRDVLQQRVIRFLLDQSKKD 448
> hsa:10131 TRAP1, HSP75, HSP90L; TNF receptor-associated protein
1; K09488 TNF receptor-associated protein 1
Length=704
Score = 190 bits (483), Expect = 3e-48, Method: Compositional matrix adjust.
Identities = 98/229 (42%), Positives = 153/229 (66%), Gaps = 12/229 (5%)
Query 1 RVEVFTRSREGGKV-LRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHR 59
RVEV++RS G + +W SDG G F ++ A +GTKI+ HLK DC EF++ R
Sbjct 214 RVEVYSRSAAPGSLGYQWLSDGSGVFEIAEASG----VRTGTKIIIHLKSDCKEFSSEAR 269
Query 60 VKECAKKFSSFVNFPVYMEENGKEVQISSQEALWL-KPSPTPE-EHKQFFRHLTNQSWGE 117
V++ K+S+FV+FP+Y+ NG+ ++++ +A+W+ P E +H++F+R++ Q+ +
Sbjct 270 VRDVVTKYSNFVSFPLYL--NGR--RMNTLQAIWMMDPKDVREWQHEEFYRYVA-QAHDK 324
Query 118 PFYSIMFSADAPLCIRSVLYFPADPPNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWL 177
P Y++ + DAPL IRS+ Y P P+ + L S V+L +R+VL++ ATD+LPKWL
Sbjct 325 PRYTLHYKTDAPLNIRSIFYVPDMKPSMFDVSRELGSSVALYSRKVLIQTKATDILPKWL 384
Query 178 WFLRGIVDCEDLPLNVSREHMQDSVLQRKLSTVIVKRILKFLNDQVKSD 226
F+RG+VD ED+PLN+SRE +Q+S L RKL V+ +R++KF DQ K D
Sbjct 385 RFIRGVVDSEDIPLNLSRELLQESALIRKLRDVLQQRLIKFFIDQSKKD 433
> mmu:68015 Trap1, 2410002K23Rik, HSP75; TNF receptor-associated
protein 1; K09488 TNF receptor-associated protein 1
Length=706
Score = 184 bits (468), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 96/229 (41%), Positives = 150/229 (65%), Gaps = 12/229 (5%)
Query 1 RVEVFTRSRE-GGKVLRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHR 59
+VEV++RS +W SDG G F ++ A GTKI+ HLK DC +FA+ R
Sbjct 216 KVEVYSRSAAPESPGYQWLSDGSGVFEIAEASG----VRPGTKIIIHLKSDCKDFASESR 271
Query 60 VKECAKKFSSFVNFPVYMEENGKEVQISSQEALWL-KPSPTPE-EHKQFFRHLTNQSWGE 117
V++ K+S+FV+FP+Y+ NGK +I++ +A+W+ P E +H++F+R++ Q++ +
Sbjct 272 VQDVVTKYSNFVSFPLYL--NGK--RINTLQAIWMMDPKDISEFQHEEFYRYIA-QAYDK 326
Query 118 PFYSIMFSADAPLCIRSVLYFPADPPNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWL 177
P +++ + DAPL IRS+ Y P P+ + L S V+L +R+VL++ A D+LPKWL
Sbjct 327 PRFTLHYKTDAPLNIRSIFYVPEMKPSMFDVSRELGSSVALYSRKVLIQTKAADILPKWL 386
Query 178 WFLRGIVDCEDLPLNVSREHMQDSVLQRKLSTVIVKRILKFLNDQVKSD 226
F+RG+VD ED+PLN+SRE +Q+S L RKL V+ +R++KF DQ K D
Sbjct 387 RFIRGVVDSEDIPLNLSRELLQESALIRKLRDVLQQRLIKFFIDQSKKD 435
> pfa:PF11_0188 heat shock protein 90, putative
Length=930
Score = 144 bits (363), Expect = 2e-34, Method: Composition-based stats.
Identities = 68/143 (47%), Positives = 102/143 (71%), Gaps = 6/143 (4%)
Query 82 KEVQISSQEALWLKPSPTPEEHKQFFRHLT-NQSWGEP----FYSIMFSADAPLCIRSVL 136
+E+ ++ Q+ LW K + T EEH+ FF L N+S+ E Y++++ DAPL I+SV
Sbjct 448 EEILVNKQKPLWCKDNVTEEEHRHFFHFLNKNKSYNEDNKSYLYNMLYKTDAPLSIKSVF 507
Query 137 YFPADPPNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSRE 196
Y P + P+RLFQ + +SL ++VLVKK+A +++PKWL+F++G++DCED+PLN+SRE
Sbjct 508 YIPEEAPSRLFQQSN-DIEISLYCKKVLVKKNADNIIPKWLYFVKGVIDCEDMPLNISRE 566
Query 197 HMQDSVLQRKLSTVIVKRILKFL 219
+MQDS L KLS V+V +ILK L
Sbjct 567 NMQDSSLINKLSRVVVSKILKTL 589
Score = 75.5 bits (184), Expect = 2e-13, Method: Composition-based stats.
Identities = 41/87 (47%), Positives = 55/87 (63%), Gaps = 6/87 (6%)
Query 1 RVEVFTRSRE--GGKVLRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPH 58
+VEVFTRS + K W S G GTF+L D++ GTKI+CHLK C EF+N
Sbjct 235 KVEVFTRSYDNNSSKGYHWVSYGNGTFTLKEV--DNIPK--GTKIICHLKDSCKEFSNIQ 290
Query 59 RVKECAKKFSSFVNFPVYMEENGKEVQ 85
V++ +KFSSF+NFPVY+ + K +Q
Sbjct 291 NVQKIVEKFSSFINFPVYVLKKKKILQ 317
> cpv:cgd7_3670 heat shock protein 90 (Hsp90), signal peptide
plus ER retention motif
Length=787
Score = 143 bits (361), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 79/240 (32%), Positives = 138/240 (57%), Gaps = 24/240 (10%)
Query 1 RVEVFTRSREGGKVLRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRV 60
RV V +++ E + + W S G+F +S P + + GT IV LK+D EF N ++
Sbjct 216 RVTVISKNNEDKQYV-WESSADGSFRVSLDPRGN-TIKRGTTIVLSLKEDATEFMNFSKL 273
Query 61 KECAKKFSSFVNFPVYM------------------EENGKEVQISSQEALWLKPSP--TP 100
K+ ++S F+NFP+Y+ E G+ Q++ ++A+WL+P T
Sbjct 274 KDLVLRYSQFINFPIYIYNPEGVNKSEKDESGEKKESKGRWEQVNVEKAIWLRPREEITK 333
Query 101 EEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPNRLFQTGPLESG-VSLL 159
EE+ F++ +T+ + EP + FSA+ + +S+L+ P+ PP +F T +SG +
Sbjct 334 EEYNGFYKSITHD-YSEPLRYLHFSAEGEIEFKSLLFIPSHPPFDMFDTYMGKSGNIKFY 392
Query 160 ARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVLQRKLSTVIVKRILKFL 219
RRVL+ DLLPK+L F++G+VD +D+ LNV+REH+Q S + + +S +V+++L+ +
Sbjct 393 VRRVLITDHIEDLLPKYLNFIKGVVDSDDISLNVAREHVQQSRIIKVISKKMVRKVLEMI 452
> cel:R151.7 hypothetical protein
Length=479
Score = 132 bits (332), Expect = 9e-31, Method: Compositional matrix adjust.
Identities = 79/221 (35%), Positives = 124/221 (56%), Gaps = 17/221 (7%)
Query 15 LRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLK-QDCLEFANPHRVKECAKKFSSFVNF 73
L+W +G ++ + A++ +GTKI LK D +A R+KE K+S FV+
Sbjct 184 LQWTWNGDNSYEI----AETSGLQTGTKIEIRLKVGDSATYAEEDRIKEVINKYSYFVSA 239
Query 74 PVYMEENGKEVQISSQEALWLKPSP--TPEEHKQFFRHLT-----NQSWGEPFYSIMFSA 126
P+ + NG+ V ++ A+W + E H+ FF+ L + + P Y+I F
Sbjct 240 PILV--NGERV--NNLNAIWTMQAREVNKEMHETFFKQLVKTQGKQEMYTRPQYTIHFQT 295
Query 127 DAPLCIRSVLYFPADPPNRL-FQTGPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVD 185
D P+ +RSV+Y P N+L F G+SL ARRVL+K A +L+P +L F+ G+VD
Sbjct 296 DTPVSLRSVIYIPQTQFNQLTFMAQQTMCGLSLYARRVLIKPDAQELIPNYLRFVIGVVD 355
Query 186 CEDLPLNVSREHMQDSVLQRKLSTVIVKRILKFLNDQVKSD 226
ED+PLN+SRE +Q++ + RKL +I +IL L ++K D
Sbjct 356 SEDIPLNLSREMLQNNPVLRKLRKIITDKILGSLQSEMKKD 396
> cel:T05E11.3 hypothetical protein; K09487 heat shock protein
90kDa beta
Length=760
Score = 123 bits (309), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 79/259 (30%), Positives = 133/259 (51%), Gaps = 44/259 (16%)
Query 1 RVEVFTRSREGGKVLRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRV 60
RV V T++ + + + W SD +F++S P + + GT+I +LK++ +F P +
Sbjct 196 RVVVTTKNNDDDQYI-WESDS-ASFTISKDPRGN-TLKRGTQITLYLKEEAADFLEPDTL 252
Query 61 KECAKKFSSFVNFPVYM------------------EENGKEV------------------ 84
K K+S F+NF +++ E+G
Sbjct 253 KNLVHKYSQFINFDIFLWQSKTEMVEEAVEEEPATTEDGAVEEEKEEKKTKKVEKTTWDW 312
Query 85 -QISSQEALWL-KPSPTPE-EHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPAD 141
++++ + +W+ KP+ E E+KQF++ +T S EP + FSA+ + RS+LY P
Sbjct 313 EKVNNVKPIWMRKPNQVEEDEYKQFYKSITKDS-EEPLSHVHFSAEGEVSFRSILYVPKK 371
Query 142 PPNRLFQT-GPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQD 200
PN +FQ G + + L RRV + D+LPK+L F+RGIVD +DLPLNVSRE++Q
Sbjct 372 SPNDMFQNYGKVIENIKLYVRRVFITDDFADMLPKYLSFIRGIVDSDDLPLNVSRENLQQ 431
Query 201 SVLQRKLSTVIVKRILKFL 219
L + + +V+++L L
Sbjct 432 HKLLKVIKKKLVRKVLDML 450
> tgo:TGME49_088380 heat shock protein 90 ; K04079 molecular chaperone
HtpG
Length=708
Score = 121 bits (303), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 81/268 (30%), Positives = 128/268 (47%), Gaps = 57/268 (21%)
Query 1 RVEVFTRSREGGKVLRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRV 60
+V V +R + ++ W S G+F++S A + GT+I+ H+K+D E+ R+
Sbjct 134 KVTVVSRHNDD-EMYVWESSAGGSFTVSKAEGQFENIVRGTRIILHMKEDQTEYLEDRRL 192
Query 61 KECAKKFSSFVNFPVYM---EENGKEV--------------------------------- 84
K+ KK S F++FP+ + + KE+
Sbjct 193 KDLVKKHSEFISFPIELAVEKSVDKEITESEDEEKPAEDAEEKKEEGEEEKKEEGAEKKK 252
Query 85 -------------QISSQEALWL-KPSP-TPEEHKQFFRHLTNQSWGEPFYSIMFSADAP 129
Q++ Q+ LW+ KP T EE+ F++ LTN W +P FS +
Sbjct 253 KTKKVKEVVVEYEQLNKQKPLWMRKPEDVTWEEYCAFYKSLTN-DWEDPLAVKHFSVEGQ 311
Query 130 LCIRSVLYFPADPPNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDL 189
L +++L+ P P LF+T + V L RRV + DL+P+WL F+RG+VD EDL
Sbjct 312 LEFKALLFLPKRAPFDLFETRKKRNNVRLYVRRVFIMDDCEDLIPEWLNFVRGVVDSEDL 371
Query 190 PLNVSREHMQDSVLQRKLSTVIVKRILK 217
PLN+SRE +Q Q K+ VI K ++K
Sbjct 372 PLNISRESLQ----QNKILKVIKKNLVK 395
> tpv:TP01_0934 heat shock protein 90
Length=1009
Score = 116 bits (290), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 61/216 (28%), Positives = 118/216 (54%), Gaps = 10/216 (4%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPVY 76
W S ++ L +SL GT I L++D ++ ++ KK+S FV +P+
Sbjct 225 WRSSAANSYELYEDTDNSLGD-HGTLITLELREDATDYLKTDVLENLVKKYSQFVKYPIQ 283
Query 77 MEENGKE------VQISSQEALWL--KPSPTPEEHKQFFRHLTNQSWGEPFYSIMFSADA 128
+ + K+ V+++ + +W K + T +E+ +F++ ++ ++ EP + F+A+
Sbjct 284 LYKKLKDKQELGWVKVNETQQIWTRNKNTITEQEYNEFYKTISGKT-DEPLAHVHFTAEG 342
Query 129 PLCIRSVLYFPADPPNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCED 188
+ +++LY P+ PP F + + V L +RRVLV + D +P++L+ + G+VD +
Sbjct 343 DVDFKALLYIPSSPPAMYFSSESVGHNVKLYSRRVLVSQEMRDFIPRYLFSVYGVVDSDS 402
Query 189 LPLNVSREHMQDSVLQRKLSTVIVKRILKFLNDQVK 224
PLNVSRE++Q S L + + +V+ +L L D +K
Sbjct 403 FPLNVSREYLQQSKLVKLIGKKVVRTVLDTLYDVMK 438
> pfa:PF14_0417 HSP90
Length=927
Score = 116 bits (290), Expect = 7e-26, Method: Composition-based stats.
Identities = 77/254 (30%), Positives = 133/254 (52%), Gaps = 46/254 (18%)
Query 1 RVEVFTRSREGGKVLRWCSDGFGTFSLSSAPA-----DSLSSASGTKIVCHLKQDCLEFA 55
RVEV+T+ + ++ RW SD G+FS++ D + SGTKI+ HLK++C E+
Sbjct 266 RVEVYTKKED--QIYRWSSDLKGSFSVNEIKKYDQEYDDIK-GSGTKIILHLKEECDEYL 322
Query 56 NPHRVKECAKKFSSFVNFPVYM----------------EENGKEV-------------QI 86
+++KE KK+S F+ FP+ + ++G ++ +I
Sbjct 323 EDYKLKELIKKYSEFIKFPIEIWSEKIDYERVPDDSVSLKDGDKMKMKTITKRYHEWEKI 382
Query 87 SSQEALWLK--PSPTPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPN 144
+ Q +W + S T ++ F+++ T +++ +P + F+ + + S+LY P P
Sbjct 383 NVQLPIWKQDEKSLTENDYYSFYKN-TFKAYDDPLAYVHFNVEGQISFNSILYIPGSLPW 441
Query 145 RLFQTGPLES--GVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSV 202
L + E G+ L +RV + ++ +P+WL FLRGIVD E+LPLNV RE +Q S
Sbjct 442 ELSKNMFDEESRGIRLYVKRVFINDKFSESIPRWLTFLRGIVDSENLPLNVGREILQKS- 500
Query 203 LQRKLSTVIVKRIL 216
K+ ++I KRI+
Sbjct 501 ---KMLSIINKRIV 511
> eco:b0473 htpG, ECK0467, JW0462; molecular chaperone HSP90 family;
K04079 molecular chaperone HtpG
Length=624
Score = 116 bits (290), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 74/237 (31%), Positives = 121/237 (51%), Gaps = 20/237 (8%)
Query 4 VFTRSREGGKV----LRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHR 59
V R+R G+ + W S G G +++ AD GT+I HL++ EF + R
Sbjct 137 VTVRTRAAGEKPENGVFWESAGEGEYTV----ADITKEDRGTEITLHLREGEDEFLDDWR 192
Query 60 VKECAKKFSSFVNFPVYMEE----NGKEV----QISSQEALWL--KPSPTPEEHKQFFRH 109
V+ K+S + PV +E+ +G+ V +I+ +ALW K T EE+K+F++H
Sbjct 193 VRSIISKYSDHIALPVEIEKREEKDGETVISWEKINKAQALWTRNKSEITDEEYKEFYKH 252
Query 110 LTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPNRLFQTGPLESGVSLLARRVLVKKSA 169
+ + + +P + S+LY P+ P ++ + G+ L +RV + A
Sbjct 253 IAH-DFNDPLTWSHNRVEGKQEYTSLLYIPSQAPWDMWNRDH-KHGLKLYVQRVFIMDDA 310
Query 170 TDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVLQRKLSTVIVKRILKFLNDQVKSD 226
+P +L F+RG++D DLPLNVSRE +QDS + R L + KR+L+ L K D
Sbjct 311 EQFMPNYLRFVRGLIDSSDLPLNVSREILQDSTVTRNLRNALTKRVLQMLEKLAKDD 367
> sce:YMR186W HSC82, HSP90; Cytoplasmic chaperone of the Hsp90
family, redundant in function and nearly identical with Hsp82p,
and together they are essential; expressed constitutively
at 10-fold higher basal levels than HSP82 and induced 2-3
fold by heat shock; K04079 molecular chaperone HtpG
Length=705
Score = 114 bits (286), Expect = 2e-25, Method: Composition-based stats.
Identities = 76/273 (27%), Positives = 133/273 (48%), Gaps = 55/273 (20%)
Query 1 RVEVFTRSREGGKVLRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRV 60
RV+V +++ E + + W S+ G+F+++ + GT + LK D LE+ R+
Sbjct 133 RVQVISKNNEDEQYI-WESNAGGSFTVTLDEVNE-RIGRGTVLRLFLKDDQLEYLEEKRI 190
Query 61 KECAKKFSSFVNFPVYM---EENGKEV--------------------------------- 84
KE K+ S FV +P+ + +E KEV
Sbjct 191 KEVIKRHSEFVAYPIQLLVTKEVEKEVPIPEEEKKDEEKKDEDDKKPKLEEVDEEEEEKK 250
Query 85 --------------QISSQEALWLK-PSP-TPEEHKQFFRHLTNQSWGEPFYSIMFSADA 128
+++ + LW + PS T EE+ F++ ++N W +P Y FS +
Sbjct 251 PKTKKVKEEVQELEELNKTKPLWTRNPSDITQEEYNAFYKSISN-DWEDPLYVKHFSVEG 309
Query 129 PLCIRSVLYFPADPPNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCED 188
L R++L+ P P LF++ ++ + L RRV + A DL+P+WL F++G+VD ED
Sbjct 310 QLEFRAILFIPKRAPFDLFESKKKKNNIKLYVRRVFITDEAEDLIPEWLSFVKGVVDSED 369
Query 189 LPLNVSREHMQDSVLQRKLSTVIVKRILKFLND 221
LPLN+SRE +Q + + + + IVK++++ N+
Sbjct 370 LPLNLSREMLQQNKIMKVIRKNIVKKLIEAFNE 402
> bbo:BBOV_IV008400 23.m06066; heat shock protein 90; K09487 heat
shock protein 90kDa beta
Length=795
Score = 114 bits (284), Expect = 4e-25, Method: Composition-based stats.
Identities = 67/216 (31%), Positives = 112/216 (51%), Gaps = 9/216 (4%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPVY 76
W S + L P + GT+I L++D E+ +++E KK S FV FP+Y
Sbjct 233 WKSSADTKYELYEDPKGNTLGEHGTQITLFLREDATEYLEIDKIEELIKKHSQFVRFPIY 292
Query 77 M------EENGKEVQISSQEALWL--KPSPTPEEHKQFFRHLTNQSWGEPFYSIMFSADA 128
+ E K ++ + +W K T +E+ F++ ++ S +P I F A+
Sbjct 293 VLKAVKGEPEAKWQHVNDIKPIWARDKSEITEDEYTAFYKAISG-STSKPLAHIHFVAEG 351
Query 129 PLCIRSVLYFPADPPNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCED 188
+ R++LY P P + F + V + ARRVLV S + LP++L+ L G+VD ++
Sbjct 352 DIDFRALLYIPERPKSAYFDNEDVGHHVKIYARRVLVSDSLPNFLPRYLYSLHGVVDSDN 411
Query 189 LPLNVSREHMQDSVLQRKLSTVIVKRILKFLNDQVK 224
PLNVSREH+Q S + + ++ IV+ +L L + +K
Sbjct 412 FPLNVSREHLQQSKMIKIIAKKIVRSVLTTLENLMK 447
> sce:YPL240C HSP82, HSP90; Hsp82p; K04079 molecular chaperone
HtpG
Length=709
Score = 112 bits (281), Expect = 8e-25, Method: Composition-based stats.
Identities = 76/277 (27%), Positives = 133/277 (48%), Gaps = 59/277 (21%)
Query 1 RVEVFTRSREGGKVLRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRV 60
RV+V ++S + + + W S+ G+F+++ + GT + LK D LE+ R+
Sbjct 133 RVQVISKSNDDEQYI-WESNAGGSFTVTLDEVNE-RIGRGTILRLFLKDDQLEYLEEKRI 190
Query 61 KECAKKFSSFVNFPVYM---EENGKEV--------------------------------- 84
KE K+ S FV +P+ + +E KEV
Sbjct 191 KEVIKRHSEFVAYPIQLVVTKEVEKEVPIPEEEKKDEEKKDEEKKDEDDKKPKLEEVDEE 250
Query 85 ------------------QISSQEALWLK-PSP-TPEEHKQFFRHLTNQSWGEPFYSIMF 124
+++ + LW + PS T EE+ F++ ++N W +P Y F
Sbjct 251 EEKKPKTKKVKEEVQEIEELNKTKPLWTRNPSDITQEEYNAFYKSISN-DWEDPLYVKHF 309
Query 125 SADAPLCIRSVLYFPADPPNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIV 184
S + L R++L+ P P LF++ ++ + L RRV + A DL+P+WL F++G+V
Sbjct 310 SVEGQLEFRAILFIPKRAPFDLFESKKKKNNIKLYVRRVFITDEAEDLIPEWLSFVKGVV 369
Query 185 DCEDLPLNVSREHMQDSVLQRKLSTVIVKRILKFLND 221
D EDLPLN+SRE +Q + + + + IVK++++ N+
Sbjct 370 DSEDLPLNLSREMLQQNKIMKVIRKNIVKKLIEAFNE 406
> tpv:TP04_0646 heat shock protein 90
Length=913
Score = 112 bits (279), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 77/263 (29%), Positives = 131/263 (49%), Gaps = 44/263 (16%)
Query 1 RVEVFTRS--REGGKVLRWCSDGFGTFSLSSAPADSLSSA---SGTKIVCHLKQDCLEFA 55
+VEVF+R+ +E G V RW SD GT+++ L+ SGT+IV HLK +C ++
Sbjct 259 KVEVFSRAYGQEAGPVYRWKSDSNGTYTIGRVENQELNDKFMKSGTRIVLHLKPECDDYL 318
Query 56 NPHRVKECAKKFSSFVNFPVYM-----------------------------EENGKEVQI 86
+++KE +K+S F+ FP+ + + N EV +
Sbjct 319 EDYKLKELLRKYSEFIRFPIQVWVERIEYERVPDDATIVDGKPGRYKTVTKKRNEWEV-V 377
Query 87 SSQEALWLKPSPT--PEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPN 144
++Q +W + T PE++ F++ T +++ +P I F + + +L+ P P
Sbjct 378 NTQLPIWRRDQSTIKPEDYISFYKS-TFKAYEDPLSYIHFKVEGQVEFTCLLFVPGTLPW 436
Query 145 RLFQT--GPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSV 202
L + G+ L +RV + ++ +P+WL F+RG+VD ++L LNV RE++Q S
Sbjct 437 ELSRNMFDDESRGIRLYVKRVFINDKFSESIPRWLTFVRGVVDSDELSLNVGREYLQRS- 495
Query 203 LQRKLSTVIVKRILKFLNDQVKS 225
K TVI KRI D +K+
Sbjct 496 ---KALTVINKRIASKAIDMLKN 515
> bbo:BBOV_III007380 17.m07646; heat shock protein 90
Length=795
Score = 109 bits (272), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 71/252 (28%), Positives = 124/252 (49%), Gaps = 42/252 (16%)
Query 1 RVEVFTRS--REGGKVLRWCSDGFGTFSLSSAPADSLSSA---SGTKIVCHLKQDCLEFA 55
+VEV++R+ E G + RW S+ GTFS++ D L GT+IV H+K +C ++
Sbjct 250 KVEVYSRAYGHEDGGIFRWKSETNGTFSVAQVNDDELQKGFMKCGTRIVLHIKPECDDYL 309
Query 56 NPHRVKECAKKFSSFVNFPVYM------------EENGKEVQ----------------IS 87
+++KE +K+S FV FP+ + E E + ++
Sbjct 310 EDYKIKELLRKYSEFVRFPIQVWVEKVEYERVPDESTAVEGKPGRYKTISKKRHEWEHVN 369
Query 88 SQEALWLKPSP--TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPNR 145
+Q +W + PE++ F++ T +++ +P I F + + +L+ P P
Sbjct 370 TQIPIWRRDQADVKPEDYVSFYKS-TFKAYDDPLSYIHFKVEGQVEFSCLLFVPGSLPWE 428
Query 146 LFQT--GPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVL 203
L + G+ L +RV + ++ +P+WL F+RG+VD ++L LNV RE++Q S
Sbjct 429 LSRNMFDDQSRGIRLYVKRVFINDKFSEAVPRWLTFVRGVVDSDELALNVGREYLQRS-- 486
Query 204 QRKLSTVIVKRI 215
K T+I KRI
Sbjct 487 --KALTIINKRI 496
> cel:C47E8.5 daf-21; abnormal DAuer Formation family member (daf-21);
K04079 molecular chaperone HtpG
Length=702
Score = 107 bits (266), Expect = 5e-23, Method: Composition-based stats.
Identities = 68/258 (26%), Positives = 126/258 (48%), Gaps = 52/258 (20%)
Query 16 RWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPV 75
+W S G+F + P + GTKIV H+K+D ++F ++KE KK S F+ +P+
Sbjct 149 QWESSAGGSFVVR--PFNDPEVTRGTKIVMHIKEDQIDFLEERKIKEIVKKHSQFIGYPI 206
Query 76 YM-------------------EENGKEVQISS-------------------------QEA 91
+ +E KE ++ + +
Sbjct 207 KLVVEKEREKEVEDEEAVEAKDEEKKEGEVENVADDADKKKTKKIKEKYFEDEELNKTKP 266
Query 92 LWLKPSP---TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPNRLFQ 148
+W + +P + EE+ +F++ L+N W + FS + L R++L+ P P LF+
Sbjct 267 IWTR-NPDDISNEEYAEFYKSLSN-DWEDHLAVKHFSVEGQLEFRALLFVPQRAPFDLFE 324
Query 149 TGPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVLQRKLS 208
++ + L RRV + ++ +L+P++L F++G+VD EDLPLN+SRE +Q S + + +
Sbjct 325 NKKSKNSIKLYVRRVFIMENCEELMPEYLNFIKGVVDSEDLPLNISREMLQQSKILKVIR 384
Query 209 TVIVKRILKFLNDQVKSD 226
+VK+ ++ + D+V D
Sbjct 385 KNLVKKCMELI-DEVAED 401
> xla:446459 hsp90ab1, hsp90b, hsp90beta; heat shock protein 90kDa
alpha (cytosolic), class B member 1; K04079 molecular chaperone
HtpG
Length=722
Score = 105 bits (261), Expect = 2e-22, Method: Composition-based stats.
Identities = 73/262 (27%), Positives = 121/262 (46%), Gaps = 63/262 (24%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPV- 75
W S G+F++ + + GTK++ HLK+D E+ RVKE KK S F+ +P+
Sbjct 157 WESSAGGSFTVKVDTGEPI--GRGTKVILHLKEDQTEYLEEKRVKETVKKHSQFIGYPIT 214
Query 76 -YME-ENGKEV------------------------------------------------- 84
Y+E E KE+
Sbjct 215 LYLEKEREKEISDDEAEEEKEEKKEEEGENDKPKIEDVGSDEEEEGKDKKKKTKKIKEKY 274
Query 85 ----QISSQEALWLKPSP---TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLY 137
+++ + +W + +P T EE+ +F++ LTN W + FS + L R++L+
Sbjct 275 IDQEELNKTKPIWTR-NPDDITQEEYGEFYKSLTN-DWEDHLAVKHFSVEGQLEFRALLF 332
Query 138 FPADPPNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREH 197
P P LF+ ++ + L RRV + S +L+P++L F+RG+VD EDLPLN+SRE
Sbjct 333 IPRRAPFDLFENKKKKNNIKLYVRRVFIMDSCDELIPEYLNFVRGVVDSEDLPLNISREM 392
Query 198 MQDSVLQRKLSTVIVKRILKFL 219
+Q S + + + IVK+ L+
Sbjct 393 LQQSKILKVIRKNIVKKCLELF 414
> pfa:PFL1070c endoplasmin homolog precursor, putative
Length=821
Score = 104 bits (260), Expect = 2e-22, Method: Composition-based stats.
Identities = 70/262 (26%), Positives = 126/262 (48%), Gaps = 50/262 (19%)
Query 1 RVEVFTRSREGGKVLRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRV 60
+V V+T++ + + + W S F++ P + + GT+I HLK+D N ++
Sbjct 203 KVIVYTKNNDDEQYI-WESTADAKFTIYKDPRGA-TLKRGTRISLHLKEDATNLLNDKKL 260
Query 61 KECAKKFSSFVNFPVYM--------------------EENGKEVQI-------------- 86
+ K+S F+ FP+Y+ + N V++
Sbjct 261 MDLISKYSQFIQFPIYLLHENVYTEEVLADIAKDMVNDPNYDSVKVEETDDPNKKTRTVE 320
Query 87 ---------SSQEALWLKPSPTP---EEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRS 134
+ Q +WL+ SP E++KQFF L+ + +P Y I F A+ + +
Sbjct 321 KKVKKWTLMNEQRPIWLR-SPKELKDEDYKQFFSVLSGYN-DQPLYHIHFFAEGEIEFKC 378
Query 135 VLYFPADPPNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVS 194
++Y P+ P+ Q ++ + L RRVLV + LP+++ F++G+VD +DLPLNVS
Sbjct 379 LIYIPSKAPSMNDQLYSKQNSLKLYVRRVLVADEFVEFLPRYMSFVKGVVDSDDLPLNVS 438
Query 195 REHMQDSVLQRKLSTVIVKRIL 216
RE +Q + + + +S IV++IL
Sbjct 439 REQLQQNKILKAVSKRIVRKIL 460
> ath:AT5G56030 HSP81-2 (HEAT SHOCK PROTEIN 81-2); ATP binding;
K04079 molecular chaperone HtpG
Length=699
Score = 102 bits (255), Expect = 8e-22, Method: Composition-based stats.
Identities = 66/249 (26%), Positives = 120/249 (48%), Gaps = 48/249 (19%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPV- 75
W S G+F+++ + + GTK+V +LK+D LE+ R+K+ KK S F+++P+
Sbjct 149 WESQAGGSFTVTRDTSGE-TLGRGTKMVLYLKEDQLEYLEERRLKDLVKKHSEFISYPIS 207
Query 76 -YMEENGKEVQ------------------------------------------ISSQEAL 92
++E+ ++ ++ Q+ +
Sbjct 208 LWIEKTIEKEISDDEEEEEKKDEEGKVEEVDEEKEKEEKKKKKIKEVSHEWDLVNKQKPI 267
Query 93 WL-KPSP-TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPNRLFQTG 150
W+ KP EE+ F++ L+N W E FS + L +++L+ P P LF T
Sbjct 268 WMRKPEEINKEEYAAFYKSLSN-DWEEHLAVKHFSVEGQLEFKAILFVPKRAPFDLFDTK 326
Query 151 PLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVLQRKLSTV 210
+ + L RRV + + D++P++L F++GIVD EDLPLN+SRE +Q + + + +
Sbjct 327 KKPNNIKLYVRRVFIMDNCEDIIPEYLGFVKGIVDSEDLPLNISRETLQQNKILKVIRKN 386
Query 211 IVKRILKFL 219
+VK+ L+
Sbjct 387 LVKKCLELF 395
> tgo:TGME49_044560 heat shock protein 90, putative (EC:2.7.13.3);
K09487 heat shock protein 90kDa beta
Length=847
Score = 102 bits (255), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 73/275 (26%), Positives = 130/275 (47%), Gaps = 65/275 (23%)
Query 1 RVEVFTRSREGGKVLRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRV 60
+V V +++ E + + W S F ++ P + + GT + HLK+D EF N ++
Sbjct 215 KVTVVSKNVEDDQHI-WESSADAKFHVAKDPRGN-TLGRGTCVTLHLKEDATEFLNEWKL 272
Query 61 KECAKKFSSFVNFPVYMEEN---GKEVQISSQE--------------------------- 90
K+ +FS F+++P+Y+ + +EV I +E
Sbjct 273 KDLTTRFSQFMSYPIYVRTSRTVTEEVPIEDEEAETKDEDKEKDEDKDKDDVEVTEGDKD 332
Query 91 -------------------------ALWLKPS--PTPEEHKQFFRHLTNQSWGEPFYSIM 123
A+WL+P +E+ +F++ ++ + W +P I
Sbjct 333 EKKDKPKTKKVEKKKDEWEQVNTQKAIWLRPKEEIEEKEYNEFYKSVS-KDWSDPLAHIH 391
Query 124 FSADAPLCIRSVLYFPADPPNRLFQTG-PLESGVSLLARRVLVKKSATDLLPKWLWFLRG 182
FSA+ + +++LY P P+ ++ ++ V + RRVLV DLLPK+L F++G
Sbjct 392 FSAEGEVEFKALLYIPKRAPSDIYSNYFDKQTSVKVYVRRVLVADQFDDLLPKYLHFVKG 451
Query 183 IVDCEDLPLNVSREHMQDSVLQRKLSTVIVKRILK 217
+VD +DLPLNVSRE +Q Q K+ VI K++++
Sbjct 452 VVDSDDLPLNVSREQLQ----QHKILNVISKKLVR 482
> ath:AT5G56010 HSP81-3; ATP binding / unfolded protein binding;
K04079 molecular chaperone HtpG
Length=699
Score = 102 bits (254), Expect = 1e-21, Method: Composition-based stats.
Identities = 65/249 (26%), Positives = 120/249 (48%), Gaps = 48/249 (19%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPV- 75
W S G+F+++ + + GTK+V +LK+D +E+ R+K+ KK S F+++P+
Sbjct 149 WESQAGGSFTVTRDTSGE-ALGRGTKMVLYLKEDQMEYIEERRLKDLVKKHSEFISYPIS 207
Query 76 -YMEENGKEVQ------------------------------------------ISSQEAL 92
++E+ ++ ++ Q+ +
Sbjct 208 LWIEKTIEKEISDDEEEEEKKDEEGKVEEVDEEKEKEEKKKKKIKEVSHEWDLVNKQKPI 267
Query 93 WL-KPSP-TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPNRLFQTG 150
W+ KP EE+ F++ L+N W E FS + L +++L+ P P LF T
Sbjct 268 WMRKPEEINKEEYAAFYKSLSN-DWEEHLAVKHFSVEGQLEFKAILFVPKRAPFDLFDTK 326
Query 151 PLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVLQRKLSTV 210
+ + L RRV + + D++P++L F++GIVD EDLPLN+SRE +Q + + + +
Sbjct 327 KKPNNIKLYVRRVFIMDNCEDIIPEYLGFVKGIVDSEDLPLNISRETLQQNKILKVIRKN 386
Query 211 IVKRILKFL 219
+VK+ L+
Sbjct 387 LVKKCLELF 395
> cpv:cgd3_3770 Hsp90 ; K04079 molecular chaperone HtpG
Length=711
Score = 102 bits (254), Expect = 1e-21, Method: Composition-based stats.
Identities = 67/250 (26%), Positives = 118/250 (47%), Gaps = 52/250 (20%)
Query 1 RVEVFTRSREGGKVLRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRV 60
+V V T+ G + W S G+F++++ +D+ GT+I+ HLK+D L++ +
Sbjct 144 KVTVITK-HNGDEQYIWESSAGGSFTITNDTSDN-KLQRGTRIILHLKEDQLDYLEERTL 201
Query 61 KECAKKFSSFVNFPVYME-ENGKEVQISSQEA---------------------------- 91
++ KK S F++FP+ + E E +I+ +
Sbjct 202 RDLVKKHSEFISFPIELSVEKTTEKEITDSDVDEEEEKKEGEDGEDAPKIEEVKEKEPKK 261
Query 92 ------------------LWL-KPSP-TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLC 131
+W+ KP T EE+ F++ ++N W +P FS + L
Sbjct 262 KKITEVTQSWDLLNKNKPIWMRKPEEVTFEEYSSFYKSISN-DWEDPLAVKHFSVEGQLE 320
Query 132 IRSVLYFPADPPNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPL 191
+++L+ P P LF+T + + L RRV + +L+P++L F+RG+VD EDLPL
Sbjct 321 FKAILFIPRRAPFDLFETRKKRNNIKLYVRRVFIMDDCEELIPEFLGFVRGVVDSEDLPL 380
Query 192 NVSREHMQDS 201
N+SRE +Q +
Sbjct 381 NISRESLQQN 390
> ath:AT5G52640 ATHSP90.1 (HEAT SHOCK PROTEIN 90.1); ATP binding
/ unfolded protein binding; K04079 molecular chaperone HtpG
Length=705
Score = 102 bits (253), Expect = 1e-21, Method: Composition-based stats.
Identities = 68/252 (26%), Positives = 118/252 (46%), Gaps = 49/252 (19%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPV- 75
W S G+F+++ D GTKI LK D LE+ R+K+ KK S F+++P+
Sbjct 154 WESQAGGSFTVTR-DVDGEPLGRGTKITLFLKDDQLEYLEERRLKDLVKKHSEFISYPIY 212
Query 76 -YMEENGKEVQ-------------------------------------------ISSQEA 91
+ E+ ++ I+ Q+
Sbjct 213 LWTEKTTEKEISDDEDEDEPKKENEGEVEEVDEEKEKDGKKKKKIKEVSHEWELINKQKP 272
Query 92 LWL-KPSP-TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPNRLFQT 149
+WL KP T EE+ F++ LTN W + FS + L +++L+ P P LF T
Sbjct 273 IWLRKPEEITKEEYAAFYKSLTN-DWEDHLAVKHFSVEGQLEFKAILFVPKRAPFDLFDT 331
Query 150 GPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVLQRKLST 209
+ + L RRV + + +L+P++L F++G+VD +DLPLN+SRE +Q + + + +
Sbjct 332 RKKLNNIKLYVRRVFIMDNCEELIPEYLSFVKGVVDSDDLPLNISRETLQQNKILKVIRK 391
Query 210 VIVKRILKFLND 221
+VK+ ++ N+
Sbjct 392 NLVKKCIEMFNE 403
> ath:AT5G56000 heat shock protein 81-4 (HSP81-4); K04079 molecular
chaperone HtpG
Length=699
Score = 101 bits (251), Expect = 2e-21, Method: Composition-based stats.
Identities = 64/249 (25%), Positives = 119/249 (47%), Gaps = 48/249 (19%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPV- 75
W S G+F+++ + + GTK++ +LK+D +E+ R+K+ KK S F+++P+
Sbjct 149 WESQAGGSFTVTRDTSGE-ALGRGTKMILYLKEDQMEYIEERRLKDLVKKHSEFISYPIS 207
Query 76 -YMEENGKEVQ------------------------------------------ISSQEAL 92
++E+ ++ ++ Q+ +
Sbjct 208 LWIEKTIEKEISDDEEEEEKKDEEGKVEEIDEEKEKEEKKKKKIKEVTHEWDLVNKQKPI 267
Query 93 WL-KPSP-TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPNRLFQTG 150
W+ KP EE+ F++ L+N W E FS + L +++L+ P P LF T
Sbjct 268 WMRKPEEINKEEYAAFYKSLSN-DWEEHLAVKHFSVEGQLEFKAILFVPKRAPFDLFDTK 326
Query 151 PLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVLQRKLSTV 210
+ + L RRV + + D++P +L F++GIVD EDLPLN+SRE +Q + + + +
Sbjct 327 KKPNNIKLYVRRVFIMDNCEDIIPDYLGFVKGIVDSEDLPLNISRETLQQNKILKVIRKN 386
Query 211 IVKRILKFL 219
+VK+ L+
Sbjct 387 LVKKCLELF 395
> dre:30591 hsp90a.1, fb17b01, hsp90, hsp90a, hsp90alpha, wu:fb17b01,
zgc:86652; heat shock protein 90-alpha 1; K04079 molecular
chaperone HtpG
Length=726
Score = 101 bits (251), Expect = 2e-21, Method: Composition-based stats.
Identities = 75/280 (26%), Positives = 127/280 (45%), Gaps = 63/280 (22%)
Query 1 RVEVFTRSREGGKVLRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRV 60
+V V T+ + + + W S G+F++ +S+ GTK++ HLK+D E+ R+
Sbjct 145 KVTVITKHNDDEQYI-WESAAGGSFTVKPDFGESI--GRGTKVILHLKEDQSEYVEEKRI 201
Query 61 KECAKKFSSFVNFPV--YME-ENGKEVQ-------------------------------- 85
KE KK S F+ +P+ Y+E + KEV
Sbjct 202 KEVVKKHSQFIGYPITLYIEKQREKEVDLEEGEKQEEEEVAAGEDKDKPKIEDLGADEDE 261
Query 86 -----------------ISSQEALWLKP----SP---TPEEHKQFFRHLTNQSWGEPFYS 121
I +QE KP +P T EE+ +F++ L+N W +
Sbjct 262 DSKDGKNKRKKKVKEKYIDAQELNKTKPIWTRNPDDITNEEYGEFYKSLSN-DWEDHLAV 320
Query 122 IMFSADAPLCIRSVLYFPADPPNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWLWFLR 181
FS + L R++L+ P LF+ + + L RRV + + +L+P++L F++
Sbjct 321 KHFSVEGQLEFRALLFVPRRAAFDLFENKKKRNNIKLYVRRVFIMDNCEELIPEYLNFIK 380
Query 182 GIVDCEDLPLNVSREHMQDSVLQRKLSTVIVKRILKFLND 221
G+VD EDLPLN+SRE +Q S + + + +VK+ L +
Sbjct 381 GVVDSEDLPLNISREMLQQSKILKVIRKNLVKKCLDLFTE 420
> hsa:3326 HSP90AB1, D6S182, FLJ26984, HSP90-BETA, HSP90B, HSPC2,
HSPCB; heat shock protein 90kDa alpha (cytosolic), class
B member 1; K04079 molecular chaperone HtpG
Length=724
Score = 100 bits (250), Expect = 3e-21, Method: Composition-based stats.
Identities = 74/265 (27%), Positives = 120/265 (45%), Gaps = 63/265 (23%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPV- 75
W S G+F++ + + + GTK++ HLK+D E+ RVKE KK S F+ +P+
Sbjct 157 WESSAGGSFTVRADHGEPI--GRGTKVILHLKEDQTEYLEERRVKEVVKKHSQFIGYPIT 214
Query 76 -YME-ENGKEVQ------------------------------------------------ 85
Y+E E KE+
Sbjct 215 LYLEKEREKEISDDEAEEEKGEKEEEDKDDEEKPKIEDVGSDEEDDSGKDKKKKTKKIKE 274
Query 86 --ISSQEALWLKP--SPTPEEHKQ-----FFRHLTNQSWGEPFYSIMFSADAPLCIRSVL 136
I +E KP + P++ Q F++ LTN W + FS + L R++L
Sbjct 275 KYIDQEELNKTKPIWTRNPDDITQEEYGEFYKSLTN-DWEDHLAVKHFSVEGQLEFRALL 333
Query 137 YFPADPPNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSRE 196
+ P P LF+ ++ + L RRV + S +L+P++L F+RG+VD EDLPLN+SRE
Sbjct 334 FIPRRAPFDLFENKKKKNNIKLYVRRVFIMDSCDELIPEYLNFIRGVVDSEDLPLNISRE 393
Query 197 HMQDSVLQRKLSTVIVKRILKFLND 221
+Q S + + + IVK+ L+ ++
Sbjct 394 MLQQSKILKVIRKNIVKKCLELFSE 418
> xla:444024 hsp90aa1.1, MGC82579, hsp86, hsp89, hsp90, hsp90a,
hspc1, hspca, hspn, lap2; heat shock protein 90kDa alpha (cytosolic),
class A member 1, gene 1; K04079 molecular chaperone
HtpG
Length=729
Score = 100 bits (248), Expect = 5e-21, Method: Composition-based stats.
Identities = 69/264 (26%), Positives = 119/264 (45%), Gaps = 62/264 (23%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPVY 76
W S G+F++ ++ L GTK++ HLK+D E+ R+KE KK S F+ +P+
Sbjct 163 WESSAGGSFTVRVDNSEPL--GRGTKVILHLKEDQSEYFEEKRIKEIVKKHSQFIGYPIT 220
Query 77 M---EENGKEVQ------------------------------------------------ 85
+ +E KE+
Sbjct 221 LFVEKERDKEISDDEAEEEKEEKKDEPKDEEKPEIEDVGSDDEEDKKEGDKKKKKKIKEK 280
Query 86 -ISSQEALWLKP--SPTPEE-----HKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLY 137
I +E KP + P++ + +F++ LTN W + FS + L R++L+
Sbjct 281 YIDQEELNKTKPIWTRNPDDITNEEYGEFYKSLTN-DWEDHLAVKHFSVEGQLEFRALLF 339
Query 138 FPADPPNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREH 197
P P LF+ ++ + L RRV + + +L+P++L F+RG+VD EDLPLN+SRE
Sbjct 340 VPRRAPFDLFENRKKKNNIKLYVRRVFIMDNCDELIPEYLNFMRGVVDSEDLPLNISREM 399
Query 198 MQDSVLQRKLSTVIVKRILKFLND 221
+Q S + + + +VK+ L+ +
Sbjct 400 LQQSKILKVIRKNLVKKCLELFTE 423
> ath:AT3G07770 ATP binding
Length=799
Score = 97.4 bits (241), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 70/262 (26%), Positives = 120/262 (45%), Gaps = 44/262 (16%)
Query 1 RVEVFTRSREGGKVLRWCSDG-FGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHR 59
RV V T+S + K W + +F++ GT+I HLKQ+ FA+P R
Sbjct 228 RVIVSTKSPKSDKQYVWEGEANSSSFTIQEDTDPQSLIPRGTRITLHLKQEAKNFADPER 287
Query 60 VKECAKKFSSFVNFPVYM-EENG--KEVQI------------------------------ 86
+++ K +S FV+FP+Y +E G KEV++
Sbjct 288 IQKLVKNYSQFVSFPIYTWQEKGYTKEVEVEDDPTETKKDDQDDQTEKKKKTKKVVERYW 347
Query 87 -----SSQEALWLK--PSPTPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFP 139
+ + +WL+ T E+ +F+R N+ + +P S F+ + + RS+LY P
Sbjct 348 DWELTNETQPIWLRNPKEVTTAEYNEFYRKAFNE-YLDPLASSHFTTEGEVEFRSILYVP 406
Query 140 -ADPPNRLFQTGPLESGVSLLARRVLVKKS-ATDLLPKWLWFLRGIVDCEDLPLNVSREH 197
P + + L +RV + +L P++L F++G+VD DLPLNVSRE
Sbjct 407 PVSPSGKDDIVNQKTKNIRLYVKRVFISDDFDGELFPRYLSFVKGVVDSHDLPLNVSREI 466
Query 198 MQDSVLQRKLSTVIVKRILKFL 219
+Q+S + R + +V++ +
Sbjct 467 LQESRIVRIMKKRLVRKAFDMI 488
> xla:399408 hsp90b1, ecgp, gp96, grp94, tra1; heat shock protein
90kDa beta (Grp94), member 1
Length=804
Score = 97.1 bits (240), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 71/257 (27%), Positives = 118/257 (45%), Gaps = 56/257 (21%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPVY 76
W SD F D+L G+ I LK++ ++ VK +K+S F+NFP+Y
Sbjct 223 WESDSNEFFVTDDPRGDTL--GRGSTITLVLKEEATDYLELETVKNLVRKYSQFINFPIY 280
Query 77 MEENGKEV---------------------------------QISSQEALW-------LKP 96
+ + E ++ +W +KP
Sbjct 281 VWSSKTETVEEPLDEEEAKEKDEDTDEEAAVEEEDEEKKPKTKKVEKTVWDWELMNDIKP 340
Query 97 ---SPTPE----EHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPNRLF-Q 148
P+ E E+K F++ + +S EP I F+A+ + +S+L+ P+ P LF +
Sbjct 341 IWQRPSKEIEEDEYKAFYKSFSKES-DEPMAYIHFTAEGEVTFKSILFIPSSAPRGLFDE 399
Query 149 TGPLESG-VSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVLQRKL 207
G +S + L RRV + D++PK+L F++G+VD +DLPLNVSRE++ Q KL
Sbjct 400 YGSKKSDFIKLFVRRVFITDDFHDMMPKYLNFVKGVVDSDDLPLNVSRENLH----QHKL 455
Query 208 STVIVKRILKFLNDQVK 224
VI K++++ D +K
Sbjct 456 LKVIRKKLVRKTLDMIK 472
> xla:398753 hypothetical protein MGC68448; K09487 heat shock
protein 90kDa beta
Length=805
Score = 94.7 bits (234), Expect = 2e-19, Method: Composition-based stats.
Identities = 69/261 (26%), Positives = 115/261 (44%), Gaps = 64/261 (24%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPVY 76
W SD F D+L GT I LK++ ++ +K +K+S F+NFP+Y
Sbjct 223 WESDSNEFFVTDDPRGDTL--GRGTTITLVLKEEATDYLELETIKNLVRKYSQFMNFPIY 280
Query 77 MEENGKEV---------------------------------------------QISSQEA 91
+ + E ++ +
Sbjct 281 VWSSKTETVEEPLDEEEAKEKDEETDEEAAVEEEDEEKKPKTKKVEKTIWDWELMNDIKP 340
Query 92 LWLKPSPTPEE--HKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPNRLFQT 149
+W +P+ EE +K F++ + +S +P I F+A+ + +S+L+ P+ P LF
Sbjct 341 IWQRPAKEIEEDEYKAFYKSFSKES-DDPMAHIHFTAEGEVTFKSILFIPSTAPRGLFD- 398
Query 150 GPLESG------VSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVL 203
E G + L RRV + D++PK+L F++G+VD +DLPLNVSRE +Q
Sbjct 399 ---EYGSKKIDFIKLFVRRVFITDDFNDMMPKYLNFVKGVVDSDDLPLNVSRETLQ---- 451
Query 204 QRKLSTVIVKRILKFLNDQVK 224
Q KL VI K++++ D +K
Sbjct 452 QHKLLKVIRKKLVRKTLDMIK 472
> ath:AT2G04030 CR88; CR88; ATP binding
Length=780
Score = 94.0 bits (232), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 60/224 (26%), Positives = 107/224 (47%), Gaps = 38/224 (16%)
Query 40 GTKIVCHLKQDC-LEFANPHRVKECAKKFSSFVNFPVYM-EENGKEVQISSQEA------ 91
GT+I +L++D EFA R+K K +S FV FP+Y +E + +++ E
Sbjct 250 GTQITLYLREDDKYEFAESTRIKNLVKNYSQFVGFPIYTWQEKSRTIEVEEDEPVKEGEE 309
Query 92 -------------------------LWLKPSPTPE--EHKQFFRHLTNQSWGEPFYSIMF 124
LW++ S E E+ +F++ N+ + +P F
Sbjct 310 GEPKKKKTTKTEKYWDWELANETKPLWMRNSKEVEKGEYNEFYKKAFNE-FLDPLAHTHF 368
Query 125 SADAPLCIRSVLYFPA-DPPNRLFQTGPLESGVSLLARRVLVKKS-ATDLLPKWLWFLRG 182
+ + + RS+LY P P N T P + L +RV + +L P++L F++G
Sbjct 369 TTEGEVEFRSILYIPGMGPLNNEDVTNPKTKNIRLYVKRVFISDDFDGELFPRYLSFVKG 428
Query 183 IVDCEDLPLNVSREHMQDSVLQRKLSTVIVKRILKFLNDQVKSD 226
+VD +DLPLNVSRE +Q+S + R + ++++ + + +S+
Sbjct 429 VVDSDDLPLNVSREILQESRIVRIMRKRLIRKTFDMIQEISESE 472
> mmu:22027 Hsp90b1, ERp99, GRP94, TA-3, Targ2, Tra-1, Tra1, endoplasmin,
gp96; heat shock protein 90, beta (Grp94), member
1; K09487 heat shock protein 90kDa beta
Length=802
Score = 93.6 bits (231), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 71/258 (27%), Positives = 120/258 (46%), Gaps = 57/258 (22%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPVY 76
W SD FS+ + P + + GT I LK++ ++ +K +K+S F+NFP+Y
Sbjct 223 WESDS-NEFSVIADPRGN-TLGRGTTITLVLKEEASDYLELDTIKNLVRKYSQFINFPIY 280
Query 77 MEENGKEV----------------------------------QISSQEALW-------LK 95
+ + E ++ +W +K
Sbjct 281 VWSSKTETVEEPLEEDEAAKEEKEESDDEAAVEEEEEEKKPKTKKVEKTVWDWELMNDIK 340
Query 96 P---SPTPE----EHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPNRLF- 147
P P+ E E+K F++ + +S +P I F+A+ + +S+L+ P P LF
Sbjct 341 PIWQRPSKEVEEDEYKAFYKSFSKES-DDPMAYIHFTAEGEVTFKSILFVPTSAPRGLFD 399
Query 148 QTGPLESG-VSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVLQRK 206
+ G +S + L RRV + D++PK+L F++G+VD +DLPLNVSRE +Q Q K
Sbjct 400 EYGSKKSDYIKLYVRRVFITDDFHDMMPKYLNFVKGVVDSDDLPLNVSRETLQ----QHK 455
Query 207 LSTVIVKRILKFLNDQVK 224
L VI K++++ D +K
Sbjct 456 LLKVIRKKLVRKTLDMIK 473
> dre:386590 hsp90b1, GP96, GRP94, fb61d09, tra-1, tra1, wu:fb61d09,
wu:fq25g01; heat shock protein 90, beta (grp94), member
1; K09487 heat shock protein 90kDa beta
Length=793
Score = 93.6 bits (231), Expect = 6e-19, Method: Composition-based stats.
Identities = 69/259 (26%), Positives = 115/259 (44%), Gaps = 59/259 (22%)
Query 17 WCSDGFGTFSLSSAP-ADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPV 75
W SD FS+ P D+L GT I +K++ ++ +K KK+S F+NFP+
Sbjct 223 WESDS-NQFSVIEDPRGDTL--GRGTTITLVMKEEASDYLELETIKNLVKKYSQFINFPI 279
Query 76 YMEENGKEV----------------------------------------------QISSQ 89
Y+ + E ++
Sbjct 280 YVWSSKTETVEEPIEDEAEAEKEEATEDEAEVEEEEEDKDKPKTKKVEKTVWDWELMNDI 339
Query 90 EALWLKPSPTPEE--HKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPNRLF 147
+ +W +P+ EE + F++ + + EP I F+A+ + +S+L+ PA P LF
Sbjct 340 KPIWQRPAKEVEEDEYTAFYKTFSRDT-DEPLSHIHFTAEGEVTFKSILFVPASAPRGLF 398
Query 148 QTGPLESG--VSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVLQR 205
+ + L RRV + D++PK+L F++G+VD +DLPLNVSRE +Q Q
Sbjct 399 DEYGTKKNDFIKLFVRRVFITDDFHDMMPKYLNFIKGVVDSDDLPLNVSRETLQ----QH 454
Query 206 KLSTVIVKRILKFLNDQVK 224
KL VI K++++ D +K
Sbjct 455 KLLKVIRKKLVRKTLDMIK 473
> tpv:TP02_0244 heat shock protein 90; K04079 molecular chaperone
HtpG
Length=721
Score = 91.3 bits (225), Expect = 2e-18, Method: Composition-based stats.
Identities = 45/138 (32%), Positives = 84/138 (60%), Gaps = 3/138 (2%)
Query 86 ISSQEALWLK-PSP-TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPP 143
++ Q+ +W++ PS T EE+ F+++LTN W + FS + L +++L+ P P
Sbjct 285 LNKQKPIWMRLPSEVTNEEYAAFYKNLTN-DWEDHLAVKHFSVEGQLEFKALLFVPRRAP 343
Query 144 NRLFQTGPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVL 203
+F++ ++ + L RRV + +L+P+WL F++G+VD EDLPLN+SRE +Q + +
Sbjct 344 FDMFESRKKKNNIKLYVRRVFIMDDCEELIPEWLSFVKGVVDSEDLPLNISRETLQQNKI 403
Query 204 QRKLSTVIVKRILKFLND 221
+ + +VK+ L+ N+
Sbjct 404 LKVIRKNLVKKCLELFNE 421
Score = 49.3 bits (116), Expect = 1e-05, Method: Composition-based stats.
Identities = 25/66 (37%), Positives = 39/66 (59%), Gaps = 2/66 (3%)
Query 40 GTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPVYME-ENGKEVQISSQEALWLKPSP 98
GT+++ HLK+D E+ R+KE KK S F++FP+ + E +E +++ EA L
Sbjct 177 GTRLILHLKEDQTEYLEERRLKELVKKHSEFISFPISLSVEKTQETEVTDDEA-ELDEDK 235
Query 99 TPEEHK 104
PEE K
Sbjct 236 KPEEEK 241
> pfa:PF07_0029 heat shock protein 86; K04079 molecular chaperone
HtpG
Length=745
Score = 89.4 bits (220), Expect = 9e-18, Method: Composition-based stats.
Identities = 47/135 (34%), Positives = 78/135 (57%), Gaps = 7/135 (5%)
Query 85 QISSQEALWL-KPSP-TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADP 142
+++ Q+ LW+ KP T EE+ F++ LTN W + FS + L +++L+ P
Sbjct 308 ELNKQKPLWMRKPEEVTNEEYASFYKSLTN-DWEDHLAVKHFSVEGQLEFKALLFIPKRA 366
Query 143 PNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSV 202
P +F+ + + L RRV + +++P+WL F++G+VD EDLPLN+SRE +Q
Sbjct 367 PFDMFENRKKRNNIKLYVRRVFIMDDCEEIIPEWLNFVKGVVDSEDLPLNISRESLQ--- 423
Query 203 LQRKLSTVIVKRILK 217
Q K+ VI K ++K
Sbjct 424 -QNKILKVIKKNLIK 437
Score = 50.1 bits (118), Expect = 6e-06, Method: Composition-based stats.
Identities = 26/74 (35%), Positives = 42/74 (56%), Gaps = 4/74 (5%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPVY 76
W S G+F+++ + GTKI+ HLK+D LE+ R+K+ KK S F++FP+
Sbjct 148 WESAAGGSFTVTKDETNE-KLGRGTKIILHLKEDQLEYLEEKRIKDLVKKHSEFISFPIK 206
Query 77 M---EENGKEVQIS 87
+ +N KE+ S
Sbjct 207 LYCERQNEKEITAS 220
> mmu:15519 Hsp90aa1, 86kDa, 89kDa, AL024080, AL024147, Hsp86-1,
Hsp89, Hsp90, Hspca, hsp4; heat shock protein 90, alpha (cytosolic),
class A member 1; K04079 molecular chaperone HtpG
Length=733
Score = 89.4 bits (220), Expect = 1e-17, Method: Composition-based stats.
Identities = 44/123 (35%), Positives = 73/123 (59%), Gaps = 1/123 (0%)
Query 99 TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPNRLFQTGPLESGVSL 158
T EE+ +F++ LTN W E FS + L R++L+ P P LF+ ++ + L
Sbjct 306 TNEEYGEFYKSLTN-DWEEHLAVKHFSVEGQLEFRALLFVPRRAPFDLFENRKKKNNIKL 364
Query 159 LARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVLQRKLSTVIVKRILKF 218
RRV + + +L+P++L F+RG+VD EDLPLN+SRE +Q S + + + +VK+ L+
Sbjct 365 YVRRVFIMDNCEELIPEYLNFIRGVVDSEDLPLNISREMLQQSKILKVIRKNLVKKCLEL 424
Query 219 LND 221
+
Sbjct 425 FTE 427
Score = 50.1 bits (118), Expect = 7e-06, Method: Composition-based stats.
Identities = 24/76 (31%), Positives = 43/76 (56%), Gaps = 3/76 (3%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPVY 76
W S G+F++ + + + GTK++ HLK+D E+ R+KE KK S F+ +P+
Sbjct 162 WESSAGGSFTVRTDTGEPM--GRGTKVILHLKEDQTEYLEERRIKEIVKKHSQFIGYPIT 219
Query 77 M-EENGKEVQISSQEA 91
+ E ++ ++S EA
Sbjct 220 LFVEKERDKEVSDDEA 235
> hsa:7184 HSP90B1, ECGP, GP96, GRP94, TRA1; heat shock protein
90kDa beta (Grp94), member 1; K09487 heat shock protein 90kDa
beta
Length=803
Score = 89.4 bits (220), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 50/137 (36%), Positives = 81/137 (59%), Gaps = 9/137 (6%)
Query 92 LWLKPSPTPEE--HKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPNRLF-Q 148
+W +PS EE +K F++ + +S +P I F+A+ + +S+L+ P P LF +
Sbjct 342 IWQRPSKEVEEDEYKAFYKSFSKES-DDPMAYIHFTAEGEVTFKSILFVPTSAPRGLFDE 400
Query 149 TGPLESG-VSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVLQRKL 207
G +S + L RRV + D++PK+L F++G+VD +DLPLNVSRE +Q Q KL
Sbjct 401 YGSKKSDYIKLYVRRVFITDDFHDMMPKYLNFVKGVVDSDDLPLNVSRETLQ----QHKL 456
Query 208 STVIVKRILKFLNDQVK 224
VI K++++ D +K
Sbjct 457 LKVIRKKLVRKTLDMIK 473
Score = 39.7 bits (91), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 33/61 (54%), Gaps = 2/61 (3%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPVY 76
W SD FS+ + P + + GT I LK++ ++ +K KK+S F+NFP+Y
Sbjct 223 WESDS-NEFSVIADPRGN-TLGRGTTITLVLKEEASDYLELDTIKNLVKKYSQFINFPIY 280
Query 77 M 77
+
Sbjct 281 V 281
> mmu:15516 Hsp90ab1, 90kDa, AL022974, C81438, Hsp84, Hsp84-1,
Hsp90, Hspcb, MGC115780; heat shock protein 90 alpha (cytosolic),
class B member 1; K04079 molecular chaperone HtpG
Length=724
Score = 88.6 bits (218), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 45/123 (36%), Positives = 74/123 (60%), Gaps = 1/123 (0%)
Query 99 TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPNRLFQTGPLESGVSL 158
T EE+ +F++ LTN W + FS + L R++L+ P P LF+ ++ + L
Sbjct 297 TQEEYGEFYKSLTN-DWEDHLAVKHFSVEGQLEFRALLFIPRRAPFDLFENKKKKNNIKL 355
Query 159 LARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVLQRKLSTVIVKRILKF 218
RRV + S +L+P++L F+RG+VD EDLPLN+SRE +Q S + + + IVK+ L+
Sbjct 356 YVRRVFIMDSCDELIPEYLNFIRGVVDSEDLPLNISREMLQQSKILKVIRKNIVKKCLEL 415
Query 219 LND 221
++
Sbjct 416 FSE 418
Score = 50.8 bits (120), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 41/71 (57%), Gaps = 5/71 (7%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPV- 75
W S G+F++ + + + GTK++ HLK+D E+ RVKE KK S F+ +P+
Sbjct 157 WESSAGGSFTVRADHGEPI--GRGTKVILHLKEDQTEYLEERRVKEVVKKHSQFIGYPIT 214
Query 76 -YME-ENGKEV 84
Y+E E KE+
Sbjct 215 LYLEKEREKEI 225
> hsa:3320 HSP90AA1, FLJ31884, HSP86, HSP89A, HSP90A, HSP90N,
HSPC1, HSPCA, HSPCAL1, HSPCAL4, HSPN, Hsp89, Hsp90, LAP2; heat
shock protein 90kDa alpha (cytosolic), class A member 1;
K04079 molecular chaperone HtpG
Length=854
Score = 87.8 bits (216), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 43/123 (34%), Positives = 73/123 (59%), Gaps = 1/123 (0%)
Query 99 TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPNRLFQTGPLESGVSL 158
T EE+ +F++ LTN W + FS + L R++L+ P P LF+ ++ + L
Sbjct 427 TNEEYGEFYKSLTN-DWEDHLAVKHFSVEGQLEFRALLFVPRRAPFDLFENRKKKNNIKL 485
Query 159 LARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVLQRKLSTVIVKRILKF 218
RRV + + +L+P++L F+RG+VD EDLPLN+SRE +Q S + + + +VK+ L+
Sbjct 486 YVRRVFIMDNCEELIPEYLNFIRGVVDSEDLPLNISREMLQQSKILKVIRKNLVKKCLEL 545
Query 219 LND 221
+
Sbjct 546 FTE 548
Score = 50.8 bits (120), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 47/87 (54%), Gaps = 6/87 (6%)
Query 1 RVEVFTRSREGGKVLRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRV 60
+V V T+ + + W S G+F++ + + + GTK++ HLK+D E+ R+
Sbjct 269 KVTVITKHNDDEQYA-WESSAGGSFTVRTDTGEPM--GRGTKVILHLKEDQTEYLEERRI 325
Query 61 KECAKKFSSFVNFPVYM---EENGKEV 84
KE KK S F+ +P+ + +E KEV
Sbjct 326 KEIVKKHSQFIGYPITLFVEKERDKEV 352
> bbo:BBOV_III004230 17.m07381; hsp90 protein; K04079 molecular
chaperone HtpG
Length=712
Score = 87.4 bits (215), Expect = 4e-17, Method: Composition-based stats.
Identities = 42/138 (30%), Positives = 83/138 (60%), Gaps = 3/138 (2%)
Query 86 ISSQEALWLK-PSP-TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPP 143
++ Q+ +W++ P+ T EE+ F+++L+N W + FS + L +++L+ P P
Sbjct 277 LNKQKPIWMRLPTEVTNEEYASFYKNLSN-DWEDHLAVKHFSVEGQLEFKAILFVPKRAP 335
Query 144 NRLFQTGPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVL 203
+F+ ++ + L RRV + +L+P+WL F++G+VD EDLPLN+SRE +Q + +
Sbjct 336 FDMFENRKKKNNIKLYVRRVFIMDDCDELIPEWLGFVKGVVDSEDLPLNISREVLQQNKI 395
Query 204 QRKLSTVIVKRILKFLND 221
+ + +VK+ L+ ++
Sbjct 396 LKVIRKNLVKKCLELFSE 413
Score = 50.1 bits (118), Expect = 6e-06, Method: Composition-based stats.
Identities = 27/87 (31%), Positives = 46/87 (52%), Gaps = 2/87 (2%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPVY 76
W S+ G F+++ ++ GT+++ HLK D E+ R+KE KK S F++FP+
Sbjct 151 WESNASGHFTVTKDESED-QLKRGTRLILHLKDDQSEYLEERRLKELVKKHSEFISFPIR 209
Query 77 ME-ENGKEVQISSQEALWLKPSPTPEE 102
+ E E +++ EA + PEE
Sbjct 210 LSVEKTTETEVTDDEAEPTEAESKPEE 236
> dre:565155 hsp90a.2, cb820, hsp90a2; heat shock protein 90-alpha
2; K04079 molecular chaperone HtpG
Length=734
Score = 84.7 bits (208), Expect = 2e-16, Method: Composition-based stats.
Identities = 45/140 (32%), Positives = 82/140 (58%), Gaps = 5/140 (3%)
Query 85 QISSQEALWLKPSP---TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPAD 141
+++ + LW + +P T EE+ +F++ LTN W + FS + L R++L+ P
Sbjct 291 ELNKTKPLWTR-NPDDITNEEYGEFYKSLTN-DWEDHLAVKHFSVEGQLEFRALLFVPRR 348
Query 142 PPNRLFQTGPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDS 201
P LF+ ++ + L RRV + + +L+P++L F++G+VD EDLPLN+SRE +Q S
Sbjct 349 APFDLFENKKKKNNIKLYVRRVFIMDNCDELIPEYLNFIKGVVDSEDLPLNISREMLQQS 408
Query 202 VLQRKLSTVIVKRILKFLND 221
+ + + +VK+ L+ +
Sbjct 409 KILKVIRKNLVKKCLELFTE 428
Score = 47.8 bits (112), Expect = 3e-05, Method: Composition-based stats.
Identities = 24/71 (33%), Positives = 40/71 (56%), Gaps = 5/71 (7%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPVY 76
W S G+F++ ++ + GTK++ HLK+D E+ R+KE KK S F+ +P+
Sbjct 160 WESSAGGSFTVKVDNSEPI--GRGTKVILHLKEDQTEYIEERRIKEIVKKHSQFIGYPIT 217
Query 77 M---EENGKEV 84
+ +E KEV
Sbjct 218 LFVEKERDKEV 228
> dre:30573 hsp90ab1, hsp90b, hsp90beta, wu:fa29f01, wu:fa91e11,
wu:fd59e11, wu:gcd22h07; heat shock protein 90, alpha (cytosolic),
class B member 1; K04079 molecular chaperone HtpG
Length=725
Score = 84.3 bits (207), Expect = 3e-16, Method: Composition-based stats.
Identities = 43/119 (36%), Positives = 71/119 (59%), Gaps = 1/119 (0%)
Query 101 EEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADPPNRLFQTGPLESGVSLLA 160
EE+ +F++ LTN W + FS + L R++L+ P P LF+ ++ + L
Sbjct 298 EEYGEFYKSLTN-DWEDHLAVKHFSVEGQLEFRALLFIPRRAPFDLFENKKKKNNIKLYV 356
Query 161 RRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVLQRKLSTVIVKRILKFL 219
RRV + + +L+P++L F+RG+VD EDLPLN+SRE +Q S + + + IVK+ L+
Sbjct 357 RRVFIMDNCEELIPEYLNFIRGVVDSEDLPLNISREMLQQSKILKVIRKNIVKKCLELF 415
Score = 47.0 bits (110), Expect = 5e-05, Method: Composition-based stats.
Identities = 26/71 (36%), Positives = 40/71 (56%), Gaps = 5/71 (7%)
Query 17 WCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPV- 75
W S G+F++ + + GTK++ HLK+D E+ RVKE KK S F+ +P+
Sbjct 156 WESSAGGSFTVKVDHGEPI--GRGTKVILHLKEDQTEYIEEKRVKEVVKKHSQFIGYPIT 213
Query 76 -YME-ENGKEV 84
Y+E E KE+
Sbjct 214 LYVEKERDKEI 224
> ath:AT4G24190 SHD; SHD (SHEPHERD); ATP binding / unfolded protein
binding
Length=823
Score = 75.5 bits (184), Expect = 1e-13, Method: Composition-based stats.
Identities = 45/143 (31%), Positives = 79/143 (55%), Gaps = 7/143 (4%)
Query 90 EALWLKPSP---TPEEHKQFFRHLTNQSWGE-PFYSIMFSADAPLCIRSVLYFPADPPNR 145
+A+WL+ SP T EE+ +F+ L+ E P F+A+ + ++VLY P P+
Sbjct 345 KAIWLR-SPKEVTEEEYTKFYHSLSKDFTDEKPMAWSHFNAEGDVEFKAVLYVPPKAPHD 403
Query 146 LFQT--GPLESGVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVL 203
L+++ ++ + L RRV + +LLPK+L FL+G+VD + LPLNVSRE +Q
Sbjct 404 LYESYYNSNKANLKLYVRRVFISDEFDELLPKYLSFLKGLVDSDTLPLNVSREMLQQHSS 463
Query 204 QRKLSTVIVKRILKFLNDQVKSD 226
+ + ++++ L + + D
Sbjct 464 LKTIKKKLIRKALDMIRKLAEED 486
Score = 40.0 bits (92), Expect = 0.006, Method: Composition-based stats.
Identities = 21/76 (27%), Positives = 41/76 (53%), Gaps = 2/76 (2%)
Query 2 VEVFTRSREGGKVLRWCSDGFGTFSLSSAPADSLSSASGTKIVCHLKQDCLEFANPHRVK 61
+EV ++ + + + W S G F++S + GT+I HL+ + E+ ++K
Sbjct 209 IEVISKHNDDSQYV-WESKANGKFAVSEDTWNE-PLGRGTEIRLHLRDEAGEYLEESKLK 266
Query 62 ECAKKFSSFVNFPVYM 77
E K++S F+NFP+ +
Sbjct 267 ELVKRYSEFINFPISL 282
> tgo:TGME49_110430 heat shock protein 90, putative (EC:3.2.1.3
2.7.13.3)
Length=1100
Score = 62.4 bits (150), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 38/135 (28%), Positives = 71/135 (52%), Gaps = 9/135 (6%)
Query 85 QISSQEALWLKPSP--TPEEHKQFFRHLTNQSWGEPFYSIMFSADAPLCIRSVLYFPADP 142
+++Q +W + T +++ F++ T +++ +P + + + ++L+ P
Sbjct 607 HLNTQPPIWRRDEKLLTDKDYVDFYKS-TFKAYDDPLGFVHMKVEGQVDFNALLFIPGAL 665
Query 143 PNRLFQTGPLES--GVSLLARRVLVKKSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQD 200
P L + E G+ L +RV + D +P+WL F+RG+VD ++LPLNV RE +Q
Sbjct 666 PWELARNMFDEESRGIRLYVKRVFINDKFADAVPRWLTFIRGVVDSDELPLNVGREILQK 725
Query 201 SVLQRKLSTVIVKRI 215
S ++ VI KRI
Sbjct 726 S----RMLQVINKRI 736
Score = 42.7 bits (99), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 28/39 (71%), Gaps = 0/39 (0%)
Query 39 SGTKIVCHLKQDCLEFANPHRVKECAKKFSSFVNFPVYM 77
SGT++V HLK+D ++ +++KE +K+S F+ P+++
Sbjct 483 SGTRVVLHLKEDSDDYLEDYKLKELMRKYSEFIQLPIHI 521
> dre:100007321 fe05a04; wu:fe05a04
Length=675
Score = 35.4 bits (80), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 34/136 (25%), Positives = 48/136 (35%), Gaps = 21/136 (15%)
Query 57 PHRVKECAKKFSSFVNFPVYMEENGKEVQISSQEALWLKPSPTPEEHKQFFRHLTNQSWG 116
P+R EC+K+F VN+ ++ + ++ I + S T E +Q HL +
Sbjct 7 PYRCDECSKQFCQLVNYRTHLRSHSQKASIHCRVC-----SNTFETEEQLQHHLDTNHFE 61
Query 117 EPFYSIMFS----ADAPLCIRSVLYFPADPPNRLFQTGPLESGVSLLARRVLVKKSATDL 172
+ FY F D LC V L L+ G S R L
Sbjct 62 KEFYQCDFCKQIFTDLELCKDHVEAHRQQAKRHLC----LKCGSSFRHR--------NSL 109
Query 173 LPKWLWFLRGIVDCED 188
L W RG C D
Sbjct 110 LLHLEWHSRGFFSCPD 125
> dre:560161 nucleotide-binding oligomerization domain containing
2-like
Length=961
Score = 33.1 bits (74), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 34/60 (56%), Gaps = 6/60 (10%)
Query 167 KSATDLLPKWLWFLRGIVDCEDLPLNVSREHMQDSVLQRKLSTVIVKRILKFLNDQVKSD 226
+SA L +L FL GI LN ++ +QD + R+ S+ I+KRI +++ Q+K D
Sbjct 514 QSANGHLNLFLRFLLGI------SLNSNQRLLQDLLPNRENSSEIIKRITQYIKVQIKGD 567
Lambda K H
0.321 0.135 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7504534908
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40