bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1069_orf1
Length=359
Score E
Sequences producing significant alignments: (Bits) Value
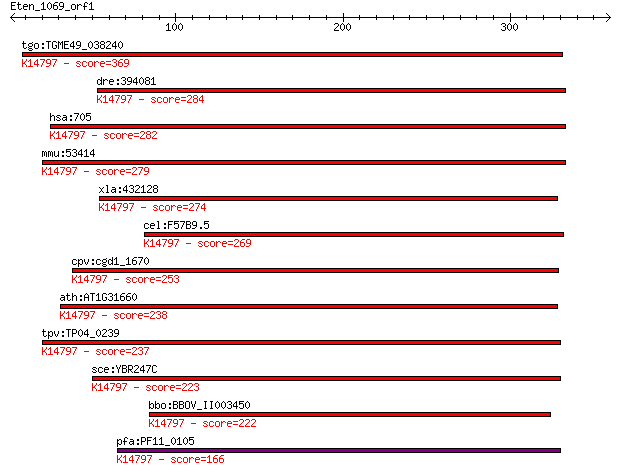
tgo:TGME49_038240 bystin, putative (EC:3.4.24.61); K14797 esse... 369 9e-102
dre:394081 bysl, MGC63889, zgc:63889; bystin-like; K14797 esse... 284 4e-76
hsa:705 BYSL, BYSTIN; bystin-like; K14797 essential nuclear pr... 282 1e-75
mmu:53414 Bysl, Bys, Enp1; bystin-like; K14797 essential nucle... 279 1e-74
xla:432128 bysl, MGC81422; bystin-like; K14797 essential nucle... 274 5e-73
cel:F57B9.5 byn-1; mammalian BYstiN (adhesion protein) related... 269 2e-71
cpv:cgd1_1670 bystin/ S.cerevisiae En1p like adaptor domain ; ... 253 1e-66
ath:AT1G31660 hypothetical protein; K14797 essential nuclear p... 238 4e-62
tpv:TP04_0239 hypothetical protein; K14797 essential nuclear p... 237 4e-62
sce:YBR247C ENP1, MEG1; Enp1p; K14797 essential nuclear protein 1 223 6e-58
bbo:BBOV_II003450 18.m06289; bystin family protein; K14797 ess... 222 2e-57
pfa:PF11_0105 rRNA processing protein, putative; K14797 essent... 166 1e-40
> tgo:TGME49_038240 bystin, putative (EC:3.4.24.61); K14797 essential
nuclear protein 1
Length=558
Score = 369 bits (948), Expect = 9e-102, Method: Compositional matrix adjust.
Identities = 202/351 (57%), Positives = 246/351 (70%), Gaps = 33/351 (9%)
Query 8 DAEGGGPDEETADKDGFVVLDCEEDEEDLVFEK---QRTGSSATRATVNLADLVMEQLKK 64
D + GG D D +GFVV+D E DEED ++ + QR G++A T LAD ++E+L++
Sbjct 187 DRDSGGED---VDSEGFVVIDGEADEEDELYVQRVQQRQGTAAAAPT--LADFILEKLRQ 241
Query 65 QTENKDKSQ----EERVESSLSPKVVEVYTAMAPFLARYRSGKMPKAFKIIPRLHAWEEI 120
+ E + E S+L PKVVEVYTAM FL +YRSGKMPKAFK++PRL WEE+
Sbjct 242 KEERESSGAAAEAPEEDCSALPPKVVEVYTAMGSFLQKYRSGKMPKAFKVLPRLQRWEEV 301
Query 121 LVLTNPTTWSKQATREATRIFCSNLREAMAQRFLCTVLLPAVRDDIAENKKLNYHLYMAL 180
L+LT P +WS+QA EAT+IF SNL A AQRFLC VLLPAVR DI+ NKKLNYHLY AL
Sbjct 302 LLLTQPESWSRQAMFEATKIFTSNLSSAGAQRFLCLVLLPAVRSDISTNKKLNYHLYQAL 361
Query 181 KKALFKPGAFFKGIYLPLALDGCTIREAVIVGSVVTKVSIPVLHGAAALVRLAAVTPDQW 240
KKALFKP AFFKGI+LPLAL+GC+ REA+IVGSVV KVSIPVLH AAAL+RLA V P QW
Sbjct 362 KKALFKPAAFFKGIFLPLALEGCSNREAIIVGSVVAKVSIPVLHAAAALMRLALVPPPQW 421
Query 241 IPSVSHMMTVLIDKKYSLPLQAINECVDHFHRFAPSEVKVTVAWHKCLLVLIQRYK---- 296
+P+VS +M +LI+KKYSLP++A+ CV HFHRFA + VAWH+ LLV +QRYK
Sbjct 422 LPAVSVLMGLLINKKYSLPVKAVQACVAHFHRFAERADLLPVAWHQALLVFVQRYKYSGV 481
Query 297 -----------------VNLSASHRMKLKAVLQSHFHAKVGPEIRRELAAQ 330
V LS R LK VL+ HFH K+GPEIRREL A+
Sbjct 482 SRVIVFQEMTPFEQFNSVCLSEDERSMLKEVLRVHFHEKIGPEIRRELLAK 532
> dre:394081 bysl, MGC63889, zgc:63889; bystin-like; K14797 essential
nuclear protein 1
Length=422
Score = 284 bits (726), Expect = 4e-76, Method: Compositional matrix adjust.
Identities = 143/284 (50%), Positives = 198/284 (69%), Gaps = 7/284 (2%)
Query 53 NLADLVMEQL-KKQTENKDKSQE--ERVESSLSPKVVEVYTAMAPFLARYRSGKMPKAFK 109
LAD++ME++ +KQTE E + P+VVEVY ++ L++YRSGK+PKAFK
Sbjct 129 TLADIIMEKITEKQTEVGTVLSEVSGHAMPQMDPRVVEVYRGVSKVLSKYRSGKLPKAFK 188
Query 110 IIPRLHAWEEILVLTNPTTWSKQATREATRIFCSNLREAMAQRFLCTVLLPAVRDDIAEN 169
IIP L WE++L LT P TWS A +ATRIF SNL+E MAQRF VLLP +RDDIAE
Sbjct 189 IIPALSNWEQVLYLTEPETWSAAAMYQATRIFSSNLKERMAQRFYNLVLLPRIRDDIAEY 248
Query 170 KKLNYHLYMALKKALFKPGAFFKGIYLPLALDG-CTIREAVIVGSVVTKVSIPVLHGAAA 228
K+LN+HLY ALKKALFKP A+FKGI LPL G CT+REA+I+GS++TK SIPVLH +AA
Sbjct 249 KRLNFHLYSALKKALFKPAAWFKGILLPLCESGSCTLREAIIIGSIITKCSIPVLHSSAA 308
Query 229 LVRLAAVTPDQWIPSVSHMMTVLIDKKYSLPLQAINECVDHFHRFAPSEVKVTVAWHKCL 288
+++LA + ++ + S + +++DKKY+LP + ++ V HF F + + V WH+ L
Sbjct 309 MLKLAEM---EYNGANSIFLRLMLDKKYALPFRVLDALVAHFLSFRTDKRILPVLWHQSL 365
Query 289 LVLIQRYKVNLSASHRMKLKAVLQSHFHAKVGPEIRRELAAQQP 332
L L+QRYK +LS+ + L +L++H H ++ EIRREL + +P
Sbjct 366 LTLVQRYKADLSSEQKEALLELLKAHTHPQISSEIRRELQSAEP 409
> hsa:705 BYSL, BYSTIN; bystin-like; K14797 essential nuclear
protein 1
Length=437
Score = 282 bits (722), Expect = 1e-75, Method: Compositional matrix adjust.
Identities = 150/312 (48%), Positives = 210/312 (67%), Gaps = 12/312 (3%)
Query 25 VVLDCEEDEEDLVFEKQRTGSSATRATVNLADLVMEQL-KKQTENKDKSQEER--VESSL 81
VV+D E++ +F + + T LAD++ME+L +KQTE + E L
Sbjct 123 VVVDPEDERAIEMFMNKNPPARRT-----LADIIMEKLTEKQTEVETVMSEVSGFPMPQL 177
Query 82 SPKVVEVYTAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTTWSKQATREATRIF 141
P+V+EVY + L++YRSGK+PKAFKIIP L WE+IL +T P W+ A +ATRIF
Sbjct 178 DPRVLEVYRGVREVLSKYRSGKLPKAFKIIPALSNWEQILYVTEPEAWTAAAMYQATRIF 237
Query 142 CSNLREAMAQRFLCTVLLPAVRDDIAENKKLNYHLYMALKKALFKPGAFFKGIYLPLALD 201
SNL+E MAQRF VLLP VRDD+AE K+LN+HLYMALKKALFKPGA+FKGI +PL
Sbjct 238 ASNLKERMAQRFYNLVLLPRVRDDVAEYKRLNFHLYMALKKALFKPGAWFKGILIPLCES 297
Query 202 G-CTIREAVIVGSVVTKVSIPVLHGAAALVRLAAVTPDQWIPSVSHMMTVLIDKKYSLPL 260
G CT+REA+IVGS++TK SIPVLH +AA++++A + ++ + S + +L+DKKY+LP
Sbjct 298 GTCTLREAIIVGSIITKCSIPVLHSSAAMLKIAEM---EYSGANSIFLRLLLDKKYALPY 354
Query 261 QAINECVDHFHRFAPSEVKVTVAWHKCLLVLIQRYKVNLSASHRMKLKAVLQSHFHAKVG 320
+ ++ V HF F + ++ V WH+CLL L+QRYK +L+ + L +L+ H ++
Sbjct 355 RVLDALVFHFLGFRTEKRELPVLWHQCLLTLVQRYKADLATDQKEALLELLRLQPHPQLS 414
Query 321 PEIRRELAAQQP 332
PEIRREL + P
Sbjct 415 PEIRRELQSAVP 426
> mmu:53414 Bysl, Bys, Enp1; bystin-like; K14797 essential nuclear
protein 1
Length=436
Score = 279 bits (713), Expect = 1e-74, Method: Compositional matrix adjust.
Identities = 150/317 (47%), Positives = 208/317 (65%), Gaps = 12/317 (3%)
Query 20 DKDGFVVLDCEEDEEDLVFEKQRTGSSATRATVNLADLVMEQL-KKQTENKDKSQEER-- 76
D V++D E++ +F + T LAD++ME+L +KQTE + E
Sbjct 117 DHQAEVIVDPEDERAIEMFMNKNPPVRRT-----LADIIMEKLTEKQTEVETVMSEVSGF 171
Query 77 VESSLSPKVVEVYTAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTTWSKQATRE 136
L P+V+EVY + L +YRSGK+PKAFK+IP L WE+IL +T P W+ A +
Sbjct 172 PMPQLDPRVLEVYRGVREVLCKYRSGKLPKAFKVIPALSNWEQILYVTEPEAWTAAAMYQ 231
Query 137 ATRIFCSNLREAMAQRFLCTVLLPAVRDDIAENKKLNYHLYMALKKALFKPGAFFKGIYL 196
ATRIF SNL+E MAQRF VLLP VRDDIAE K+LN+HLYMALKKALFKPGA+FKGI +
Sbjct 232 ATRIFASNLKERMAQRFYNLVLLPRVRDDIAEYKRLNFHLYMALKKALFKPGAWFKGILI 291
Query 197 PLALDG-CTIREAVIVGSVVTKVSIPVLHGAAALVRLAAVTPDQWIPSVSHMMTVLIDKK 255
PL G CT+REA+IVGS++TK SIPVLH +AA++++A + ++ + S + +L+DKK
Sbjct 292 PLCESGTCTLREAIIVGSIITKCSIPVLHSSAAMLKIAEM---EYSGANSIFLRLLLDKK 348
Query 256 YSLPLQAINECVDHFHRFAPSEVKVTVAWHKCLLVLIQRYKVNLSASHRMKLKAVLQSHF 315
Y+LP + ++ V HF F + ++ V WH+CLL L QRYK +L+ + L +L+
Sbjct 349 YALPYRVLDALVFHFLAFRTEKRQLPVLWHQCLLTLAQRYKADLATEQKEALLELLRLQP 408
Query 316 HAKVGPEIRRELAAQQP 332
H ++ PEIRREL + P
Sbjct 409 HPQLSPEIRRELQSAVP 425
> xla:432128 bysl, MGC81422; bystin-like; K14797 essential nuclear
protein 1
Length=431
Score = 274 bits (700), Expect = 5e-73, Method: Compositional matrix adjust.
Identities = 140/278 (50%), Positives = 193/278 (69%), Gaps = 7/278 (2%)
Query 54 LADLVMEQL-KKQTENKDKSQE--ERVESSLSPKVVEVYTAMAPFLARYRSGKMPKAFKI 110
LAD++ME++ +KQTE + E R L P+++EVY + L+ YRSGK+PKAFKI
Sbjct 144 LADIIMEKITEKQTEVETMMSEVSGRPMPQLDPRILEVYKGVKEVLSSYRSGKLPKAFKI 203
Query 111 IPRLHAWEEILVLTNPTTWSKQATREATRIFCSNLREAMAQRFLCTVLLPAVRDDIAENK 170
+P L WE+IL +T P W+ A +ATRIF SNL E MAQRF VLLP VRDDIAE K
Sbjct 204 VPALSNWEQILYITEPEAWTAAAVYQATRIFSSNLAERMAQRFYNLVLLPRVRDDIAEYK 263
Query 171 KLNYHLYMALKKALFKPGAFFKGIYLPLALDG-CTIREAVIVGSVVTKVSIPVLHGAAAL 229
+LN+HLYMALKKALFKPGA+FKGI +PL G C++REAVI+GS++TK SIPVLH +AA+
Sbjct 264 RLNFHLYMALKKALFKPGAWFKGILIPLCESGTCSLREAVIIGSILTKCSIPVLHSSAAM 323
Query 230 VRLAAVTPDQWIPSVSHMMTVLIDKKYSLPLQAINECVDHFHRFAPSEVKVTVAWHKCLL 289
+++A + ++ + S + +LIDKKY+LP + ++ + HF F + + V WH+ LL
Sbjct 324 LKIAEM---EYNGANSIFLRLLIDKKYALPFRVLDALLFHFLAFRSDKRTLPVLWHQSLL 380
Query 290 VLIQRYKVNLSASHRMKLKAVLQSHFHAKVGPEIRREL 327
L QRYK ++S+ + L +L+ H H ++ EIRREL
Sbjct 381 TLCQRYKDDMSSEQKEALLDLLRIHSHPQISNEIRREL 418
> cel:F57B9.5 byn-1; mammalian BYstiN (adhesion protein) related
family member (byn-1); K14797 essential nuclear protein 1
Length=449
Score = 269 bits (687), Expect = 2e-71, Method: Compositional matrix adjust.
Identities = 127/252 (50%), Positives = 176/252 (69%), Gaps = 4/252 (1%)
Query 81 LSPKVVEVYTAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTTWSKQATREATRI 140
+ P+VVE+Y + ++++YRSGK+PKAFKIIP++ WE+IL LT P TW+ A +ATR+
Sbjct 179 MDPEVVEMYEQIGQYMSKYRSGKVPKAFKIIPKMINWEQILFLTKPETWTAAAMYQATRL 238
Query 141 FCSNLREAMAQRFLCTVLLPAVRDDIAENKKLNYHLYMALKKALFKPGAFFKGIYLPLAL 200
F SN+ M QRF VLLP +RDDI E KKLNYHLY AL KA++KP AFFKG+ LPL
Sbjct 239 FASNMNPKMCQRFYTLVLLPRLRDDIDEFKKLNYHLYQALCKAIYKPAAFFKGLILPLLE 298
Query 201 DG-CTIREAVIVGSVVTKVSIPVLHGAAALVRLAAVTPDQWIPSVSHMMTVLIDKKYSLP 259
G CT+REAVI SV+TKV IP+ H AAA++R+A + ++ + S + LIDKKY+LP
Sbjct 299 SGTCTLREAVIFSSVLTKVPIPIFHSAAAMLRIAEM---EYTGANSVFLRALIDKKYALP 355
Query 260 LQAINECVDHFHRFAPSEVKVTVAWHKCLLVLIQRYKVNLSASHRMKLKAVLQSHFHAKV 319
+A++ V+HF R E + V WH+CLL L QRYK +L+A + + +++ H H +
Sbjct 356 YRAVDGVVNHFIRLKTDERDMPVLWHQCLLALCQRYKNDLNAEQKAAIYELIRFHGHYLI 415
Query 320 GPEIRRELAAQQ 331
PEIRREL +++
Sbjct 416 SPEIRRELESKE 427
> cpv:cgd1_1670 bystin/ S.cerevisiae En1p like adaptor domain
; K14797 essential nuclear protein 1
Length=461
Score = 253 bits (645), Expect = 1e-66, Method: Compositional matrix adjust.
Identities = 131/320 (40%), Positives = 195/320 (60%), Gaps = 31/320 (9%)
Query 38 FEKQRTGSSATRATVNLADLVMEQLKKQTENKD--KSQEERVESSLSPKVVEVYTAMAPF 95
+E + S + R D ++ +L++++ ++ + E S++ KV +VYT + +
Sbjct 136 YENENHSSQSIRNQTGFIDTIVSKLREKSSDRKPLEISESCQNSNIPEKVTQVYTLIGEW 195
Query 96 LARYRSGKMPKAFKIIPRLHAWEEILVLTNPTTWSKQATREATRIFCSNLREAMAQRFLC 155
L +Y+SGK+PKAF IIP+L WEE++ LTNP WS A EA RIF SNL + +F
Sbjct 196 LTKYKSGKLPKAFSIIPKLENWEEVVFLTNPHNWSPNAMNEAVRIFSSNLSPKDSLKFYS 255
Query 156 TVLLPAVRDDIAEN-KKLNYHLYMALKKALFKPGAFFKGIYLPLALD-GCTIREAVIVGS 213
+L P +R +I++N KLNYH Y ALKKA+FKP A+FKGI LPLA D CTI+EA+I+GS
Sbjct 256 KILYPTIRTNISQNYGKLNYHYYQALKKAMFKPAAWFKGILLPLAEDESCTIKEAIIIGS 315
Query 214 VVTKVSIPVLHGAAALVRLAAVTPDQWIPSVSHMMTVLIDKKYSLPLQAINECVDHFHRF 273
V++KVSIPVLH AAA+++L+ + W +H + VL+ KKYS+P + I+E VD+F +F
Sbjct 316 VLSKVSIPVLHTAAAIIKLSQIK--VWNTCQTHFIMVLLCKKYSMPKKVIDELVDNFTKF 373
Query 274 ----------AP-----------SEVKVT----VAWHKCLLVLIQRYKVNLSASHRMKLK 308
+P + V+ V WHK LLV +QRYK S+ +L
Sbjct 374 DQKFLHYNDSSPLFSQNNINLNSTNVQTNSILPVTWHKTLLVFVQRYKYEFSSDQINRLN 433
Query 309 AVLQSHFHAKVGPEIRRELA 328
++++ +H + PEI REL+
Sbjct 434 DLVKNQYHYLISPEIIRELS 453
> ath:AT1G31660 hypothetical protein; K14797 essential nuclear
protein 1
Length=444
Score = 238 bits (606), Expect = 4e-62, Method: Compositional matrix adjust.
Identities = 124/298 (41%), Positives = 185/298 (62%), Gaps = 9/298 (3%)
Query 31 EDEEDLVFEKQRTGSSATRATVNLADLVMEQLKKQTENKDKSQEERVESSLSPKVVEVYT 90
ED+E L FE ++ + T L D+++++LK + + D ++EER + + P + ++Y
Sbjct 118 EDDEKL-FESFLNKNAPPQRT--LTDIIIKKLKDK--DADLAEEERPDPKMDPAITKLYK 172
Query 91 AMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTTWSKQATREATRIFCSNLREAMA 150
+ F++ Y GK+PKAFK++ + WE++L LT P WS A +ATRIF SNL++
Sbjct 173 GVGKFMSEYTVGKLPKAFKLVTSMEHWEDVLYLTEPEKWSPNALYQATRIFASNLKDRQV 232
Query 151 QRFLCTVLLPAVRDDIAENKKLNYHLYMALKKALFKPGAFFKGIYLPLALDG-CTIREAV 209
QRF VLLP VR+DI ++KKL++ LY ALKK+L+KP AF +GI PL G C +REAV
Sbjct 233 QRFYNYVLLPRVREDIRKHKKLHFALYQALKKSLYKPSAFNQGILFPLCKSGTCNLREAV 292
Query 210 IVGSVVTKVSIPVLHGAAALVRLAAVTPDQWIPSVSHMMTVLIDKKYSLPLQAINECVDH 269
I+GS++ K SIP+LH AL RLA + + + S+ + VL++KKY +P + ++ V H
Sbjct 293 IIGSILEKCSIPMLHSCVALNRLAEM---DYCGTTSYFIKVLLEKKYCMPYRVLDALVAH 349
Query 270 FHRFAPSEVKVTVAWHKCLLVLIQRYKVNLSASHRMKLKAVLQSHFHAKVGPEIRREL 327
F RF + V WH+ LL +QRYK + + L+ +LQ H V PEI REL
Sbjct 350 FMRFVDDIRVMPVIWHQSLLTFVQRYKYEILKEDKEHLQTLLQRQKHHLVTPEILREL 407
> tpv:TP04_0239 hypothetical protein; K14797 essential nuclear
protein 1
Length=384
Score = 237 bits (605), Expect = 4e-62, Method: Compositional matrix adjust.
Identities = 121/310 (39%), Positives = 178/310 (57%), Gaps = 19/310 (6%)
Query 20 DKDGFVVLDCEEDEEDLVFEKQRTGSSATRATVNLADLVMEQLKKQTENKDKSQEERVES 79
+ D V D ED + E Q T S+ + L DL +E V
Sbjct 86 ENDPLVSFDFSEDSAGFLPESQTT-SNTSEIWSKLNDL------------STKPKESVMD 132
Query 80 SLSPKVVEVYTAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTTWSKQATREATR 139
L P VY + +L+RY+SG +PKAFK++P++ W EI+ LTNP W+ A E TR
Sbjct 133 RLRP----VYNEIGLYLSRYKSGGLPKAFKVLPKMTNWAEIIQLTNPQNWTPNAMYEVTR 188
Query 140 IFCSNLREAMAQRFLCTVLLPAVRDDIAENKKLNYHLYMALKKALFKPGAFFKGIYLPLA 199
+F SN+ E+ A+RF +LLPAVR +++ NK LN+H Y +L KA+FKP A+FKGI LPL
Sbjct 189 LFSSNMNESNAERFYLCILLPAVRANLSSNKTLNHHYYESLMKAMFKPTAWFKGILLPLV 248
Query 200 LDGCTIREAVIVGSVVTKVSIPVLHGAAALVRLAAVTPDQWIPSVSHMMTVLIDKKYSLP 259
+GCT RE I+GSV+ K+SIPV H +A ++++ +W S S ++T L KKY LP
Sbjct 249 QEGCTYREGAIIGSVLKKISIPVFHVSAFIIQICQYP--KWFGSTSFILTTLFQKKYKLP 306
Query 260 LQAINECVDHFHRFAPSEVKVTVAWHKCLLVLIQRYKVNLSASHRMKLKAVLQSHFHAKV 319
+ I EC+ +F+RF + V WH+ L + + Y+ L+ L+A+L+ H H ++
Sbjct 307 RKVIGECLQYFYRFINFPDHLPVIWHQSLYLFLYHYQHMLNEDEYGLLRALLERHKHPQI 366
Query 320 GPEIRRELAA 329
GP I + L+
Sbjct 367 GPAIEKLLSC 376
> sce:YBR247C ENP1, MEG1; Enp1p; K14797 essential nuclear protein
1
Length=483
Score = 223 bits (569), Expect = 6e-58, Method: Compositional matrix adjust.
Identities = 117/311 (37%), Positives = 175/311 (56%), Gaps = 34/311 (10%)
Query 50 ATVNLADLVMEQLK-KQTENKDKSQEERVES-------------------SLSPKVVEVY 89
+ NLAD +M ++ K+++ +D +E + + +L KV++ Y
Sbjct 155 GSYNLADKIMASIREKESQVEDMQDDEPLANEQNTSRGNISSGLKSGEGVALPEKVIKAY 214
Query 90 TAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTTWSKQATREATRIFCSNLREAM 149
T + L + GK+PK FK+IP L W++++ +TNP WS EAT++F SNL
Sbjct 215 TTVGSILKTWTHGKLPKLFKVIPSLRNWQDVIYVTNPEEWSPHVVYEATKLFVSNLTAKE 274
Query 150 AQRFLCTVLLPAVRDDI--AENKKLNYHLYMALKKALFKPGAFFKGIYLPLALDGCTIRE 207
+Q+F+ +LL RD+I +E+ LNYH+Y A+KK+L+KP AFFKG PL GC +RE
Sbjct 275 SQKFINLILLERFRDNIETSEDHSLNYHIYRAVKKSLYKPSAFFKGFLFPLVETGCNVRE 334
Query 208 AVIVGSVVTKVSIPVLHGAAALVRLAAVTPDQWIPSVSHMMTVLIDKKYSLPLQAINECV 267
A I GSV+ KVS+P LH +AAL L + + P + + +L+DKKY+LP Q +++CV
Sbjct 335 ATIAGSVLAKVSVPALHSSAALSYLLRLP---FSPPTTVFIKILLDKKYALPYQTVDDCV 391
Query 268 DHFHRF--------APSEVKV-TVAWHKCLLVLIQRYKVNLSASHRMKLKAVLQSHFHAK 318
+F RF +V V WHK L QRYK +++ R L ++ H
Sbjct 392 YYFMRFRILDDGSNGEDATRVLPVIWHKAFLTFAQRYKNDITQDQRDFLLETVRQRGHKD 451
Query 319 VGPEIRRELAA 329
+GPEIRREL A
Sbjct 452 IGPEIRRELLA 462
> bbo:BBOV_II003450 18.m06289; bystin family protein; K14797 essential
nuclear protein 1
Length=383
Score = 222 bits (565), Expect = 2e-57, Method: Compositional matrix adjust.
Identities = 100/240 (41%), Positives = 156/240 (65%), Gaps = 2/240 (0%)
Query 84 KVVEVYTAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLTNPTTWSKQATREATRIFCS 143
K+ V++ + ++++Y SG +PKAFK +PR+ WEE L LT+P W+ A EATRI S
Sbjct 133 KMRAVFSEIGVYMSKYTSGGLPKAFKFLPRMSNWEEFLELTHPENWTPNAMYEATRILAS 192
Query 144 NLREAMAQRFLCTVLLPAVRDDIAENKKLNYHLYMALKKALFKPGAFFKGIYLPLALDGC 203
N+ ++ ++F VLLP VR DI ++KLNYHLYMALKKA++KP A+FKGI +PL +GC
Sbjct 193 NMTDSTVEKFYSKVLLPTVRKDIRGSRKLNYHLYMALKKAMYKPTAWFKGIMVPLVEEGC 252
Query 204 TIREAVIVGSVVTKVSIPVLHGAAALVRLAAVTPDQWIPSVSHMMTVLIDKKYSLPLQAI 263
REA I+G+++ ++SIP+LH +A ++RL +W S S +M++++ K+Y+LP +
Sbjct 253 LYREAAIIGNILRRMSIPILHASAFILRLCQC--QRWYGSSSFIMSIMLQKQYNLPKNVV 310
Query 264 NECVDHFHRFAPSEVKVTVAWHKCLLVLIQRYKVNLSASHRMKLKAVLQSHFHAKVGPEI 323
ECV +F +F + V WH+ LL+L+ YK + + +K + + H H + P I
Sbjct 311 QECVQYFCKFENFGDLLPVIWHQSLLILVTSYKHYFTPADFASIKRLTKVHVHPHITPAI 370
> pfa:PF11_0105 rRNA processing protein, putative; K14797 essential
nuclear protein 1
Length=436
Score = 166 bits (421), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 86/265 (32%), Positives = 147/265 (55%), Gaps = 3/265 (1%)
Query 65 QTENKDKSQEERVESSLSPKVVEVYTAMAPFLARYRSGKMPKAFKIIPRLHAWEEILVLT 124
+ EN + E ++ V + Y + LA Y+SGK+ +A I+ + W E L+LT
Sbjct 155 KMENTTQEPNPSTEDKVNLLVEKCYRTIGEDLAHYKSGKLHRALTILTKSSKWYEYLLLT 214
Query 125 NPTTWSKQATREATRIFCSNLREAMAQRFLCTVLLPAVRDDIAENKKLNYHLYMALKKAL 184
P W+ AT E T++F S L+E +F +LLP V ++I +NKKL+ LY AL KAL
Sbjct 215 KPKKWTAHATFEITKLFSSGLKEKEVSKFYEFILLPIVLENIDKNKKLDAFLYKALIKAL 274
Query 185 FKPGAFFKGIYLPLALDGCTIREAVIVGSVVTKVSIPVLHGAAALVRLAAVTPDQWIPSV 244
+K ++FKGI P+ CT ++ +I+GSV+ K+SI + ++ L + W +
Sbjct 275 YKSKSWFKGILFPILHRECTKKQMIIIGSVIQKMSISI---NCVVLALDEIFSFSWNSII 331
Query 245 SHMMTVLIDKKYSLPLQAINECVDHFHRFAPSEVKVTVAWHKCLLVLIQRYKVNLSASHR 304
S+++ +L +KKY+ Q I+ CV++F RF + +++ WHK LL L+ Y+ + +
Sbjct 332 SYILILLFNKKYAFTKQCIDNCVNYFLRFENYQDVLSINWHKSLLTLVHNYRGLMCDTQV 391
Query 305 MKLKAVLQSHFHAKVGPEIRRELAA 329
L +++ H ++ EI + L +
Sbjct 392 EALTHLVKKKNHHQISSEIMKHLYS 416
Lambda K H
0.319 0.132 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 15513116780
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40