bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1085_orf1
Length=134
Score E
Sequences producing significant alignments: (Bits) Value
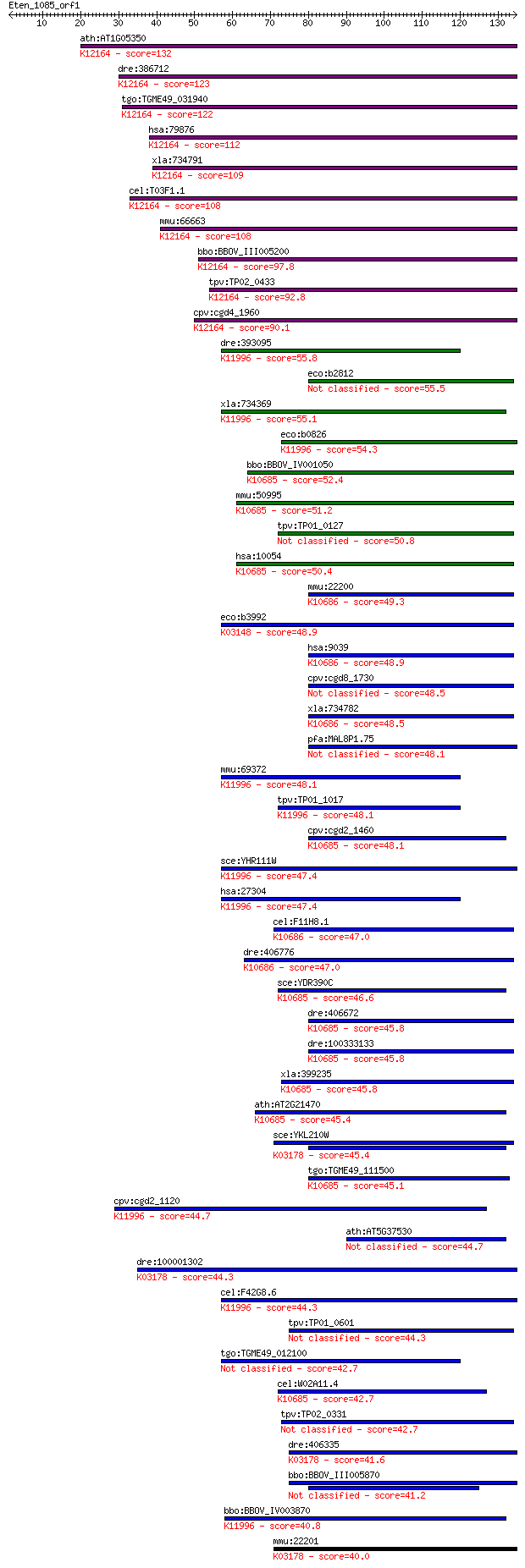
ath:AT1G05350 thiF family protein; K12164 ubiquitin-like modif... 132 2e-31
dre:386712 uba5, cb972, ube1dc1; ubiquitin-like modifier activ... 123 2e-28
tgo:TGME49_031940 thiF family domain-containing protein ; K121... 122 3e-28
hsa:79876 UBA5, FLJ17281, FLJ23251, FLJ23251UBA5, THIFP1, UBE1... 112 4e-25
xla:734791 uba5, MGC131082, ube1dc1; ubiquitin-like modifier a... 109 3e-24
cel:T03F1.1 hypothetical protein; K12164 ubiquitin-like modifi... 108 4e-24
mmu:66663 Uba5, 5730525G14Rik, AW240750, Ube1dc1; ubiquitin-li... 108 5e-24
bbo:BBOV_III005200 17.m07465; ThiF family protein; K12164 ubiq... 97.8 7e-21
tpv:TP02_0433 hypothetical protein; K12164 ubiquitin-like modi... 92.8 3e-19
cpv:cgd4_1960 ThiF/moeB family ; K12164 ubiquitin-like modifie... 90.1 2e-18
dre:393095 mocs3, MGC55696, zgc:55696; molybdenum cofactor syn... 55.8 4e-08
eco:b2812 csdL, ECK2808, JW2783, ygdL; sulfur acceptor for CsdA 55.5 5e-08
xla:734369 mocs3, MGC84877, uba4; molybdenum cofactor synthesi... 55.1 6e-08
eco:b0826 moeB, chlN, ECK0816, JW0810; molybdopterin synthase ... 54.3 1e-07
bbo:BBOV_IV001050 21.m02728; ubiquitin-activating enzyme; K106... 52.4 4e-07
mmu:50995 Uba2, AA986091, Arx, Sae2, UBA1, Ubl1a2, Uble1b; ubi... 51.2 8e-07
tpv:TP01_0127 ubiquitin-protein ligase 50.8 1e-06
hsa:10054 UBA2, ARX, FLJ13058, SAE2; ubiquitin-like modifier a... 50.4 1e-06
mmu:22200 Uba3, A830034N06Rik, AI256736, AI848246, AW546539, U... 49.3 3e-06
eco:b3992 thiF, ECK3984, JW3956, thiA; adenylyltransferase, mo... 48.9 4e-06
hsa:9039 UBA3, DKFZp566J164, MGC22384, UBE1C, hUBA3; ubiquitin... 48.9 5e-06
cpv:cgd8_1730 Uba3p like ubiquitin activating enzyme E1 48.5 5e-06
xla:734782 uba3, MGC131020, ube1c; ubiquitin-like modifier act... 48.5 6e-06
pfa:MAL8P1.75 ubiquitin-activating enzyme, putative 48.1 7e-06
mmu:69372 Mocs3, 1700020H17Rik, Uba4; molybdenum cofactor synt... 48.1 7e-06
tpv:TP01_1017 hypothetical protein; K11996 adenylyltransferase... 48.1 8e-06
cpv:cgd2_1460 SUMO-1 activating enzyme subunit 2 ; K10685 ubiq... 48.1 8e-06
sce:YHR111W UBA4, YHR1; Protein that activates Urm1p before it... 47.4 1e-05
hsa:27304 MOCS3, MGC9252, UBA4, dJ914P20.3; molybdenum cofacto... 47.4 1e-05
cel:F11H8.1 rfl-1; ectopic membrane RuFfLes in embryo family m... 47.0 2e-05
dre:406776 uba3, ube1c, wu:fb75e04, wu:fc37b11, zgc:55528; ubi... 47.0 2e-05
sce:YDR390C UBA2, UAL1; Nuclear protein that acts as a heterod... 46.6 2e-05
dre:406672 uba2, sae2, sae2b, uble1b, wu:fi17g06, zgc:66354; u... 45.8 4e-05
dre:100333133 ubiquitin-like modifier activating enzyme 2-like... 45.8 4e-05
xla:399235 uba2, MGC84651, Uble1b, sae2, sae2-B, uba2-a, uba2-... 45.8 4e-05
ath:AT2G21470 SAE2; SAE2 (SUMO-ACTIVATING ENZYME 2); SUMO acti... 45.4 4e-05
sce:YKL210W UBA1; Uba1p; K03178 ubiquitin-activating enzyme E1... 45.4 5e-05
tgo:TGME49_111500 ubiquitin-activating enzyme, putative (EC:1.... 45.1 7e-05
cpv:cgd2_1120 molybdopterin synthase sulphurylase ; K11996 ade... 44.7 7e-05
ath:AT5G37530 thiF family protein 44.7 9e-05
dre:100001302 ubiquitin-like modifier-activating enzyme 1-like... 44.3 9e-05
cel:F42G8.6 hypothetical protein; K11996 adenylyltransferase a... 44.3 1e-04
tpv:TP01_0601 hypothetical protein 44.3 1e-04
tgo:TGME49_012100 molybdenum cofactor synthesis protein, putative 42.7 3e-04
cel:W02A11.4 uba-2; UBA (human ubiquitin) related family membe... 42.7 3e-04
tpv:TP02_0331 ubiquitin activating enzyme, putatuve 42.7 3e-04
dre:406335 uba1, ube1, wu:fa01e08, wu:fb30f01, wu:fi21c11, wu:... 41.6 8e-04
bbo:BBOV_III005870 17.m07520; ThiF family protein 41.2 0.001
bbo:BBOV_IV003870 23.m05802; molybdenum cofactor synthesis pro... 40.8 0.001
mmu:22201 Uba1, A1S9, AA989744, Sbx, Ube-1, Ube1x; ubiquitin-l... 40.0 0.002
> ath:AT1G05350 thiF family protein; K12164 ubiquitin-like modifier-activating
enzyme 5
Length=445
Score = 132 bits (333), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 70/120 (58%), Positives = 88/120 (73%), Gaps = 7/120 (5%)
Query 20 PAVLINLKMRKDAE-----SQTEGGLRPKVLSMSAEVRADNPYSRLMALQRMGVVRDYQA 74
PA LIN K+R E S+ R KV +S+EV NPYSRLMALQRMG+V +Y+
Sbjct 22 PA-LIN-KLRSHVENLATLSKCNPHRRSKVKELSSEVVDSNPYSRLMALQRMGIVDNYER 79
Query 75 ITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLFFTPKDVGMTKVEA 134
I + +V +VG+GGVGSVAAEML RCGIG+++L D+D+VEL+NMNRLFF P VGMTK +A
Sbjct 80 IREFSVAIVGIGGVGSVAAEMLTRCGIGRLLLYDYDTVELANMNRLFFRPDQVGMTKTDA 139
> dre:386712 uba5, cb972, ube1dc1; ubiquitin-like modifier activating
enzyme 5; K12164 ubiquitin-like modifier-activating
enzyme 5
Length=398
Score = 123 bits (308), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 67/105 (63%), Positives = 81/105 (77%), Gaps = 0/105 (0%)
Query 30 KDAESQTEGGLRPKVLSMSAEVRADNPYSRLMALQRMGVVRDYQAITKKAVLLVGVGGVG 89
K +S E +RPK+ MSAEV NPYSRLMAL+RMG+V+DY+ I AV +VGVGGVG
Sbjct 22 KQKQSDAEHNIRPKIEQMSAEVVDSNPYSRLMALKRMGIVQDYEKIRSFAVAVVGVGGVG 81
Query 90 SVAAEMLVRCGIGKIILLDFDSVELSNMNRLFFTPKDVGMTKVEA 134
SV AEML RCGIGK++L D+D VEL+NMNRLFF P G++KVEA
Sbjct 82 SVTAEMLTRCGIGKLLLFDYDKVELANMNRLFFQPHQAGLSKVEA 126
> tgo:TGME49_031940 thiF family domain-containing protein ; K12164
ubiquitin-like modifier-activating enzyme 5
Length=401
Score = 122 bits (306), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 72/104 (69%), Positives = 85/104 (81%), Gaps = 1/104 (0%)
Query 31 DAESQTEGGLRPKVLSMSAEVRADNPYSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGS 90
D+ S T G +R K+ MSA VR DNPYSRLMALQRMGVV +Y+AI K VL+VGVGGVGS
Sbjct 2 DSSSGTPG-VRRKIPEMSAVVRDDNPYSRLMALQRMGVVDNYRAIRHKCVLVVGVGGVGS 60
Query 91 VAAEMLVRCGIGKIILLDFDSVELSNMNRLFFTPKDVGMTKVEA 134
VA++ML RCG+GK+IL D+D VEL+NMNRLFFTPK G +KVEA
Sbjct 61 VASDMLARCGVGKLILFDYDKVELANMNRLFFTPKQCGQSKVEA 104
> hsa:79876 UBA5, FLJ17281, FLJ23251, FLJ23251UBA5, THIFP1, UBE1DC1;
ubiquitin-like modifier activating enzyme 5; K12164 ubiquitin-like
modifier-activating enzyme 5
Length=404
Score = 112 bits (279), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 62/97 (63%), Positives = 76/97 (78%), Gaps = 0/97 (0%)
Query 38 GGLRPKVLSMSAEVRADNPYSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLV 97
GG R ++ MS+EV NPYSRLMAL+RMG+V DY+ I AV +VGVGGVGSV AEML
Sbjct 34 GGGRVRIEKMSSEVVDSNPYSRLMALKRMGIVSDYEKIRTFAVAIVGVGGVGSVTAEMLT 93
Query 98 RCGIGKIILLDFDSVELSNMNRLFFTPKDVGMTKVEA 134
RCGIGK++L D+D VEL+NMNRLFF P G++KV+A
Sbjct 94 RCGIGKLLLFDYDKVELANMNRLFFQPHQAGLSKVQA 130
> xla:734791 uba5, MGC131082, ube1dc1; ubiquitin-like modifier
activating enzyme 5; K12164 ubiquitin-like modifier-activating
enzyme 5
Length=397
Score = 109 bits (272), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 62/96 (64%), Positives = 74/96 (77%), Gaps = 0/96 (0%)
Query 39 GLRPKVLSMSAEVRADNPYSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVR 98
G R K+ MSAEV NPYSRLMAL+RMG+V +Y+ I V +VGVGGVGSV AEML R
Sbjct 29 GHRTKIEKMSAEVVDSNPYSRLMALKRMGIVENYEKIRTFTVAVVGVGGVGSVTAEMLTR 88
Query 99 CGIGKIILLDFDSVELSNMNRLFFTPKDVGMTKVEA 134
CGIGK++L D+D VEL+NMNRLFF P G++KVEA
Sbjct 89 CGIGKLLLFDYDKVELANMNRLFFQPHQAGLSKVEA 124
> cel:T03F1.1 hypothetical protein; K12164 ubiquitin-like modifier-activating
enzyme 5
Length=419
Score = 108 bits (270), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 61/102 (59%), Positives = 78/102 (76%), Gaps = 0/102 (0%)
Query 33 ESQTEGGLRPKVLSMSAEVRADNPYSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVA 92
+ ++ R K+ +SAEV NPYSRLMALQRMG+V +Y+ I +K V +VGVGGVGSV
Sbjct 40 QPKSPAPYRQKIEKLSAEVVDSNPYSRLMALQRMGIVNEYERIREKTVAVVGVGGVGSVV 99
Query 93 AEMLVRCGIGKIILLDFDSVELSNMNRLFFTPKDVGMTKVEA 134
AEML RCGIGK+IL D+D VE++NMNRLF+ P G++KVEA
Sbjct 100 AEMLTRCGIGKLILFDYDKVEIANMNRLFYQPNQAGLSKVEA 141
> mmu:66663 Uba5, 5730525G14Rik, AW240750, Ube1dc1; ubiquitin-like
modifier activating enzyme 5; K12164 ubiquitin-like modifier-activating
enzyme 5
Length=403
Score = 108 bits (269), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 60/94 (63%), Positives = 72/94 (76%), Gaps = 0/94 (0%)
Query 41 RPKVLSMSAEVRADNPYSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCG 100
R ++ MS EV NPYSRLMAL+RMG+V DY+ I AV +VGVGGVGSV AEML RCG
Sbjct 35 RTRIQEMSDEVLDSNPYSRLMALKRMGIVSDYKKIRTYAVAIVGVGGVGSVTAEMLTRCG 94
Query 101 IGKIILLDFDSVELSNMNRLFFTPKDVGMTKVEA 134
IGK++L D+D VEL+NMNRLFF P G++KV A
Sbjct 95 IGKLLLFDYDKVELANMNRLFFQPYQAGLSKVHA 128
> bbo:BBOV_III005200 17.m07465; ThiF family protein; K12164 ubiquitin-like
modifier-activating enzyme 5
Length=342
Score = 97.8 bits (242), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 56/84 (66%), Positives = 68/84 (80%), Gaps = 0/84 (0%)
Query 51 VRADNPYSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFD 110
+ D+P SRL AL+RMGVV DY I +V++VGVGGVG+V AEML RCGIGK+IL D+D
Sbjct 3 TQKDDPVSRLKALERMGVVEDYSKIYGCSVIIVGVGGVGAVVAEMLTRCGIGKLILFDYD 62
Query 111 SVELSNMNRLFFTPKDVGMTKVEA 134
VEL+NMNRLF+TP VGM+KVEA
Sbjct 63 DVELANMNRLFYTPSQVGMSKVEA 86
> tpv:TP02_0433 hypothetical protein; K12164 ubiquitin-like modifier-activating
enzyme 5
Length=348
Score = 92.8 bits (229), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 52/81 (64%), Positives = 65/81 (80%), Gaps = 0/81 (0%)
Query 54 DNPYSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVE 113
D+P SRL AL++MGVV D+ I V +VGVGGVG+V AEML RCGIGK+IL D+D +E
Sbjct 6 DDPSSRLKALEKMGVVEDFDRIHSCTVAIVGVGGVGAVVAEMLTRCGIGKLILFDYDDIE 65
Query 114 LSNMNRLFFTPKDVGMTKVEA 134
L+NMNRLF+TPK +G+ KVEA
Sbjct 66 LANMNRLFYTPKQLGLPKVEA 86
> cpv:cgd4_1960 ThiF/moeB family ; K12164 ubiquitin-like modifier-activating
enzyme 5
Length=370
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 50/85 (58%), Positives = 62/85 (72%), Gaps = 0/85 (0%)
Query 50 EVRADNPYSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDF 109
E D YSRL AL+ MGVV DY I KK + ++GVGGVGSV EML RCG+GK+I++DF
Sbjct 21 ENSIDERYSRLTALENMGVVDDYSLIMKKTIFVIGVGGVGSVVVEMLTRCGVGKLIIVDF 80
Query 110 DSVELSNMNRLFFTPKDVGMTKVEA 134
D VELSNMNR+F+ +GM K +A
Sbjct 81 DIVELSNMNRMFYNMNHIGMYKTDA 105
> dre:393095 mocs3, MGC55696, zgc:55696; molybdenum cofactor synthesis
3 (EC:2.8.1.-); K11996 adenylyltransferase and sulfurtransferase
Length=459
Score = 55.8 bits (133), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 30/63 (47%), Positives = 43/63 (68%), Gaps = 1/63 (1%)
Query 57 YSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSN 116
YSR + L +GV + AI+ +VL+VG GG+G A+ L GIG++ LLD+D VELSN
Sbjct 63 YSRQLLLPELGV-KGQIAISNISVLVVGCGGLGCPLAQYLAAAGIGRLGLLDYDVVELSN 121
Query 117 MNR 119
++R
Sbjct 122 LHR 124
> eco:b2812 csdL, ECK2808, JW2783, ygdL; sulfur acceptor for CsdA
Length=268
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 28/55 (50%), Positives = 36/55 (65%), Gaps = 1/55 (1%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLFFTPKD-VGMTKVE 133
+ +VG+GGVGS AAE L R GIG I L+D D V ++N NR +D VG+ K E
Sbjct 33 ICVVGIGGVGSWAAEALARTGIGAITLIDMDDVCVTNTNRQIHALRDNVGLAKAE 87
> xla:734369 mocs3, MGC84877, uba4; molybdenum cofactor synthesis
3 (EC:2.7.7.- 2.8.1.-); K11996 adenylyltransferase and sulfurtransferase
Length=451
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 30/76 (39%), Positives = 50/76 (65%), Gaps = 2/76 (2%)
Query 57 YSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSN 116
YSR + L +GV + ++K +VL++G GG+G A+ L GIG++ LLD+D VE+SN
Sbjct 57 YSRQLVLPDLGVQGQLK-LSKASVLVIGCGGLGCPVAQYLAASGIGRLGLLDYDVVEMSN 115
Query 117 MNRLFFTPKD-VGMTK 131
++R ++ +GM+K
Sbjct 116 LHRQVLHGENRLGMSK 131
> eco:b0826 moeB, chlN, ECK0816, JW0810; molybdopterin synthase
sulfurylase; K11996 adenylyltransferase and sulfurtransferase
Length=249
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 27/63 (42%), Positives = 40/63 (63%), Gaps = 1/63 (1%)
Query 73 QAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNR-LFFTPKDVGMTK 131
+A+ VL+VG+GG+G A++ L G+G + LLDFD+V LSN+ R + VG K
Sbjct 27 EALKDSRVLIVGLGGLGCAASQYLASAGVGNLTLLDFDTVSLSNLQRQTLHSDATVGQPK 86
Query 132 VEA 134
VE+
Sbjct 87 VES 89
> bbo:BBOV_IV001050 21.m02728; ubiquitin-activating enzyme; K10685
ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=630
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 25/71 (35%), Positives = 43/71 (60%), Gaps = 1/71 (1%)
Query 64 QRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-F 122
Q V Y + ++L+VG GG+G + LV CG+ ++++D D++++SN+NR F +
Sbjct 32 QTTAVDNYYDLLRNVSLLVVGAGGIGCELIKNLVLCGVRNLVIVDIDTIDVSNLNRQFLY 91
Query 123 TPKDVGMTKVE 133
+DVG K E
Sbjct 92 RAEDVGRYKAE 102
> mmu:50995 Uba2, AA986091, Arx, Sae2, UBA1, Ubl1a2, Uble1b; ubiquitin-like
modifier activating enzyme 2 (EC:6.3.2.-); K10685
ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=638
Score = 51.2 bits (121), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 31/75 (41%), Positives = 47/75 (62%), Gaps = 3/75 (4%)
Query 61 MALQRMGVVRDY-QAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNR 119
MAL R G+ R+ +A++ VL+VG GG+G + LV G I L+D D++++SN+NR
Sbjct 1 MALSR-GLPRELAEAVSGGRVLVVGAGGIGCELLKNLVLTGFSHIDLIDLDTIDVSNLNR 59
Query 120 LF-FTPKDVGMTKVE 133
F F K VG +K +
Sbjct 60 QFLFQKKHVGRSKAQ 74
> tpv:TP01_0127 ubiquitin-protein ligase
Length=543
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 24/63 (38%), Positives = 41/63 (65%), Gaps = 1/63 (1%)
Query 72 YQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMT 130
Y+ ++ +VLLVG GG+G + L+ G+ K+ ++D D+V++SN+NR F + P+ V
Sbjct 21 YEYLSDVSVLLVGAGGIGCELIKTLLLTGVKKLTIVDMDTVDVSNLNRQFLYLPEHVNKY 80
Query 131 KVE 133
K E
Sbjct 81 KAE 83
> hsa:10054 UBA2, ARX, FLJ13058, SAE2; ubiquitin-like modifier
activating enzyme 2; K10685 ubiquitin-like 1-activating enzyme
E1 B [EC:6.3.2.19]
Length=640
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 31/75 (41%), Positives = 46/75 (61%), Gaps = 3/75 (4%)
Query 61 MALQRMGVVRDY-QAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNR 119
MAL R G+ R+ +A+ VL+VG GG+G + LV G I L+D D++++SN+NR
Sbjct 1 MALSR-GLPRELAEAVAGGRVLVVGAGGIGCELLKNLVLTGFSHIDLIDLDTIDVSNLNR 59
Query 120 LF-FTPKDVGMTKVE 133
F F K VG +K +
Sbjct 60 QFLFQKKHVGRSKAQ 74
> mmu:22200 Uba3, A830034N06Rik, AI256736, AI848246, AW546539,
Ube1c; ubiquitin-like modifier activating enzyme 3; K10686
ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=448
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 24/55 (43%), Positives = 36/55 (65%), Gaps = 1/55 (1%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMTKVE 133
VL++G GG+G + L G +I ++D D++++SN+NR F F PKDVG K E
Sbjct 58 VLVIGAGGLGCELLKNLALSGFRQIHVIDMDTIDVSNLNRQFLFRPKDVGRPKAE 112
> eco:b3992 thiF, ECK3984, JW3956, thiA; adenylyltransferase,
modifies ThiS C-terminus (EC:2.7.7.-); K03148 adenylyltransferase
[EC:2.7.7.-]
Length=251
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 28/78 (35%), Positives = 45/78 (57%), Gaps = 2/78 (2%)
Query 57 YSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSN 116
YSR + L + + Q + VL++G+GG+G+ AA L G+G ++L D D V LSN
Sbjct 9 YSRQILLDDI-ALDGQQKLLDSQVLIIGLGGLGTPAALYLAGAGVGTLVLADDDDVHLSN 67
Query 117 MNR-LFFTPKDVGMTKVE 133
+ R + FT +D+ K +
Sbjct 68 LQRQILFTTEDIDRPKSQ 85
> hsa:9039 UBA3, DKFZp566J164, MGC22384, UBE1C, hUBA3; ubiquitin-like
modifier activating enzyme 3; K10686 ubiquitin-activating
enzyme E1 C [EC:6.3.2.19]
Length=463
Score = 48.9 bits (115), Expect = 5e-06, Method: Composition-based stats.
Identities = 23/55 (41%), Positives = 36/55 (65%), Gaps = 1/55 (1%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMTKVE 133
VL++G GG+G + L G +I ++D D++++SN+NR F F PKD+G K E
Sbjct 72 VLVIGAGGLGCELLKNLALSGFRQIHVIDMDTIDVSNLNRQFLFRPKDIGRPKAE 126
> cpv:cgd8_1730 Uba3p like ubiquitin activating enzyme E1
Length=346
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 26/55 (47%), Positives = 37/55 (67%), Gaps = 1/55 (1%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNR-LFFTPKDVGMTKVE 133
VLLVGVGG+G+ L+ G +I ++D+D VE+SN++R LFF D G +KV
Sbjct 54 VLLVGVGGIGTEILRCLIFSGFRRIDIVDYDYVEVSNISRQLFFNLGDEGKSKVH 108
> xla:734782 uba3, MGC131020, ube1c; ubiquitin-like modifier activating
enzyme 3; K10686 ubiquitin-activating enzyme E1 C
[EC:6.3.2.19]
Length=461
Score = 48.5 bits (114), Expect = 6e-06, Method: Composition-based stats.
Identities = 24/55 (43%), Positives = 36/55 (65%), Gaps = 1/55 (1%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMTKVE 133
+L+VG GG+G + L G +I ++D D++++SN+NR F F PKDVG K E
Sbjct 71 LLVVGAGGLGCELLKNLALSGFRQIHVIDMDTIDVSNLNRQFLFRPKDVGRPKAE 125
> pfa:MAL8P1.75 ubiquitin-activating enzyme, putative
Length=389
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 23/56 (41%), Positives = 37/56 (66%), Gaps = 1/56 (1%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNR-LFFTPKDVGMTKVEA 134
+L+VG GG+G+ + L+ I I ++D+D VE+SN+ R LFF+ D+G KV+
Sbjct 15 ILVVGCGGLGNEVIKNLIFLHIKNICIVDYDIVEVSNLQRQLFFSHDDIGKYKVDV 70
> mmu:69372 Mocs3, 1700020H17Rik, Uba4; molybdenum cofactor synthesis
3; K11996 adenylyltransferase and sulfurtransferase
Length=460
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 27/63 (42%), Positives = 38/63 (60%), Gaps = 1/63 (1%)
Query 57 YSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSN 116
YSR + L +GV R + AVL+VG GG+G A+ L G+G++ L+D D VE SN
Sbjct 63 YSRQLLLPELGV-RGQLRLAAAAVLVVGCGGLGCPLAQYLAAAGVGRLGLVDHDVVETSN 121
Query 117 MNR 119
+ R
Sbjct 122 LAR 124
> tpv:TP01_1017 hypothetical protein; K11996 adenylyltransferase
and sulfurtransferase
Length=554
Score = 48.1 bits (113), Expect = 8e-06, Method: Composition-based stats.
Identities = 23/48 (47%), Positives = 34/48 (70%), Gaps = 0/48 (0%)
Query 72 YQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNR 119
Y++I+ AVL++G GG+GS + L GIG I ++D D VELSN++R
Sbjct 116 YRSISSCAVLVIGAGGLGSPLLQYLASSGIGLIGIMDGDVVELSNLHR 163
> cpv:cgd2_1460 SUMO-1 activating enzyme subunit 2 ; K10685 ubiquitin-like
1-activating enzyme E1 B [EC:6.3.2.19]
Length=637
Score = 48.1 bits (113), Expect = 8e-06, Method: Composition-based stats.
Identities = 22/53 (41%), Positives = 33/53 (62%), Gaps = 1/53 (1%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNR-LFFTPKDVGMTK 131
+L+VG GG+G + L+ G I ++D D +++SN+NR FF K VGM K
Sbjct 24 ILVVGAGGIGCELVKDLILSGFSNITIIDMDGIDISNLNRQFFFRRKHVGMNK 76
> sce:YHR111W UBA4, YHR1; Protein that activates Urm1p before
its conjugation to proteins (urmylation); one target is the
thioredoxin peroxidase Ahp1p, suggesting a role of urmylation
in the oxidative stress response; K11996 adenylyltransferase
and sulfurtransferase
Length=440
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 42/79 (53%), Gaps = 1/79 (1%)
Query 57 YSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSN 116
Y R M ++ G V + VL+VG GG+G A L G+G+I ++D D VE SN
Sbjct 47 YGRQMIVEETGGVAGQVKLKNTKVLVVGAGGLGCPALPYLAGAGVGQIGIVDNDVVETSN 106
Query 117 MNR-LFFTPKDVGMTKVEA 134
++R + VGM K E+
Sbjct 107 LHRQVLHDSSRVGMLKCES 125
> hsa:27304 MOCS3, MGC9252, UBA4, dJ914P20.3; molybdenum cofactor
synthesis 3; K11996 adenylyltransferase and sulfurtransferase
Length=460
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 26/63 (41%), Positives = 39/63 (61%), Gaps = 1/63 (1%)
Query 57 YSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSN 116
YSR + L +GV + T VL+VG GG+G A+ L G+G++ L+D+D VE+SN
Sbjct 63 YSRQLVLPELGVHGQLRLGTA-CVLIVGCGGLGCPLAQYLAAAGVGRLGLVDYDVVEMSN 121
Query 117 MNR 119
+ R
Sbjct 122 LAR 124
> cel:F11H8.1 rfl-1; ectopic membrane RuFfLes in embryo family
member (rfl-1); K10686 ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=430
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 22/64 (34%), Positives = 39/64 (60%), Gaps = 1/64 (1%)
Query 71 DYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGM 129
+++A+ +L++G GG+G + L G I ++D D++++SN+NR F F DVG
Sbjct 36 NFEALQNTKILVIGAGGLGCELLKNLALSGFRTIEVIDMDTIDVSNLNRQFLFRESDVGK 95
Query 130 TKVE 133
+K E
Sbjct 96 SKAE 99
> dre:406776 uba3, ube1c, wu:fb75e04, wu:fc37b11, zgc:55528; ubiquitin-like
modifier activating enzyme 3 (EC:6.3.2.-); K10686
ubiquitin-activating enzyme E1 C [EC:6.3.2.19]
Length=462
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/82 (35%), Positives = 45/82 (54%), Gaps = 11/82 (13%)
Query 63 LQRMGVVR--DYQAITKK--------AVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSV 112
L+R G D++A T+ +L++G GG+G + L G I ++D D++
Sbjct 44 LERTGPFTHPDFEASTESLQFLLDTCKILVIGAGGLGCELLKDLALSGFRHIHVVDMDTI 103
Query 113 ELSNMNRLF-FTPKDVGMTKVE 133
++SN+NR F F PKDVG K E
Sbjct 104 DVSNLNRQFLFRPKDVGRPKAE 125
> sce:YDR390C UBA2, UAL1; Nuclear protein that acts as a heterodimer
with Aos1p to activate Smt3p (SUMO) before its conjugation
to proteins (sumoylation), which may play a role in protein
targeting; essential for viability; K10685 ubiquitin-like
1-activating enzyme E1 B [EC:6.3.2.19]
Length=636
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 23/61 (37%), Positives = 37/61 (60%), Gaps = 1/61 (1%)
Query 72 YQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMT 130
Y+ + LLVG GG+GS + ++ G+I ++D D+++LSN+NR F F KD+
Sbjct 16 YKKLRSSRCLLVGAGGIGSELLKDIILMEFGEIHIVDLDTIDLSNLNRQFLFRQKDIKQP 75
Query 131 K 131
K
Sbjct 76 K 76
> dre:406672 uba2, sae2, sae2b, uble1b, wu:fi17g06, zgc:66354;
ubiquitin-like modifier activating enzyme 2 (EC:6.3.2.-); K10685
ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=640
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/55 (41%), Positives = 35/55 (63%), Gaps = 1/55 (1%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMTKVE 133
VL+VG GG+G + LV G I ++D D++++SN+NR F F K VG +K +
Sbjct 21 VLVVGAGGIGCELLKNLVLTGFKNIEVIDLDTIDVSNLNRQFLFQKKHVGKSKAQ 75
> dre:100333133 ubiquitin-like modifier activating enzyme 2-like;
K10685 ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=642
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/55 (41%), Positives = 35/55 (63%), Gaps = 1/55 (1%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMTKVE 133
VL+VG GG+G + LV G I ++D D++++SN+NR F F K VG +K +
Sbjct 21 VLVVGAGGIGCELLKNLVLTGFKNIEVIDLDTIDVSNLNRQFLFQKKHVGKSKAQ 75
> xla:399235 uba2, MGC84651, Uble1b, sae2, sae2-B, uba2-a, uba2-b,
uble1b-B; ubiquitin-like modifier activating enzyme 2;
K10685 ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=641
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 39/62 (62%), Gaps = 1/62 (1%)
Query 73 QAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMTK 131
+A++ +L+VG GG+G + LV G + ++D D++++SN+NR F F K VG +K
Sbjct 13 EAVSASRLLVVGAGGIGCELLKNLVLTGFTNLDVIDLDTIDVSNLNRQFLFQKKHVGRSK 72
Query 132 VE 133
+
Sbjct 73 AQ 74
> ath:AT2G21470 SAE2; SAE2 (SUMO-ACTIVATING ENZYME 2); SUMO activating
enzyme; K10685 ubiquitin-like 1-activating enzyme E1
B [EC:6.3.2.19]
Length=625
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 36/67 (53%), Gaps = 1/67 (1%)
Query 66 MGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTP 124
M + AI VL+VG GG+G + L G I ++D D++E+SN+NR F F
Sbjct 1 MATQQQQSAIKGAKVLMVGAGGIGCELLKTLALSGFEDIHIIDMDTIEVSNLNRQFLFRR 60
Query 125 KDVGMTK 131
VG +K
Sbjct 61 SHVGQSK 67
> sce:YKL210W UBA1; Uba1p; K03178 ubiquitin-activating enzyme
E1 [EC:6.3.2.19]
Length=1024
Score = 45.4 bits (106), Expect = 5e-05, Method: Composition-based stats.
Identities = 29/70 (41%), Positives = 37/70 (52%), Gaps = 7/70 (10%)
Query 71 DYQA-ITKKAVLLVGVGGVGSVAAEMLVRCGIGK-----IILLDFDSVELSNMNRLF-FT 123
D+Q I V LVG G +G + G+G I++ D DS+E SN+NR F F
Sbjct 427 DFQKKIANSKVFLVGSGAIGCEMLKNWALLGLGSGSDGYIVVTDNDSIEKSNLNRQFLFR 486
Query 124 PKDVGMTKVE 133
PKDVG K E
Sbjct 487 PKDVGKNKSE 496
Score = 34.7 bits (78), Expect = 0.080, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 34/53 (64%), Gaps = 1/53 (1%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLFF-TPKDVGMTK 131
VL++G+ G+G A+ +V G+ + + D + V+L++++ FF T KD+G +
Sbjct 39 VLILGLKGLGVEIAKNVVLAGVKSMTVFDPEPVQLADLSTQFFLTEKDIGQKR 91
> tgo:TGME49_111500 ubiquitin-activating enzyme, putative (EC:1.2.1.70);
K10685 ubiquitin-like 1-activating enzyme E1 B [EC:6.3.2.19]
Length=730
Score = 45.1 bits (105), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 35/54 (64%), Gaps = 1/54 (1%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNR-LFFTPKDVGMTKV 132
VL+VG GG+G + L+ G ++ ++D D++++SN+NR FF VG++K
Sbjct 39 VLVVGAGGIGCEVCKDLLLSGFRRLCVVDLDTIDVSNLNRQFFFRNAHVGLSKA 92
> cpv:cgd2_1120 molybdopterin synthase sulphurylase ; K11996 adenylyltransferase
and sulfurtransferase
Length=495
Score = 44.7 bits (104), Expect = 7e-05, Method: Composition-based stats.
Identities = 32/98 (32%), Positives = 48/98 (48%), Gaps = 1/98 (1%)
Query 29 RKDAESQTEGGLRPKVLSMSAEVRADNPYSRLMALQRMGVVRDYQAITKKAVLLVGVGGV 88
+ D E T+ + + S S YSR +AL+ +GV + K VL++G GG+
Sbjct 32 KSDGEVNTKKDIFETINSPSLSEENVIRYSRQIALKEVGVSGQVKLKNAK-VLVIGAGGL 90
Query 89 GSVAAEMLVRCGIGKIILLDFDSVELSNMNRLFFTPKD 126
GS L GIG I ++D D+V SN++R D
Sbjct 91 GSPILLYLTGAGIGVIGVVDHDTVSTSNLHRQIIHSTD 128
> ath:AT5G37530 thiF family protein
Length=456
Score = 44.7 bits (104), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 23/43 (53%), Positives = 31/43 (72%), Gaps = 1/43 (2%)
Query 90 SVAAEMLVRCGIGKIILLDFDSVELSNMNR-LFFTPKDVGMTK 131
S AA ML+R G+GK++L+DFD V LS++NR T DVG+ K
Sbjct 105 SHAASMLLRSGVGKLLLVDFDQVSLSSLNRHAVATRADVGIPK 147
> dre:100001302 ubiquitin-like modifier-activating enzyme 1-like;
K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1016
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 37/107 (34%), Positives = 51/107 (47%), Gaps = 15/107 (14%)
Query 35 QTEGGLRPKVLSMSAEVRADNPYSRLMALQRMGVVRDYQ-AITKKAVLLVGVGGVGSVAA 93
Q EGG VLS A D+ Y +A+ D+Q + K+ LVG G +G
Sbjct 391 QEEGG----VLSEDACAPRDSRYDGQIAV----FGSDFQNKLKKQKYFLVGAGAIGCELL 442
Query 94 EMLVRCGIG-----KIILLDFDSVELSNMNRLF-FTPKDVGMTKVEA 134
+ G+G I + D DS+E SN+NR F F +D+G K EA
Sbjct 443 KNFALIGLGAGEGGSITVTDMDSIERSNLNRQFLFRSQDIGRPKSEA 489
> cel:F42G8.6 hypothetical protein; K11996 adenylyltransferase
and sulfurtransferase
Length=402
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 43/79 (54%), Gaps = 2/79 (2%)
Query 57 YSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSN 116
YSR + + GV + + VL+VG GG+G A L GIG I ++D+D + L N
Sbjct 18 YSRQLLVDDFGV-SGQKNLKNLNVLIVGAGGLGCPVATYLGAAGIGTIGIVDYDHISLDN 76
Query 117 MNRLFFTPKD-VGMTKVEA 134
++R +D VG +K +A
Sbjct 77 LHRQVAYKEDQVGKSKAQA 95
> tpv:TP01_0601 hypothetical protein
Length=343
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/60 (43%), Positives = 35/60 (58%), Gaps = 1/60 (1%)
Query 75 ITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGMTKVE 133
+ K +L+VG GG+G + LV G I ++DFD V LSN+NR F F DVG K +
Sbjct 4 VLKSRILVVGSGGLGCELLKSLVLNGFENISIVDFDKVVLSNLNRQFLFQKNDVGKFKSQ 63
> tgo:TGME49_012100 molybdenum cofactor synthesis protein, putative
Length=1082
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 26/63 (41%), Positives = 38/63 (60%), Gaps = 1/63 (1%)
Query 57 YSRLMALQRMGVVRDYQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSN 116
+SR + L G+ + T +VL+VG GG+G AA L GIG++ L+D D VE+SN
Sbjct 287 FSRQVLLPSWGLAAQRRLRTS-SVLIVGCGGLGCPAALYLCAGGIGRLGLVDGDDVEVSN 345
Query 117 MNR 119
+ R
Sbjct 346 LQR 348
> cel:W02A11.4 uba-2; UBA (human ubiquitin) related family member
(uba-2); K10685 ubiquitin-like 1-activating enzyme E1 B
[EC:6.3.2.19]
Length=582
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 17/55 (30%), Positives = 34/55 (61%), Gaps = 0/55 (0%)
Query 72 YQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRLFFTPKD 126
++ I + +L++G GG+G + L G K+ ++D D++++SN+NR F K+
Sbjct 8 HEKIVQSKILVIGAGGIGCELLKNLAVTGFRKVHVIDLDTIDISNLNRQFLFRKE 62
> tpv:TP02_0331 ubiquitin activating enzyme, putatuve
Length=1126
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 36/62 (58%), Gaps = 1/62 (1%)
Query 73 QAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNR-LFFTPKDVGMTK 131
+ + K + L+VG G +G ++L G+ + + D D+V++SN+ R + FT DVG K
Sbjct 485 EQVAKMSFLVVGAGALGCDYLKLLAEMGVSDVTVFDNDTVDVSNLTRQVLFTINDVGKPK 544
Query 132 VE 133
+
Sbjct 545 AQ 546
> dre:406335 uba1, ube1, wu:fa01e08, wu:fb30f01, wu:fi21c11, wu:fj14g11,
zgc:66143; ubiquitin-like modifier activating enzyme
1 (EC:6.3.2.19); K03178 ubiquitin-activating enzyme E1 [EC:6.3.2.19]
Length=1058
Score = 41.6 bits (96), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 35/65 (53%), Gaps = 5/65 (7%)
Query 75 ITKKAVLLVGVGGVGSV----AAEMLVRCGIGKIILLDFDSVELSNMNRLF-FTPKDVGM 129
+ K+ LVG G +G A M + G G++I+ D D++E SN+NR F F P DV
Sbjct 467 LAKQRYFLVGAGAIGCELLKNFAMMGLASGEGEVIVTDMDTIEKSNLNRQFLFRPWDVTK 526
Query 130 TKVEA 134
K E
Sbjct 527 MKSET 531
> bbo:BBOV_III005870 17.m07520; ThiF family protein
Length=1009
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 25/62 (40%), Positives = 37/62 (59%), Gaps = 2/62 (3%)
Query 75 ITKKA-VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNMNRL-FFTPKDVGMTKV 132
I K+A L+VGVG +G ++L G+ + +D DSV++SN+ R FT DVG+ K
Sbjct 386 IVKEAEYLMVGVGALGCEYLKILEAMGVEHLTAMDNDSVDVSNLTRQSLFTDADVGLNKA 445
Query 133 EA 134
A
Sbjct 446 TA 447
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 19/47 (40%), Positives = 24/47 (51%), Gaps = 2/47 (4%)
Query 80 VLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVELSNM--NRLFFTP 124
VL+VG V S A L+R G+G I ++D D NM N L P
Sbjct 4 VLIVGSTPVASNIATHLIRAGVGTIYVVDDDVYNAKNMVENVLILHP 50
> bbo:BBOV_IV003870 23.m05802; molybdenum cofactor synthesis protein
3 / molybdopterin synthase sulphurylase; K11996 adenylyltransferase
and sulfurtransferase
Length=502
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 27/79 (34%), Positives = 45/79 (56%), Gaps = 5/79 (6%)
Query 58 SRLMALQRMGVVRD----YQAITKKAVLLVGVGGVGSVAAEMLVRCGIGKIILLDFDSVE 113
++ +ALQ+ ++ A++ AVL++G GG+GS L GIG I ++D D VE
Sbjct 90 AQYIALQKCDAEKNAAHIVDAVSTCAVLVIGAGGLGSPLLLYLAAGGIGIIGVMDGDVVE 149
Query 114 LSNMNR-LFFTPKDVGMTK 131
+SN++R + + GM K
Sbjct 150 VSNLHRQIIHDEESRGMNK 168
> mmu:22201 Uba1, A1S9, AA989744, Sbx, Ube-1, Ube1x; ubiquitin-like
modifier activating enzyme 1 (EC:6.3.2.19); K03178 ubiquitin-activating
enzyme E1 [EC:6.3.2.19]
Length=1058
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 38/71 (53%), Gaps = 7/71 (9%)
Query 71 DYQA-ITKKAVLLVGVGGVGSVAAEMLVRCGIG-----KIILLDFDSVELSNMNRLF-FT 123
D+Q ++K+ LVG G +G + G+G ++++ D D++E SN+NR F F
Sbjct 461 DFQEKLSKQKYFLVGAGAIGCELLKNFAMIGLGCGEGGEVVVTDMDTIEKSNLNRQFLFR 520
Query 124 PKDVGMTKVEA 134
P DV K +
Sbjct 521 PWDVTKLKSDT 531
Lambda K H
0.321 0.137 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2231140792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40