bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1091_orf1
Length=179
Score E
Sequences producing significant alignments: (Bits) Value
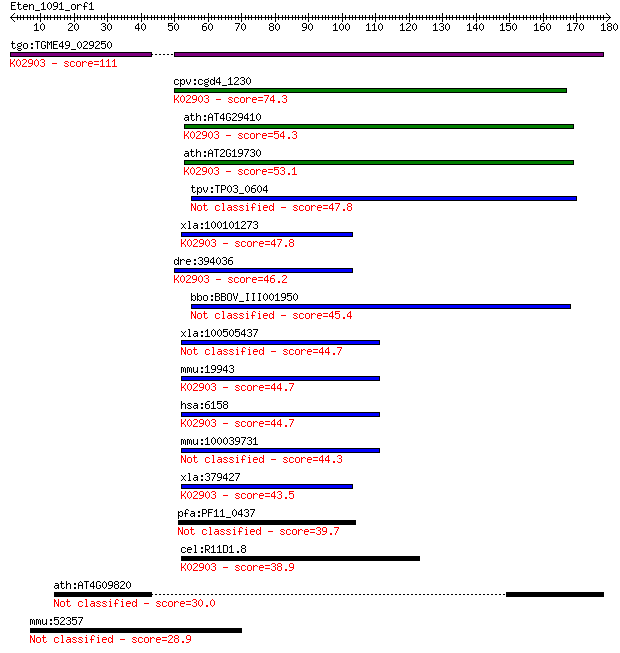
tgo:TGME49_029250 60S ribosomal protein L28, putative ; K02903... 111 1e-24
cpv:cgd4_1230 60S ribosomal protein L28 ; K02903 large subunit... 74.3 2e-13
ath:AT4G29410 60S ribosomal protein L28 (RPL28C); K02903 large... 54.3 2e-07
ath:AT2G19730 60S ribosomal protein L28 (RPL28A); K02903 large... 53.1 5e-07
tpv:TP03_0604 60S ribosomal protein L28 47.8 2e-05
xla:100101273 rpl28; ribosomal protein L28 (EC:3.6.5.3); K0290... 47.8 2e-05
dre:394036 rpl28, MGC66289, rpl28l, zgc:66289; ribosomal prote... 46.2 6e-05
bbo:BBOV_III001950 17.m07191; hypothetical protein 45.4 1e-04
xla:100505437 hypothetical protein LOC100505437 44.7 2e-04
mmu:19943 Rpl28, D7Wsu21e, MGC107666, MGC115078; ribosomal pro... 44.7 2e-04
hsa:6158 RPL28, FLJ43307; ribosomal protein L28; K02903 large ... 44.7 2e-04
mmu:100039731 Gm10051; predicted pseudogene 10051 44.3
xla:379427 hypothetical protein MGC64430; K02903 large subunit... 43.5 3e-04
pfa:PF11_0437 60S ribosomal protein L28, putative 39.7 0.005
cel:R11D1.8 rpl-28; Ribosomal Protein, Large subunit family me... 38.9 0.009
ath:AT4G09820 TT8; TT8 (TRANSPARENT TESTA 8); DNA binding / tr... 30.0 4.7
mmu:52357 Wwc2, 5330431C14Rik, AU022508, C85457, D8Ertd594e; W... 28.9 8.9
> tgo:TGME49_029250 60S ribosomal protein L28, putative ; K02903
large subunit ribosomal protein L28e
Length=129
Score = 111 bits (278), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 57/129 (44%), Positives = 82/129 (63%), Gaps = 1/129 (0%)
Query 50 MASAELLWQCVRRTSCFLKKSHGIVLTREPLNLTQKNTLRHSGLCHPRPLGLQLVG-NNA 108
M S+ELLWQCVRR CF++K +GI L+ E +NLT KNTL++SG+ H +PLGL G NN
Sbjct 1 MVSSELLWQCVRRNHCFIRKFNGITLSAERMNLTNKNTLKYSGIAHKQPLGLNRHGANNG 60
Query 109 AVKLSYRPKNPNKARFPRRALCSKKFPKSHSKTKQLQQLVAQQRPDLVRLAKKRFHKLCK 168
+ L K R P RA+ +KF KS + ++ + VA++RPDL+++A K+ KL +
Sbjct 61 CIALVTVQKCSRAMRKPTRAVQVRKFKKSKKEMSKVMKAVAERRPDLLKVANKKMKKLIR 120
Query 169 TSPKPAQAE 177
T + E
Sbjct 121 TGNSSNKKE 129
Score = 30.4 bits (67), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 26/42 (61%), Gaps = 0/42 (0%)
Query 1 KSHSKTKQLQQLVAQQRPDLVRLAKKRFHKLCKTSPKPAQAE 42
KS + ++ + VA++RPDL+++A K+ KL +T + E
Sbjct 88 KSKKEMSKVMKAVAERRPDLLKVANKKMKKLIRTGNSSNKKE 129
> cpv:cgd4_1230 60S ribosomal protein L28 ; K02903 large subunit
ribosomal protein L28e
Length=127
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 64/122 (52%), Gaps = 5/122 (4%)
Query 50 MASAELLWQCVRRTSCFLKKSHGIVLTREPLNLTQKNTLRHSGLCHPRPLGLQLVGNNAA 109
M S ELLWQCV++ + F+KK++G T EP N+ NTL++SGL R + + LV N+
Sbjct 1 MFSTELLWQCVKKNTSFIKKNNGFTFTSEPFNIMNLNTLKYSGLAARRSISMDLVPNSNG 60
Query 110 -----VKLSYRPKNPNKARFPRRALCSKKFPKSHSKTKQLQQLVAQQRPDLVRLAKKRFH 164
K++ + + R P++ + + + K+ + RPDL +A R+
Sbjct 61 KLVKNAKITKKNTGSSTIRCPKKLISKVNLSNNFKQAKKTIKNQISSRPDLKNVAIIRYR 120
Query 165 KL 166
K+
Sbjct 121 KI 122
> ath:AT4G29410 60S ribosomal protein L28 (RPL28C); K02903 large
subunit ribosomal protein L28e
Length=143
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 36/123 (29%), Positives = 62/123 (50%), Gaps = 8/123 (6%)
Query 53 AELLWQCVRRTSCFLKKSHG-----IVLTREPLNLTQKNTLRHSGLCHPRPLGLQLVGNN 107
+L+W+ V+R +CFL K G + ++E NL N+ +HSGL + + + +Q G +
Sbjct 6 GQLIWEIVKRNNCFLVKQFGRGNAKVQFSKESNNLVNINSYKHSGLANKKTVTIQAAGKD 65
Query 108 AAVKL-SYRPKNPNKARFP-RRALCSKKFPKSHSKTKQLQQLVAQQRPDLVRLAKKRFHK 165
V L + + K NK + +++ K+F + SK Q + RPDL + A R
Sbjct 66 QGVVLGTTKTKRQNKPKLSVNKSILKKEFSR-MSKVVANQVVDNYYRPDLKKAALARLSA 124
Query 166 LCK 168
+ K
Sbjct 125 ISK 127
> ath:AT2G19730 60S ribosomal protein L28 (RPL28A); K02903 large
subunit ribosomal protein L28e
Length=143
Score = 53.1 bits (126), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 36/123 (29%), Positives = 63/123 (51%), Gaps = 8/123 (6%)
Query 53 AELLWQCVRRTSCFLKKSHG-----IVLTREPLNLTQKNTLRHSGLCHPRPLGLQLVGNN 107
+L+W+ V+ +CFL K G + ++E NLT ++ +HSGL + + + +Q +
Sbjct 6 GQLIWEIVKNNNCFLVKQFGRGNSKVQFSKETNNLTNVHSYKHSGLANKKTVTIQAADKD 65
Query 108 AAVKL-SYRPKNPNKARFP-RRALCSKKFPKSHSKTKQLQQLVAQQRPDLVRLAKKRFHK 165
AV L + + K NK + +++ K+FP+ SK Q + RPDL + A R
Sbjct 66 QAVVLATTKTKKQNKPKLSVNKSILKKEFPR-MSKAVANQVVDNYYRPDLKKAALARLSA 124
Query 166 LCK 168
+ K
Sbjct 125 ISK 127
> tpv:TP03_0604 60S ribosomal protein L28
Length=140
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 33/115 (28%), Positives = 55/115 (47%), Gaps = 2/115 (1%)
Query 55 LLWQCVRRTSCFLKKSHGIVLTREPLNLTQKNTLRHSGLCHPRPLGLQLVGNNAAVKLSY 114
LLW+ +R FLKKS + E NL ++ R +G + +P +V N+ A +++
Sbjct 20 LLWELLRNNHAFLKKSRTTTFSLESHNLLSRHCPRFTGYTN-KP-SYNVVLNSTASLVTF 77
Query 115 RPKNPNKARFPRRALCSKKFPKSHSKTKQLQQLVAQQRPDLVRLAKKRFHKLCKT 169
+ N +R P + K KS ++ V + RPDL L ++F L K+
Sbjct 78 KKNNLKNSRRPSSKISVKTLIKSLKLLDSVKTTVRKDRPDLTELLARKFTMLVKS 132
> xla:100101273 rpl28; ribosomal protein L28 (EC:3.6.5.3); K02903
large subunit ribosomal protein L28e
Length=137
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 31/51 (60%), Gaps = 0/51 (0%)
Query 52 SAELLWQCVRRTSCFLKKSHGIVLTREPLNLTQKNTLRHSGLCHPRPLGLQ 102
SA L W +R S F+ K + V + EP NL +N+ R++GL H + +G+Q
Sbjct 2 SAHLQWMVIRNCSSFIVKRNHQVFSTEPNNLKARNSFRYNGLIHKKTVGVQ 52
> dre:394036 rpl28, MGC66289, rpl28l, zgc:66289; ribosomal protein
L28; K02903 large subunit ribosomal protein L28e
Length=138
Score = 46.2 bits (108), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 30/53 (56%), Gaps = 0/53 (0%)
Query 50 MASAELLWQCVRRTSCFLKKSHGIVLTREPLNLTQKNTLRHSGLCHPRPLGLQ 102
MAS L W +R +S FL K + + EP NL +N R +GL H + +G++
Sbjct 1 MASPHLQWMVIRNSSSFLIKRNKQTYSTEPNNLKARNAFRFNGLIHRKTIGIE 53
> bbo:BBOV_III001950 17.m07191; hypothetical protein
Length=146
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/113 (23%), Positives = 57/113 (50%), Gaps = 2/113 (1%)
Query 55 LLWQCVRRTSCFLKKSHGIVLTREPLNLTQKNTLRHSGLCHPRPLGLQLVGNNAAVKLSY 114
L+W+ +++ + ++ KS T+E NL ++T +SGL + L+L + + + L+
Sbjct 27 LVWELIKKNNAYIFKSGHETFTKERYNLMARHTPMYSGLMQNSVMNLELSPSGSKIVLT- 85
Query 115 RPKNPNKARFPRRALCSKKFPKSHSKTKQLQQLVAQQRPDLVRLAKKRFHKLC 167
+ AR PR +K K+ ++ + L+ RP+L + ++F K+
Sbjct 86 -SMDLQNARQPRNKTQTKNLDKNPARVEFLKNFAYPSRPELAEVLAEKFIKMT 137
> xla:100505437 hypothetical protein LOC100505437
Length=137
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 32/59 (54%), Gaps = 0/59 (0%)
Query 52 SAELLWQCVRRTSCFLKKSHGIVLTREPLNLTQKNTLRHSGLCHPRPLGLQLVGNNAAV 110
SA L W VR S FL K + + EP NL +N+ R++GL H + +G++ + V
Sbjct 2 SAHLQWMVVRNCSSFLIKRNKQTYSTEPNNLKARNSFRYNGLIHRKTVGVEPAADGKGV 60
> mmu:19943 Rpl28, D7Wsu21e, MGC107666, MGC115078; ribosomal protein
L28 (EC:3.6.5.3); K02903 large subunit ribosomal protein
L28e
Length=137
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 32/59 (54%), Gaps = 0/59 (0%)
Query 52 SAELLWQCVRRTSCFLKKSHGIVLTREPLNLTQKNTLRHSGLCHPRPLGLQLVGNNAAV 110
SA L W VR S FL K + + EP NL +N+ R++GL H + +G++ + V
Sbjct 2 SAHLQWMVVRNCSSFLIKRNKQTYSTEPNNLKARNSFRYNGLIHRKTVGVEPAADGKGV 60
> hsa:6158 RPL28, FLJ43307; ribosomal protein L28; K02903 large
subunit ribosomal protein L28e
Length=137
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 32/59 (54%), Gaps = 0/59 (0%)
Query 52 SAELLWQCVRRTSCFLKKSHGIVLTREPLNLTQKNTLRHSGLCHPRPLGLQLVGNNAAV 110
SA L W VR S FL K + + EP NL +N+ R++GL H + +G++ + V
Sbjct 2 SAHLQWMVVRNCSSFLIKRNKQTYSTEPNNLKARNSFRYNGLIHRKTVGVEPAADGKGV 60
> mmu:100039731 Gm10051; predicted pseudogene 10051
Length=137
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/59 (35%), Positives = 32/59 (54%), Gaps = 0/59 (0%)
Query 52 SAELLWQCVRRTSCFLKKSHGIVLTREPLNLTQKNTLRHSGLCHPRPLGLQLVGNNAAV 110
SA L W VR S FL K + + EP NL +N+ R++GL H + +G++ + V
Sbjct 2 SAHLQWMVVRNCSSFLIKRNKQTYSTEPNNLKARNSFRYNGLIHRKTVGVEPAADGKGV 60
> xla:379427 hypothetical protein MGC64430; K02903 large subunit
ribosomal protein L28e
Length=137
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 30/51 (58%), Gaps = 0/51 (0%)
Query 52 SAELLWQCVRRTSCFLKKSHGIVLTREPLNLTQKNTLRHSGLCHPRPLGLQ 102
SA L W VR S F+ K + V + EP NL +N+ R++G H + +G++
Sbjct 2 SAHLQWMIVRNCSSFIVKRNHQVFSTEPNNLKARNSFRYNGPIHKKTVGVE 52
> pfa:PF11_0437 60S ribosomal protein L28, putative
Length=127
Score = 39.7 bits (91), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 31/57 (54%), Gaps = 5/57 (8%)
Query 51 ASAELLWQCVRRTSCFLKKS----HGIVLTREPLNLTQKNTLRHSGLCHPRPLGLQL 103
S L+W+ R+++CF+KK+ G+ L +PLN+ KNT SGL + L
Sbjct 4 VSNALVWELTRKSNCFIKKNKAGKKGVFLC-DPLNVNYKNTPSSSGLVKSNSTNVTL 59
> cel:R11D1.8 rpl-28; Ribosomal Protein, Large subunit family
member (rpl-28); K02903 large subunit ribosomal protein L28e
Length=126
Score = 38.9 bits (89), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 32/73 (43%), Gaps = 2/73 (2%)
Query 52 SAELLWQCVRRTSCFLKKSHGIV--LTREPLNLTQKNTLRHSGLCHPRPLGLQLVGNNAA 109
S L+WQ +R S FL+ GI + E NL + N+ ++SGL + + +
Sbjct 2 SDALVWQVIRNNSAFLRTQRGIGKRFSTEKFNLKKVNSPKYSGLANKHAIDVSAAAKGVV 61
Query 110 VKLSYRPKNPNKA 122
V P KA
Sbjct 62 VSTKNEKGRPAKA 74
> ath:AT4G09820 TT8; TT8 (TRANSPARENT TESTA 8); DNA binding /
transcription factor
Length=518
Score = 30.0 bits (66), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 14 AQQRPDLVRLAKKRFHKLCKTSPKPAQAE 42
++ + V L K F+ CKT+PKPA +E
Sbjct 187 VREDVEFVELTKSFFYDHCKTNPKPALSE 215
Score = 30.0 bits (66), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 149 AQQRPDLVRLAKKRFHKLCKTSPKPAQAE 177
++ + V L K F+ CKT+PKPA +E
Sbjct 187 VREDVEFVELTKSFFYDHCKTNPKPALSE 215
> mmu:52357 Wwc2, 5330431C14Rik, AU022508, C85457, D8Ertd594e;
WW, C2 and coiled-coil domain containing 2
Length=1187
Score = 28.9 bits (63), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 39/65 (60%), Gaps = 3/65 (4%)
Query 7 KQLQQLVAQQRPDL--VRLAKKRFHKLCKTSPKPAQAEQQQQQQKMASAELLWQCVRRTS 64
++L++L AQ DL L +RF KL K + K A+ +++Q+Q + +AE L + V +
Sbjct 1090 QKLEELKAQGETDLPPGVLEDERFQKLLKQAEKQAEQTKEEQKQDL-NAERLMRQVSKDV 1148
Query 65 CFLKK 69
C L++
Sbjct 1149 CRLRE 1153
Lambda K H
0.318 0.128 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4795148792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40