bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1105_orf1
Length=338
Score E
Sequences producing significant alignments: (Bits) Value
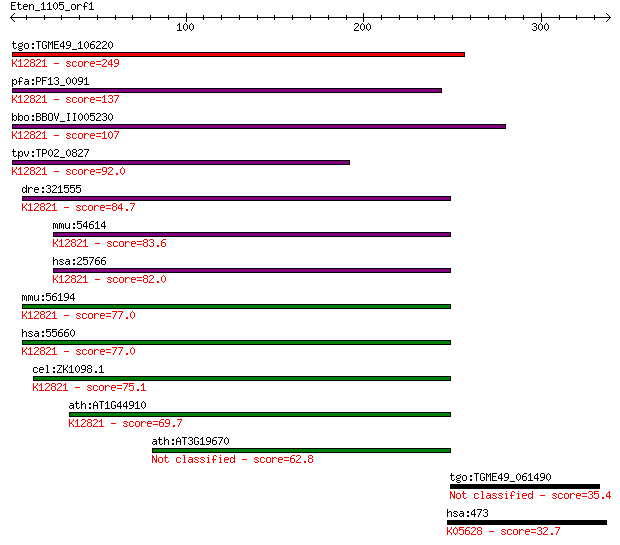
tgo:TGME49_106220 formin binding protein, putative ; K12821 pr... 249 1e-65
pfa:PF13_0091 conserved Plasmodium protein, unknown function; ... 137 7e-32
bbo:BBOV_II005230 18.m06432; WW domain containing protein; K12... 107 7e-23
tpv:TP02_0827 hypothetical protein; K12821 pre-mRNA-processing... 92.0 3e-18
dre:321555 prpf40a, fb18h07, fe47a12, fnbp3, wu:fb18h07, wu:fe... 84.7 5e-16
mmu:54614 Prpf40b, 2610317D23Rik, Hypc, MGC91243; PRP40 pre-mR... 83.6 1e-15
hsa:25766 PRPF40B, HYPC; PRP40 pre-mRNA processing factor 40 h... 82.0 3e-15
mmu:56194 Prpf40a, 2810012K09Rik, FBP11, Fnbp11, Fnbp3; PRP40 ... 77.0 9e-14
hsa:55660 PRPF40A, FBP-11, FBP11, FLAF1, FLJ20585, FNBP3, HIP-... 77.0 1e-13
cel:ZK1098.1 hypothetical protein; K12821 pre-mRNA-processing ... 75.1 4e-13
ath:AT1G44910 protein binding; K12821 pre-mRNA-processing fact... 69.7 1e-11
ath:AT3G19670 protein binding 62.8 2e-09
tgo:TGME49_061490 hypothetical protein 35.4 0.35
hsa:473 RERE, ARG, ARP, ATN1L, DNB1, FLJ38775, KIAA0458; argin... 32.7 2.1
> tgo:TGME49_106220 formin binding protein, putative ; K12821
pre-mRNA-processing factor 40
Length=601
Score = 249 bits (636), Expect = 1e-65, Method: Compositional matrix adjust.
Identities = 117/255 (45%), Positives = 178/255 (69%), Gaps = 4/255 (1%)
Query 2 LLAWKELSFKTTYIDVADRFHFEEWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRRED 61
L W+EL+ TTYI +AD+ H +EWWT++ E ERD+FFQ++M + ++ ++ K++R++D
Sbjct 262 LQNWEELAPGTTYIAMADKMHEQEWWTFLTEQERDDFFQDYMEEFDKRHRELFKKKRKKD 321
Query 62 VAMLEEILEQNPTEFAFQKRWSEVREQLFSHPKLQNMMKIDVLQVWEEWVRHGYEQERKQ 121
V +E+IL++ EF ++++W +VR++LF+ P+L ++++D+LQVWE WV HGY ER+
Sbjct 322 VETVEKILDKRSAEFDYRRKWVDVRDELFAIPELSTVLRLDILQVWENWVEHGYADERRN 381
Query 122 RRAQIFRRERKRRDAFRELLQEAVDKGELSSKSEWVGFARRVEKDSRYLAMVGQSGSTPR 181
RR +FRRERKRRDAFR LL EA KGEL++K+EW F ++ D RY MVGQ GSTPR
Sbjct 382 RRHVVFRRERKRRDAFRSLLDEAAKKGELTAKTEWPDFVAQIVNDPRYYQMVGQGGSTPR 441
Query 182 ELFSDAVQQLQQQHERLKHFFKRFADREGLDLRDKALSFTQFYETVRGLDEVKNLNMLHV 241
ELF DAV L+++++R K R GL+L +L+F +FY + + +K +++++
Sbjct 442 ELFEDAVDNLKEEYQRQKPVILDCLKRAGLELDSPSLTFEEFYSALCTCEGMKGVSLMNA 501
Query 242 KLVFESLKNADTKPA 256
KL FESL T PA
Sbjct 502 KLTFESL----TSPA 512
> pfa:PF13_0091 conserved Plasmodium protein, unknown function;
K12821 pre-mRNA-processing factor 40
Length=906
Score = 137 bits (344), Expect = 7e-32, Method: Compositional matrix adjust.
Identities = 75/248 (30%), Positives = 140/248 (56%), Gaps = 10/248 (4%)
Query 2 LLAWKELSFKTTYIDVADRFHFEEWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRRED 61
LL W +L+ TTY++ A +F+ +EWW W+ E ERDE FQ++M + FK+ +++R++
Sbjct 517 LLNWDKLNECTTYVEFASQFYKQEWWEWITEKERDEVFQDFMDGYKSKFKETRRKKRKQK 576
Query 62 VAMLEEILEQNPTEFAFQKRWSEVREQLFSHPKLQNMMKIDVLQVWEEWVRHGYEQERKQ 121
+ +L++ ++ T+ +W++V++ ++ KID L WE++ + E+ +
Sbjct 577 MEILKQKFQEYATDNKNPLKWNDVQKYFRDDEDFHSLHKIDALAAWEDFYEKYHNVEKMK 636
Query 122 RRAQIFRRERKRRDAFRELLQEAVDKGELSSKSEWVGFARRVEKDSRYLAMVGQSGSTPR 181
+ +I+R RK+R+AF ELL E + L+ K++W+ F ++ KD+RY ++G GS+PR
Sbjct 637 LKKKIYRILRKKREAFIELLNEYYENNILNMKTQWIFFVSKIYKDTRYTDILGHQGSSPR 696
Query 182 ELFSDAVQQLQQQHERLKHFFKR------FADREGLDLRDKALSFTQFYETVRGLDEVKN 235
LF + + LQ+Q+ K + K+ F E + L D F + + V+ + +
Sbjct 697 ILFDEFIDSLQEQYLIHKSYIKKAYKEMDFTIDENITLED----FLKTFSNVQSKYNIPD 752
Query 236 LNMLHVKL 243
NM + L
Sbjct 753 ANMNFIYL 760
> bbo:BBOV_II005230 18.m06432; WW domain containing protein; K12821
pre-mRNA-processing factor 40
Length=457
Score = 107 bits (267), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 79/279 (28%), Positives = 141/279 (50%), Gaps = 12/279 (4%)
Query 2 LLAWKELSFKTTYIDVADRFHFEEWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRRED 61
L W+EL+ TTY++ A+R H EWWTW E RD FQE M + K++ ++RRR
Sbjct 173 LEKWEELTPYTTYVEFAERCHTREWWTWADEKTRDGIFQETMERMDHELKERQRERRRVS 232
Query 62 VAMLEEILEQNPTEFAFQKRWSEVREQLFSHPKLQNMMKIDVLQVWEEWVRHGYEQERKQ 121
+ LE +E+ T+ Q W V+ Q + + +D+L+ + + Y K+
Sbjct 233 MEKLEAEMEKLVTDDMPQ--WPNVKRQFTG---FEGLHLLDILECHQVVFKRHYRNHVKE 287
Query 122 RRAQIFRRERKRRDAFRELLQEAVDKGELSSKSEWVGFARRVEKDSRYLAMVGQSGSTPR 181
+ +R +RKRR F L++ V KG ++ ++ + F + ++ YL +VGQ GSTP
Sbjct 288 AEKRAYRAQRKRRQHFVAFLEDCVAKGTINGRTVFEDFIKAHSTEAMYLDIVGQPGSTPY 347
Query 182 ELFSDAVQQLQQQHERLKHFFKRFADREGLDLRDKALSFTQFYETVRGLDEVKNLNMLHV 241
+LF D +++Q++ + K+ ++ L D+ S + G K + +V
Sbjct 348 DLFKDVHSPIRKQYKIERENVKQLINQGIL---DRGASLADYERICLG---NKACSPENV 401
Query 242 KLVFESL-KNADTKPAAAAAAAADGKQRSSRSSRTDPDA 279
L+ ESL + ++ + +A +G+ R+S S + D+
Sbjct 402 ALIHESLRRRLHSERRNRSLSAEEGEIRNSDVSESSRDS 440
> tpv:TP02_0827 hypothetical protein; K12821 pre-mRNA-processing
factor 40
Length=390
Score = 92.0 bits (227), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 61/190 (32%), Positives = 99/190 (52%), Gaps = 5/190 (2%)
Query 2 LLAWKELSFKTTYIDVADRFHFEEWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRRED 61
LL W+ELS+ T Y D + +FH EWW W E RD FQE+M E K K K+ +
Sbjct 171 LLNWEELSYATVYADFSKQFHTAEWWDWGDEVTRDAIFQEFMEREENKLKKKRKETKIAA 230
Query 62 VAMLEEILEQNPTEFAFQKRWSEVREQLFSHPKLQNMMKIDVLQVWEEWVRHGYEQERKQ 121
+ L +++ ++ T+ W + + + Q + IDVL + + + Q+ K+
Sbjct 231 MDTLIDVMTKDYTQELIP--WETAKAK---YLGFQGLYNIDVLNSHKYVFKEVFAQKYKE 285
Query 122 RRAQIFRRERKRRDAFRELLQEAVDKGELSSKSEWVGFARRVEKDSRYLAMVGQSGSTPR 181
+ FR +RK R F LQ V+KGE+ +++ F ++ Y+ +VGQ+GSTP
Sbjct 286 AQRTSFRLQRKIRQRFLTFLQTMVEKGEIDKNTKFSDFISNHSTEAVYVDLVGQAGSTPI 345
Query 182 ELFSDAVQQL 191
+LF++ Q L
Sbjct 346 DLFTEIQQSL 355
> dre:321555 prpf40a, fb18h07, fe47a12, fnbp3, wu:fb18h07, wu:fe47a12;
PRP40 pre-mRNA processing factor 40 homolog A (yeast);
K12821 pre-mRNA-processing factor 40
Length=851
Score = 84.7 bits (208), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 70/250 (28%), Positives = 123/250 (49%), Gaps = 11/250 (4%)
Query 8 LSFKTTYIDVADRFHFEEWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRREDVAMLEE 67
++ T Y F +E W+ + E +R E +++ +F + K++ KQ R+ + L+
Sbjct 371 MTSTTRYKKAEQMFGDQEVWSCVPERDRLEIYEDVLFYLAKKEKEQAKQLRKRNWEALKN 430
Query 68 ILEQNPTEFAFQKRWSEVREQLFSHP------KLQNMMKIDVLQVWEEWVRHGYEQERKQ 121
IL+ N ++ WSE ++ L +P +LQNM K D L +EE +R ++E +
Sbjct 431 ILD-NMANVTYRTTWSEAQQYLLDNPTFAEDEELQNMDKEDALICFEEHIRALEKEEEDE 489
Query 122 RRAQIFRRERKRR---DAFRELLQEAVDKGELSSKSEWVGFARRVEKDSRYLAMVGQSGS 178
++ + R R++R ++F++ L E D G+L S S W+ + D R+ M+GQ GS
Sbjct 490 KQKTLLRERRRQRKNRESFQKFLDELHDHGQLHSMSAWMEMYPTISADIRFSNMLGQPGS 549
Query 179 TPRELFSDAVQQLQQQHERLKHFFKRFADREGLDLRDKALSFTQFYETVRGLDEVKNLNM 238
TP +LF V+ L+ ++ K K +G L + F F + L+
Sbjct 550 TPLDLFKFYVEDLKARYHDEKRIIKDILKDKGF-LVEINTGFEDFGSVISSDKRATTLDA 608
Query 239 LHVKLVFESL 248
++KL F SL
Sbjct 609 GNIKLAFNSL 618
> mmu:54614 Prpf40b, 2610317D23Rik, Hypc, MGC91243; PRP40 pre-mRNA
processing factor 40 homolog B (yeast); K12821 pre-mRNA-processing
factor 40
Length=873
Score = 83.6 bits (205), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 71/233 (30%), Positives = 112/233 (48%), Gaps = 11/233 (4%)
Query 25 EWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRREDVAMLEEILEQNPTEFAFQKRWSE 84
E W + E ER E + + +F + K++ KQ RR ++ L+ IL+ + FQ WS+
Sbjct 377 EVWAVVPERERKEVYDDVLFFLAKKEKEQAKQLRRRNIQALKSILD-GMSSVNFQTTWSQ 435
Query 85 VREQLFSHP------KLQNMMKIDVLQVWEEWVRHGYEQERKQRRAQIFRRER---KRRD 135
++ L +P +LQNM K D L +EE +R +E ++R R R K R+
Sbjct 436 AQQYLMDNPSFAQDQQLQNMDKEDALICFEEHIRALEREEEEERERARLRERRQQRKNRE 495
Query 136 AFRELLQEAVDKGELSSKSEWVGFARRVEKDSRYLAMVGQSGSTPRELFSDAVQQLQQQH 195
AF+ L E + G+L S S W+ V D R+ M+GQ GSTP +LF V++L+ +
Sbjct 496 AFQSFLDELHETGQLHSMSTWMELYPAVSTDVRFANMLGQPGSTPLDLFKFYVEELKARF 555
Query 196 ERLKHFFKRFADREGLDLRDKALSFTQFYETVRGLDEVKNLNMLHVKLVFESL 248
K K G + + +F F + L+ ++KL F SL
Sbjct 556 HDEKKIIKDILKDRGFCV-EVNTAFEDFAHVISFDKRAAALDAGNIKLTFNSL 607
> hsa:25766 PRPF40B, HYPC; PRP40 pre-mRNA processing factor 40
homolog B (S. cerevisiae); K12821 pre-mRNA-processing factor
40
Length=871
Score = 82.0 bits (201), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 70/233 (30%), Positives = 112/233 (48%), Gaps = 11/233 (4%)
Query 25 EWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRREDVAMLEEILEQNPTEFAFQKRWSE 84
E W + E +R E + + +F + K++ KQ RR ++ L+ IL+ + FQ WS+
Sbjct 377 EVWAVVPERDRKEVYDDVLFFLAKKEKEQAKQLRRRNIQALKSILD-GMSSVNFQTTWSQ 435
Query 85 VREQLFSHP------KLQNMMKIDVLQVWEEWVRHGYEQERKQRRAQIFRRER---KRRD 135
++ L +P +LQNM K D L +EE +R +E ++R R R K R+
Sbjct 436 AQQYLMDNPSFAQDHQLQNMDKEDALICFEEHIRALEREEEEERERARLRERRQQRKNRE 495
Query 136 AFRELLQEAVDKGELSSKSEWVGFARRVEKDSRYLAMVGQSGSTPRELFSDAVQQLQQQH 195
AF+ L E + G+L S S W+ V D R+ M+GQ GSTP +LF V++L+ +
Sbjct 496 AFQTFLDELHETGQLHSMSTWMELYPAVSTDVRFANMLGQPGSTPLDLFKFYVEELKARF 555
Query 196 ERLKHFFKRFADREGLDLRDKALSFTQFYETVRGLDEVKNLNMLHVKLVFESL 248
K K G + + +F F + L+ ++KL F SL
Sbjct 556 HDEKKIIKDILKDRGFCV-EVNTAFEDFAHVISFDKRAAALDAGNIKLTFNSL 607
> mmu:56194 Prpf40a, 2810012K09Rik, FBP11, Fnbp11, Fnbp3; PRP40
pre-mRNA processing factor 40 homolog A (yeast); K12821 pre-mRNA-processing
factor 40
Length=953
Score = 77.0 bits (188), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 67/250 (26%), Positives = 120/250 (48%), Gaps = 11/250 (4%)
Query 8 LSFKTTYIDVADRFHFEEWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRREDVAMLEE 67
++ T Y F E W + E +R E +++ +F + K++ KQ R+ + L+
Sbjct 473 MTSTTRYKKAEQMFGEMEVWNAISERDRLEIYEDVLFFLSKKEKEQAKQLRKRNWEALKN 532
Query 68 ILEQNPTEFAFQKRWSEVREQLFSHP------KLQNMMKIDVLQVWEEWVRHGYEQERKQ 121
IL+ N + WSE ++ L +P +LQNM K D L +EE +R ++E ++
Sbjct 533 ILD-NMANVTYSTTWSEAQQYLMDNPTFAEDEELQNMDKEDALICFEEHIRALEKEEEEE 591
Query 122 RRAQIFRRERKR---RDAFRELLQEAVDKGELSSKSEWVGFARRVEKDSRYLAMVGQSGS 178
++ + R R++ R++F+ L E + G+L S S W+ + D R+ M+GQ GS
Sbjct 592 KQKTLLRERRRQRKNRESFQIFLDELHEHGQLHSMSSWMELYPTISSDIRFTNMLGQPGS 651
Query 179 TPRELFSDAVQQLQQQHERLKHFFKRFADREGLDLRDKALSFTQFYETVRGLDEVKNLNM 238
T +LF V+ L+ ++ K K +G + + +F F + L+
Sbjct 652 TALDLFKFYVEDLKARYHDEKKIIKDILKDKGF-VVEVNTTFEDFVAIISSTKRSTTLDA 710
Query 239 LHVKLVFESL 248
++KL F SL
Sbjct 711 GNIKLAFNSL 720
> hsa:55660 PRPF40A, FBP-11, FBP11, FLAF1, FLJ20585, FNBP3, HIP-10,
HIP10, HYPA, NY-REN-6, Prp40; PRP40 pre-mRNA processing
factor 40 homolog A (S. cerevisiae); K12821 pre-mRNA-processing
factor 40
Length=930
Score = 77.0 bits (188), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 67/250 (26%), Positives = 120/250 (48%), Gaps = 11/250 (4%)
Query 8 LSFKTTYIDVADRFHFEEWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRREDVAMLEE 67
++ T Y F E W + E +R E +++ +F + K++ KQ R+ + L+
Sbjct 450 MTSTTRYKKAEQMFGEMEVWNAISERDRLEIYEDVLFFLSKKEKEQAKQLRKRNWEALKN 509
Query 68 ILEQNPTEFAFQKRWSEVREQLFSHP------KLQNMMKIDVLQVWEEWVRHGYEQERKQ 121
IL+ N + WSE ++ L +P +LQNM K D L +EE +R ++E ++
Sbjct 510 ILD-NMANVTYSTTWSEAQQYLMDNPTFAEDEELQNMDKEDALICFEEHIRALEKEEEEE 568
Query 122 RRAQIFRRERKR---RDAFRELLQEAVDKGELSSKSEWVGFARRVEKDSRYLAMVGQSGS 178
++ + R R++ R++F+ L E + G+L S S W+ + D R+ M+GQ GS
Sbjct 569 KQKSLLRERRRQRKNRESFQIFLDELHEHGQLHSMSSWMELYPTISSDIRFTNMLGQPGS 628
Query 179 TPRELFSDAVQQLQQQHERLKHFFKRFADREGLDLRDKALSFTQFYETVRGLDEVKNLNM 238
T +LF V+ L+ ++ K K +G + + +F F + L+
Sbjct 629 TALDLFKFYVEDLKARYHDEKKIIKDILKDKGF-VVEVNTTFEDFVAIISSTKRSTTLDA 687
Query 239 LHVKLVFESL 248
++KL F SL
Sbjct 688 GNIKLAFNSL 697
> cel:ZK1098.1 hypothetical protein; K12821 pre-mRNA-processing
factor 40
Length=724
Score = 75.1 bits (183), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 58/244 (23%), Positives = 123/244 (50%), Gaps = 11/244 (4%)
Query 14 YIDVADRFHFEEWWTWMQESERDEFFQEWMFDNEQMFKDKIKQRRREDVAMLEEILEQNP 73
Y +D F E W + + +R E F++ + + K+K ++ R+ D+A +L Q+
Sbjct 318 YQKASDIFSKEPLWIAVNDEDRKEIFRDCIDFVARRDKEKKEEDRKRDIAAFSHVL-QSM 376
Query 74 TEFAFQKRWSEVREQLFSHPK------LQNMMKIDVLQVWEEWVRHGYEQERKQRRA--- 124
+ ++ W++ + L+ +P+ L M K D L V+E+ ++ ++ +++
Sbjct 377 EQITYKTTWAQAQRILYENPQFAERKDLHFMDKEDALTVFEDHIKQAEKEHDEEKEQEEK 436
Query 125 QIFRRERKRRDAFRELLQEAVDKGELSSKSEWVGFARRVEKDSRYLAMVGQSGSTPRELF 184
++ R++RK R+ +R LL+ +GEL+S S W + D+R+ M+ Q GS+P +LF
Sbjct 437 RLRRQQRKVREEYRLLLESLHKRGELTSMSLWTSLFPIISTDTRFELMLFQPGSSPLDLF 496
Query 185 SDAVQQLQQQHERLKHFFKRFADREGLDLRDKALSFTQFYETVRGLDEVKNLNMLHVKLV 244
V+ L++Q+ + K +G + + +F + V ++ ++ ++KL
Sbjct 497 KFFVEDLKEQYTEDRRLIKEILTEKGCQVI-ATTEYREFSDWVVSHEKGGKVDHGNMKLC 555
Query 245 FESL 248
+ SL
Sbjct 556 YNSL 559
> ath:AT1G44910 protein binding; K12821 pre-mRNA-processing factor
40
Length=926
Score = 69.7 bits (169), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 55/219 (25%), Positives = 114/219 (52%), Gaps = 6/219 (2%)
Query 34 ERDEFFQEWMFDNEQMFKDKIKQRRREDVAMLEEILEQNPTEFAFQKRWSEVREQLFSHP 93
+R++ F ++ + E+ ++K + R+ +A + LE +W +++++L
Sbjct 515 DREDLFDNYIVELERKEREKAAEEHRQYMADYRKFLETC-DYIKAGTQWRKIQDRLEDDD 573
Query 94 KLQNMMKIDVLQVWEEWVRHGYEQERKQRRA---QIFRRERKRRDAFRELLQEAVDKGEL 150
+ + KID L +EE++ ++E + +R + R ERK RDAFR LL+E V G L
Sbjct 574 RCSCLEKIDRLIGFEEYILDLEKEEEELKRVEKEHVRRAERKNRDAFRTLLEEHVAAGIL 633
Query 151 SSKSEWVGFARRVEKDSRYLAMVGQ-SGSTPRELFSDAVQQLQQQHERLKHFFKRFADRE 209
++K+ W+ + ++ +Y A+ SGSTP++LF D ++L++Q+ K + K
Sbjct 634 TAKTYWLDYCIELKDLPQYQAVASNTSGSTPKDLFEDVTEELEKQYHEDKSYVKDAMKSR 693
Query 210 GLDLRDKALSFTQFYETVRGLDEVKNLNMLHVKLVFESL 248
+ + L F F + + ++ +++KL+++ L
Sbjct 694 KISMVSSWL-FEDFKSAISEDLSTQQISDINLKLIYDDL 731
> ath:AT3G19670 protein binding
Length=992
Score = 62.8 bits (151), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 54/179 (30%), Positives = 98/179 (54%), Gaps = 19/179 (10%)
Query 81 RWSEVREQLFSHPKLQNMMKIDVLQVWEEWVR---HGYEQERKQRRAQIFRRERKRRDAF 137
+W +V+++L + + KID L++++E++R E+++K ++ ++ + ERK RD F
Sbjct 607 QWRKVQDRLEVDERCSRLEKIDQLEIFQEYLRDLEREEEEKKKIQKEELKKVERKHRDEF 666
Query 138 RELLQEAVDKGELSSKSEWVGFARRVEKDSRYLAMVGQ-SGSTPRELFSDAVQQLQQQHE 196
LL E + GEL++K+ W + +V+ Y A+ SG+TP++LF DAV+ L+++
Sbjct 667 HGLLDEHIATGELTAKTIWRDYLMKVKDLPVYSAIASNSSGATPKDLFEDAVEDLKKRDH 726
Query 197 RLKHFFKRFADREGLDLRDKALS----FTQFYETVR---GLDEVKNLNMLHVKLVFESL 248
LK K L LR LS F +F ++ G + ++ + KLVF+ L
Sbjct 727 ELKSQIKDV-----LKLRKVNLSAGSTFDEFKVSISEDIGFPLIPDVRL---KLVFDDL 777
> tgo:TGME49_061490 hypothetical protein
Length=2128
Score = 35.4 bits (80), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 33/88 (37%), Positives = 47/88 (53%), Gaps = 9/88 (10%)
Query 249 KNADTKPAAAAAAAADGKQRSSRSSRTDPDAGAYRSSKHDDATSSSSSSSS----SSANH 304
++ DT+PA +A +RSSR+ P A +YRS S S+ + S+ H
Sbjct 893 RDGDTQPATRSARV---NERSSRN--RSPSACSYRSRSQASEASPRSTRRARGFLGSSAH 947
Query 305 HGSHRDREREREREREKEREREKERERE 332
G RD E E EREK RE+++ERER+
Sbjct 948 AGPLRDAEEAGEMEREKAREKKRERERD 975
> hsa:473 RERE, ARG, ARP, ATN1L, DNB1, FLJ38775, KIAA0458; arginine-glutamic
acid dipeptide (RE) repeats; K05628 arginine-glutamic
acid dipeptide (RE) repeats
Length=1566
Score = 32.7 bits (73), Expect = 2.1, Method: Composition-based stats.
Identities = 24/100 (24%), Positives = 43/100 (43%), Gaps = 10/100 (10%)
Query 247 SLKNADTKPAAAAAAAADGKQRSSRSSRTDPDAGAYRSSKHDDATSSSSSSSSSSA---- 302
S++ ++ + + + KQ +S RT P RSS + +++S+SS+ S A
Sbjct 571 SMRTRRSRGSMSTLRSGRKKQPASPDGRTSPINEDIRSSGRNSPSAASTSSNDSKAETVK 630
Query 303 ------NHHGSHRDREREREREREKEREREKEREREKKKK 336
S + +R+RE+ E +R KK K
Sbjct 631 KSAKKVKEEASSPLKSNKRQREKVASDTEEADRTSSKKTK 670
Lambda K H
0.315 0.127 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 14283751500
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40