bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1111_orf3
Length=155
Score E
Sequences producing significant alignments: (Bits) Value
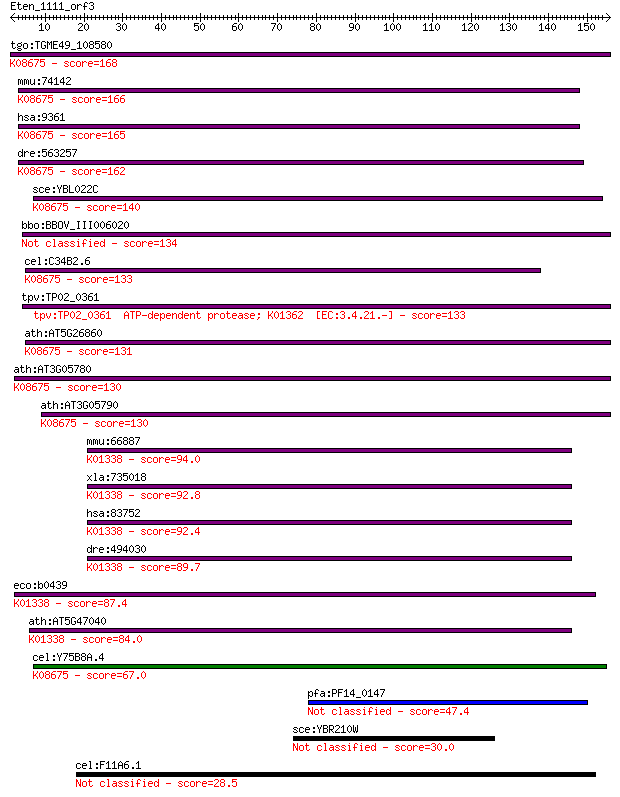
tgo:TGME49_108580 lon protease, putative (EC:3.4.21.53); K0867... 168 7e-42
mmu:74142 Lonp1, 1200017E13Rik, LON, Prss15; lon peptidase 1, ... 166 2e-41
hsa:9361 LONP1, LON, LONP, LonHS, MGC1498, PIM1, PRSS15, hLON;... 165 5e-41
dre:563257 lonp1, fc64d11, prss15, wu:fc64d11; lon peptidase 1... 162 4e-40
sce:YBL022C PIM1; ATP-dependent Lon protease, involved in degr... 140 1e-33
bbo:BBOV_III006020 17.m07531; ATP-dependent protease La family... 134 1e-31
cel:C34B2.6 hypothetical protein; K08675 Lon-like ATP-dependen... 133 2e-31
tpv:TP02_0361 ATP-dependent protease; K01362 [EC:3.4.21.-] 133 2e-31
ath:AT5G26860 LON1; LON1 (LON PROTEASE 1); ATP binding / ATP-d... 131 8e-31
ath:AT3G05780 LON3; LON3 (LON PROTEASE 3); ATP binding / ATP-d... 130 1e-30
ath:AT3G05790 LON4; LON4 (LON PROTEASE 4); ATP binding / ATP-d... 130 2e-30
mmu:66887 Lonp2, 1300002A08Rik, AU015403, Lonp; lon peptidase ... 94.0 2e-19
xla:735018 lonp2, MGC132158; lon peptidase 2, peroxisomal; K01... 92.8 4e-19
hsa:83752 LONP2, LONP, LONPL, MGC4840; lon peptidase 2, peroxi... 92.4 5e-19
dre:494030 lonp2, wu:fc51e03, zgc:92557; lon peptidase 2, pero... 89.7 3e-18
eco:b0439 lon, capR, deg, dir, ECK0433, JW0429, lonA, lopA, mu... 87.4 1e-17
ath:AT5G47040 LON2; LON2 (LON PROTEASE 2); ATP binding / ATP-d... 84.0 2e-16
cel:Y75B8A.4 hypothetical protein; K08675 Lon-like ATP-depende... 67.0 2e-11
pfa:PF14_0147 ATP-dependent protease, putative 47.4 2e-05
sce:YBR210W ERV15; Erv15p 30.0 3.4
cel:F11A6.1 kpc-1; Kex-2 Proprotein Convertase family member (... 28.5 8.3
> tgo:TGME49_108580 lon protease, putative (EC:3.4.21.53); K08675
Lon-like ATP-dependent protease [EC:3.4.21.-]
Length=1498
Score = 168 bits (425), Expect = 7e-42, Method: Compositional matrix adjust.
Identities = 90/181 (49%), Positives = 115/181 (63%), Gaps = 26/181 (14%)
Query 1 RKETDAVSVTQENLTNFVDQPPWTSSRLFERTQPGVVMGLAWTAMGGAVIFVEAVGR--- 57
RKE + V V +NL+ FV QP + S RL+ T PGVVMGLAWT MGGA ++VEA+GR
Sbjct 1240 RKEQEKVEVHMDNLSKFVGQPTFQSDRLYGETPPGVVMGLAWTQMGGATLYVEAIGRRPR 1299
Query 58 ----------------SAEQGSSKPSKGDFK-----PTG--GSLKVTGQLGGVMNESCEI 94
++ + +G + P G G LKVTGQLG VM+ES EI
Sbjct 1300 DTSKRVEMERKNWRLADDDEACRREKRGRTEQRSRSPAGAEGRLKVTGQLGNVMSESSEI 1359
Query 95 AMTFSRGFIKKQNPNNNYLFESAIHLHVPEGATPKDGPSAGVTLASALLSLALNLNPKPD 154
A+TF R F+++ P N++L + IHLHVPEGATPKDGPSAGVT+A+AL+SLALN PD
Sbjct 1360 ALTFCRVFVRRIEPRNSFLETAQIHLHVPEGATPKDGPSAGVTMATALVSLALNKPVLPD 1419
Query 155 L 155
+
Sbjct 1420 V 1420
> mmu:74142 Lonp1, 1200017E13Rik, LON, Prss15; lon peptidase 1,
mitochondrial; K08675 Lon-like ATP-dependent protease [EC:3.4.21.-]
Length=949
Score = 166 bits (421), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 84/145 (57%), Positives = 102/145 (70%), Gaps = 6/145 (4%)
Query 3 ETDAVSVTQENLTNFVDQPPWTSSRLFERTQPGVVMGLAWTAMGGAVIFVEAVGRSAEQG 62
E V VT ENL +FV +P +T R++E T PGVVMGLAWTAMGG+ +FVE R +
Sbjct 720 EAQTVQVTPENLQDFVGKPVFTVERMYEVTPPGVVMGLAWTAMGGSTLFVETSLRRPQPS 779
Query 63 SSKPSKGDFKPTGGSLKVTGQLGGVMNESCEIAMTFSRGFIKKQNPNNNYLFESAIHLHV 122
SK K GSL+VTGQLG VM ES IA T++R F+ +Q+P N++L S IHLHV
Sbjct 780 GSKEDKD------GSLEVTGQLGDVMKESARIAYTYARAFLMEQDPENDFLVTSHIHLHV 833
Query 123 PEGATPKDGPSAGVTLASALLSLAL 147
PEGATPKDGPSAG T+ +ALLSLAL
Sbjct 834 PEGATPKDGPSAGCTIVTALLSLAL 858
> hsa:9361 LONP1, LON, LONP, LonHS, MGC1498, PIM1, PRSS15, hLON;
lon peptidase 1, mitochondrial; K08675 Lon-like ATP-dependent
protease [EC:3.4.21.-]
Length=959
Score = 165 bits (418), Expect = 5e-41, Method: Compositional matrix adjust.
Identities = 84/145 (57%), Positives = 104/145 (71%), Gaps = 6/145 (4%)
Query 3 ETDAVSVTQENLTNFVDQPPWTSSRLFERTQPGVVMGLAWTAMGGAVIFVEAVGRSAEQG 62
E ++V VT ENL +FV +P +T R+++ T PGVVMGLAWTAMGG+ +FVE S +
Sbjct 731 EAESVEVTPENLQDFVGKPVFTVERMYDVTPPGVVMGLAWTAMGGSTLFVET---SLRRP 787
Query 63 SSKPSKGDFKPTGGSLKVTGQLGGVMNESCEIAMTFSRGFIKKQNPNNNYLFESAIHLHV 122
K +KGD GSL+VTGQLG VM ES IA TF+R F+ + P N+YL S IHLHV
Sbjct 788 QDKDAKGD---KDGSLEVTGQLGEVMKESARIAYTFARAFLMQHAPANDYLVTSHIHLHV 844
Query 123 PEGATPKDGPSAGVTLASALLSLAL 147
PEGATPKDGPSAG T+ +ALLSLA+
Sbjct 845 PEGATPKDGPSAGCTIVTALLSLAM 869
> dre:563257 lonp1, fc64d11, prss15, wu:fc64d11; lon peptidase
1, mitochondrial; K08675 Lon-like ATP-dependent protease [EC:3.4.21.-]
Length=966
Score = 162 bits (410), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 82/146 (56%), Positives = 102/146 (69%), Gaps = 5/146 (3%)
Query 3 ETDAVSVTQENLTNFVDQPPWTSSRLFERTQPGVVMGLAWTAMGGAVIFVEAVGRSAEQG 62
E AV VT NL ++V +P +T R+++ T PGVVMGLAWTAMGG+ +F+E S +
Sbjct 745 EETAVDVTSNNLQDYVGKPLFTVDRMYDVTPPGVVMGLAWTAMGGSTLFIET---SLRRP 801
Query 63 SSKPSKGDFKPTGGSLKVTGQLGGVMNESCEIAMTFSRGFIKKQNPNNNYLFESAIHLHV 122
P K P GSL+VTGQLG VM ES +IA TF+R F+ K+ P+N +L S IHLHV
Sbjct 802 RDPPGKDG--PKDGSLEVTGQLGDVMKESAKIAYTFARSFLMKEQPDNEFLVSSHIHLHV 859
Query 123 PEGATPKDGPSAGVTLASALLSLALN 148
PEGATPKDGPSAG T+ +ALLSLA N
Sbjct 860 PEGATPKDGPSAGCTIVTALLSLATN 885
> sce:YBL022C PIM1; ATP-dependent Lon protease, involved in degradation
of misfolded proteins in mitochondria; required for
biogenesis and maintenance of mitochondria (EC:3.4.21.-);
K08675 Lon-like ATP-dependent protease [EC:3.4.21.-]
Length=1133
Score = 140 bits (354), Expect = 1e-33, Method: Composition-based stats.
Identities = 69/147 (46%), Positives = 98/147 (66%), Gaps = 10/147 (6%)
Query 7 VSVTQENLTNFVDQPPWTSSRLFERTQPGVVMGLAWTAMGGAVIFVEAVGRSAEQGSSKP 66
VS++Q+NL ++V P +T+ RL+E T PGVVMGLAWT MGG ++VE+V P
Sbjct 899 VSISQKNLKDYVGPPVYTTDRLYETTPPGVVMGLAWTNMGGCSLYVESVLEQPLHNCKHP 958
Query 67 SKGDFKPTGGSLKVTGQLGGVMNESCEIAMTFSRGFIKKQNPNNNYLFESAIHLHVPEGA 126
+ + TGQLG VM ES +A +F++ ++ ++ P N + +++IHLH PEGA
Sbjct 959 T----------FERTGQLGDVMKESSRLAYSFAKMYLAQKFPENRFFEKASIHLHCPEGA 1008
Query 127 TPKDGPSAGVTLASALLSLALNLNPKP 153
TPKDGPSAGVT+A++ LSLALN + P
Sbjct 1009 TPKDGPSAGVTMATSFLSLALNKSIDP 1035
> bbo:BBOV_III006020 17.m07531; ATP-dependent protease La family
protein
Length=1122
Score = 134 bits (337), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 75/175 (42%), Positives = 98/175 (56%), Gaps = 23/175 (13%)
Query 4 TDAVSVTQENLTNFVDQPPWTSSRLFERTQP-GVVMGLAWTAMGGAVIFVEAVGR----- 57
D V ++ + L ++ P +T L P GVVMGLAWT GGA ++VEA G+
Sbjct 876 VDPVVISSDKLQGYLGVPTYTKDSLHPYPLPYGVVMGLAWTNAGGATMYVEARGQMVDKR 935
Query 58 -----------------SAEQGSSKPSKGDFKPTGGSLKVTGQLGGVMNESCEIAMTFSR 100
++ S + F G+LKVTG LG VM ES +IA+TF +
Sbjct 936 GNLVEPDRDTIPLSPDTQVDEVQSSDVENRFGTFHGTLKVTGHLGNVMTESSQIALTFCK 995
Query 101 GFIKKQNPNNNYLFESAIHLHVPEGATPKDGPSAGVTLASALLSLALNLNPKPDL 155
FI+K P N +L E+ IH+HVPEGATPKDGPS G+T+ASAL+SLA KP L
Sbjct 996 TFIRKHQPRNLFLDEAHIHIHVPEGATPKDGPSGGITMASALVSLAARKRLKPQL 1050
> cel:C34B2.6 hypothetical protein; K08675 Lon-like ATP-dependent
protease [EC:3.4.21.-]
Length=971
Score = 133 bits (335), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 63/134 (47%), Positives = 89/134 (66%), Gaps = 10/134 (7%)
Query 5 DAVSVTQENLTNFVDQPPWTSSRLFERTQPGVVMGLAWTAMGGAVIFVEAV-GRSAEQGS 63
+ + V ENL FV +P +TS R++E T PGV+MGLAWTAMGG+ +++E V R + +
Sbjct 758 EQIVVCTENLQKFVGRPKFTSDRMYEVTPPGVIMGLAWTAMGGSALYIETVLKRPVDLTN 817
Query 64 SKPSKGDFKPTGGSLKVTGQLGGVMNESCEIAMTFSRGFIKKQNPNNNYLFESAIHLHVP 123
K GS++ TG LG VM ES A+T ++G + ++ P+N + ++ IH+HVP
Sbjct 818 DK---------DGSIETTGNLGDVMKESVRTALTVAKGILAREQPDNKFFDKAHIHIHVP 868
Query 124 EGATPKDGPSAGVT 137
EGATPKDGPSAGVT
Sbjct 869 EGATPKDGPSAGVT 882
> tpv:TP02_0361 ATP-dependent protease; K01362 [EC:3.4.21.-]
Length=1115
Score = 133 bits (335), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 75/173 (43%), Positives = 101/173 (58%), Gaps = 21/173 (12%)
Query 4 TDAVSVTQENLTNFVDQPPWTSSRLFERTQP-GVVMGLAWTAMGGAVIFVEAVGR----- 57
++ V E++ +++ P +T L P GVV+GLAWT GGA ++VEA G+
Sbjct 872 SECTVVRSEDVQSYLGVPIYTKETLHPYPLPYGVVLGLAWTNAGGATMYVEAHGQMLDKK 931
Query 58 ---------------SAEQGSSKPSKGDFKPTGGSLKVTGQLGGVMNESCEIAMTFSRGF 102
S + S S G GGSLKVTG LG VM ES +IA+T+ + F
Sbjct 932 GNMIEPDRTPVKELSSTDTTPSSDSTGVDALKGGSLKVTGHLGTVMTESSQIALTYCKTF 991
Query 103 IKKQNPNNNYLFESAIHLHVPEGATPKDGPSAGVTLASALLSLALNLNPKPDL 155
I+K PNN +L E+ IH+HVPEGATPKDGPS G+T+ASAL+S+A KP +
Sbjct 992 IRKHQPNNIFLDEANIHIHVPEGATPKDGPSGGITMASALISIAAKKRIKPSI 1044
> ath:AT5G26860 LON1; LON1 (LON PROTEASE 1); ATP binding / ATP-dependent
peptidase/ serine-type peptidase; K08675 Lon-like
ATP-dependent protease [EC:3.4.21.-]
Length=985
Score = 131 bits (330), Expect = 8e-31, Method: Compositional matrix adjust.
Identities = 66/151 (43%), Positives = 95/151 (62%), Gaps = 12/151 (7%)
Query 5 DAVSVTQENLTNFVDQPPWTSSRLFERTQPGVVMGLAWTAMGGAVIFVEAVGRSAEQGSS 64
+ V + + NL ++V +P + + +L+E+T GVVMGLAWT+MGG+ +++E E+G
Sbjct 770 ETVMIDESNLADYVGKPVFHAEKLYEQTPVGVVMGLAWTSMGGSTLYIETT--VVEEGEG 827
Query 65 KPSKGDFKPTGGSLKVTGQLGGVMNESCEIAMTFSRGFIKKQNPNNNYLFESAIHLHVPE 124
K G L +TGQLG VM ES +IA T +R + ++ P N + S +HLHVP
Sbjct 828 K----------GGLNITGQLGDVMKESAQIAHTVARKIMLEKEPENQFFANSKLHLHVPA 877
Query 125 GATPKDGPSAGVTLASALLSLALNLNPKPDL 155
GATPKDGPSAG T+ ++LLSLA + DL
Sbjct 878 GATPKDGPSAGCTMITSLLSLATKKPVRKDL 908
> ath:AT3G05780 LON3; LON3 (LON PROTEASE 3); ATP binding / ATP-dependent
peptidase/ nucleoside-triphosphatase/ nucleotide
binding / serine-type endopeptidase/ serine-type peptidase;
K08675 Lon-like ATP-dependent protease [EC:3.4.21.-]
Length=924
Score = 130 bits (328), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 66/154 (42%), Positives = 96/154 (62%), Gaps = 12/154 (7%)
Query 2 KETDAVSVTQENLTNFVDQPPWTSSRLFERTQPGVVMGLAWTAMGGAVIFVEAVGRSAEQ 61
K + V + + NL ++V +P + +++E+T GVVMGLAWT+MGG+ +++E E+
Sbjct 709 KTFEKVMIDESNLADYVGKPVFQEEKIYEQTPVGVVMGLAWTSMGGSTLYIETT--FVEE 766
Query 62 GSSKPSKGDFKPTGGSLKVTGQLGGVMNESCEIAMTFSRGFIKKQNPNNNYLFESAIHLH 121
G K G L +TGQLG VM ES +IA T +R + ++ P N + S +HLH
Sbjct 767 GLGK----------GGLHITGQLGDVMKESAQIAHTVARRIMFEKEPENLFFANSKLHLH 816
Query 122 VPEGATPKDGPSAGVTLASALLSLALNLNPKPDL 155
VPEGATPKDGPSAG T+ ++ LSLA+ + DL
Sbjct 817 VPEGATPKDGPSAGCTMITSFLSLAMKKLVRKDL 850
> ath:AT3G05790 LON4; LON4 (LON PROTEASE 4); ATP binding / ATP-dependent
peptidase/ nucleoside-triphosphatase/ nucleotide
binding / serine-type endopeptidase/ serine-type peptidase;
K08675 Lon-like ATP-dependent protease [EC:3.4.21.-]
Length=942
Score = 130 bits (326), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 65/147 (44%), Positives = 93/147 (63%), Gaps = 12/147 (8%)
Query 9 VTQENLTNFVDQPPWTSSRLFERTQPGVVMGLAWTAMGGAVIFVEAVGRSAEQGSSKPSK 68
+ + NL+++V +P + +++E+T GVVMGLAWT+MGG+ +++E E+G K
Sbjct 734 IDESNLSDYVGKPVFQEEKIYEQTPVGVVMGLAWTSMGGSTLYIETT--FVEEGEGK--- 788
Query 69 GDFKPTGGSLKVTGQLGGVMNESCEIAMTFSRGFIKKQNPNNNYLFESAIHLHVPEGATP 128
G L +TG+LG VM ES EIA T +R + ++ P N S +HLHVP GATP
Sbjct 789 -------GGLHITGRLGDVMKESAEIAHTVARRIMLEKEPENKLFANSKLHLHVPAGATP 841
Query 129 KDGPSAGVTLASALLSLALNLNPKPDL 155
KDGPSAG T+ ++LLSLAL + DL
Sbjct 842 KDGPSAGCTMITSLLSLALKKPVRKDL 868
> mmu:66887 Lonp2, 1300002A08Rik, AU015403, Lonp; lon peptidase
2, peroxisomal; K01338 ATP-dependent Lon protease [EC:3.4.21.53]
Length=432
Score = 94.0 bits (232), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 56/131 (42%), Positives = 72/131 (54%), Gaps = 21/131 (16%)
Query 21 PPWTSSRLFER-TQPGVVMGLAWTAMGGAVIFVEAVGRSAEQGSSKPSKGDFKPTGGSLK 79
PP + ER +QPGV +GLAWT +GG ++FVEA E G L
Sbjct 220 PPLYELEVSERLSQPGVAIGLAWTPLGGKIMFVEASRMDGE---------------GQLT 264
Query 80 VTGQLGGVMNESCEIAMTFSRGFIKKQNPNNNY-----LFESAIHLHVPEGATPKDGPSA 134
+TGQLG VM ES +A+++ R KK + N + L + IHLH P GA KDGPSA
Sbjct 265 LTGQLGDVMKESAHLAISWLRSNAKKYHLTNAFGSFDLLDNTDIHLHFPAGAVTKDGPSA 324
Query 135 GVTLASALLSL 145
GVT+ + L SL
Sbjct 325 GVTIVTCLASL 335
> xla:735018 lonp2, MGC132158; lon peptidase 2, peroxisomal; K01338
ATP-dependent Lon protease [EC:3.4.21.53]
Length=856
Score = 92.8 bits (229), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 55/131 (41%), Positives = 70/131 (53%), Gaps = 21/131 (16%)
Query 21 PPWTSSRLFER-TQPGVVMGLAWTAMGGAVIFVEAVGRSAEQGSSKPSKGDFKPTGGSLK 79
PP + +F R QPGV +GLAWT +GG ++FVEA E G L
Sbjct 644 PPMYETEVFGRLNQPGVAIGLAWTPLGGEIMFVEASRMDGE---------------GQLT 688
Query 80 VTGQLGGVMNESCEIAMTFSRGFIKKQNPNN-----NYLFESAIHLHVPEGATPKDGPSA 134
+TGQLG VM ES +A+++ R KK N + L + IHLH P GA KDGPSA
Sbjct 689 LTGQLGDVMKESAHLAISWLRSNAKKYQLTNASGSFDLLDNTDIHLHFPAGAVTKDGPSA 748
Query 135 GVTLASALLSL 145
GV + + L SL
Sbjct 749 GVAIVTCLASL 759
> hsa:83752 LONP2, LONP, LONPL, MGC4840; lon peptidase 2, peroxisomal;
K01338 ATP-dependent Lon protease [EC:3.4.21.53]
Length=852
Score = 92.4 bits (228), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 55/131 (41%), Positives = 71/131 (54%), Gaps = 21/131 (16%)
Query 21 PPWTSSRLFER-TQPGVVMGLAWTAMGGAVIFVEAVGRSAEQGSSKPSKGDFKPTGGSLK 79
PP + +R +QPGV +GLAWT +GG ++FVEA E G L
Sbjct 640 PPMYEMEVSQRLSQPGVAIGLAWTPLGGEIMFVEASRMDGE---------------GQLT 684
Query 80 VTGQLGGVMNESCEIAMTFSRGFIKKQNPNNNY-----LFESAIHLHVPEGATPKDGPSA 134
+TGQLG VM ES +A+++ R KK N + L + IHLH P GA KDGPSA
Sbjct 685 LTGQLGDVMKESAHLAISWLRSNAKKYQLTNAFGSFDLLDNTDIHLHFPAGAVTKDGPSA 744
Query 135 GVTLASALLSL 145
GVT+ + L SL
Sbjct 745 GVTIVTCLASL 755
> dre:494030 lonp2, wu:fc51e03, zgc:92557; lon peptidase 2, peroxisomal
(EC:3.4.21.-); K01338 ATP-dependent Lon protease [EC:3.4.21.53]
Length=840
Score = 89.7 bits (221), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 56/129 (43%), Positives = 70/129 (54%), Gaps = 19/129 (14%)
Query 21 PPWTSSRLFER-TQPGVVMGLAWTAMGGAVIFVEAVGRSAEQGSSKPSKGDFKPTGGSLK 79
PP + ER T PGV +GLAWT MGG ++FVEA E G L
Sbjct 630 PPLFEMEVSERLTLPGVAIGLAWTPMGGEIMFVEASRMEGE---------------GQLT 674
Query 80 VTGQLGGVMNESCEIAMTFSRGFIKKQNPNN---NYLFESAIHLHVPEGATPKDGPSAGV 136
+TGQLG VM ES +A+++ R K N+ + L + IHLH P GA KDGPSAGV
Sbjct 675 LTGQLGDVMKESAHLAISWLRSNAKTYLLNDGSADLLEGTDIHLHFPAGAVTKDGPSAGV 734
Query 137 TLASALLSL 145
T+ + L SL
Sbjct 735 TIVTCLASL 743
> eco:b0439 lon, capR, deg, dir, ECK0433, JW0429, lonA, lopA,
muc; DNA-binding ATP-dependent protease La (EC:3.4.21.53); K01338
ATP-dependent Lon protease [EC:3.4.21.53]
Length=784
Score = 87.4 bits (215), Expect = 1e-17, Method: Composition-based stats.
Identities = 53/150 (35%), Positives = 76/150 (50%), Gaps = 16/150 (10%)
Query 2 KETDAVSVTQENLTNFVDQPPWTSSRLFERTQPGVVMGLAWTAMGGAVIFVEAVGRSAEQ 61
K + + +NL +++ + R + G V GLAWT +GG ++ +E
Sbjct 563 KSLKHIEINGDNLHDYLGVQRFDYGRADNENRVGQVTGLAWTEVGGDLLTIETA------ 616
Query 62 GSSKPSKGDFKPTGGSLKVTGQLGGVMNESCEIAMTFSRGFIKKQNPNNNYLFESAIHLH 121
P G L TG LG VM ES + A+T R +K N ++ + IH+H
Sbjct 617 ---------CVPGKGKLTYTGSLGEVMQESIQAALTVVRARAEKLGINPDFYEKRDIHVH 667
Query 122 VPEGATPKDGPSAGVTLASALLSLALNLNP 151
VPEGATPKDGPSAG+ + +AL+S L NP
Sbjct 668 VPEGATPKDGPSAGIAMCTALVS-CLTGNP 696
> ath:AT5G47040 LON2; LON2 (LON PROTEASE 2); ATP binding / ATP-dependent
peptidase/ nucleoside-triphosphatase/ nucleotide
binding / serine-type endopeptidase/ serine-type peptidase;
K01338 ATP-dependent Lon protease [EC:3.4.21.53]
Length=888
Score = 84.0 bits (206), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 57/145 (39%), Positives = 76/145 (52%), Gaps = 20/145 (13%)
Query 6 AVSVTQENLTNFVDQPPWTSSRLFERT-QPGVVMGLAWTAMGGAVIFVEAVGRSAEQGSS 64
A+ V + L + P + S +R GV +GL WT GG V FVEA +S
Sbjct 666 ALVVDETMLEKILGPPRFDDSEAADRVASAGVSVGLVWTTFGGEVQFVEA--------TS 717
Query 65 KPSKGDFKPTGGSLKVTGQLGGVMNESCEIAMTFSRG----FIKKQNPNNNYLFESAIHL 120
KG+ + +TGQLG V+ ES ++A+T+ R F + N L IH+
Sbjct 718 MVGKGE-------MHLTGQLGDVIKESAQLALTWVRARASDFKLALAGDMNVLDGRDIHI 770
Query 121 HVPEGATPKDGPSAGVTLASALLSL 145
H P GA PKDGPSAGVTL +AL+SL
Sbjct 771 HFPAGAVPKDGPSAGVTLVTALVSL 795
> cel:Y75B8A.4 hypothetical protein; K08675 Lon-like ATP-dependent
protease [EC:3.4.21.-]
Length=773
Score = 67.0 bits (162), Expect = 2e-11, Method: Composition-based stats.
Identities = 49/153 (32%), Positives = 75/153 (49%), Gaps = 24/153 (15%)
Query 7 VSVTQENLTNFVDQPPWTSSRLFERTQP---GVVMGLAWTAMGGAVIFVEAVGRSAEQGS 63
+ ++ N+ + ++ E+ +P GV GL+ T +GG V+ +EA
Sbjct 560 IQISASNIHKILKNKHMKRVKIVEKMRPLPAGVCFGLSVTTIGGRVMPIEA--------- 610
Query 64 SKPSKGDFKPTGGSLKVTGQLGGVMNESCEIAMTFSRGFIKKQNPNNNY--LFESAIHLH 121
SKG G + TG LG V+ ES +A +G++ + L + IH+H
Sbjct 611 -SKSKGT-----GKIVTTGHLGKVLKESILVA----KGWLSANSERLGLGTLEDQDIHVH 660
Query 122 VPEGATPKDGPSAGVTLASALLSLALNLNPKPD 154
+P GA KDGPSAG LA AL+SLA N+ + D
Sbjct 661 LPAGAVNKDGPSAGTGLACALVSLATNIPLRSD 693
> pfa:PF14_0147 ATP-dependent protease, putative
Length=1192
Score = 47.4 bits (111), Expect = 2e-05, Method: Composition-based stats.
Identities = 28/74 (37%), Positives = 43/74 (58%), Gaps = 7/74 (9%)
Query 78 LKVTGQLGGVMNESCEIAMTFSRGFIKKQNPNNNYL-FESA-IHLHVPEGATPKDGPSAG 135
+ +TG +G +M ES IA T+S + N L FES +H+++ +G KDGPSAG
Sbjct 1044 ITITGNVGRIMQESILIAYTYSMKLL-----NTIMLKFESKPLHINLSDGDLKKDGPSAG 1098
Query 136 VTLASALLSLALNL 149
+ + +LS LN+
Sbjct 1099 INFVTCILSYYLNI 1112
> sce:YBR210W ERV15; Erv15p
Length=142
Score = 30.0 bits (66), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 25/52 (48%), Gaps = 0/52 (0%)
Query 74 TGGSLKVTGQLGGVMNESCEIAMTFSRGFIKKQNPNNNYLFESAIHLHVPEG 125
TG SL VTG + +N C+I T G ++ N+ L + L VPE
Sbjct 4 TGLSLFVTGLILNCLNSICQIYFTILYGDLEADYINSIELCKRVNRLSVPEA 55
> cel:F11A6.1 kpc-1; Kex-2 Proprotein Convertase family member
(kpc-1)
Length=760
Score = 28.5 bits (62), Expect = 8.3, Method: Composition-based stats.
Identities = 36/140 (25%), Positives = 60/140 (42%), Gaps = 22/140 (15%)
Query 18 VDQPPWTSSRLFERTQPGVVMGLAWTAMGGAVIFVEAVGRSAEQGSSKPSKGDFKPTGGS 77
VD P + FE+ G+ MG G IFV A G + S G + + +
Sbjct 331 VDGPAKLTRSAFEK---GITMG----RKGKGSIFVWASGNGGKDADSCNCDG-YTNSIYT 382
Query 78 LKVTG--QLGGV--MNESCE--IAMTFSRGFIKKQNPNNNYLFESAIHLHVPEGATPKDG 131
L ++ + G + +E+C +A T+S G ++ L + ++H G
Sbjct 383 LSISSATENGNIPWYSEACSSTLATTYSSGATGEKMILTTDLHHACTNMHT--------G 434
Query 132 PSAGVTLASALLSLALNLNP 151
SA LA+ +++LAL NP
Sbjct 435 TSASAPLAAGIVALALEANP 454
Lambda K H
0.312 0.130 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3386671600
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40