bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1123_orf3
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
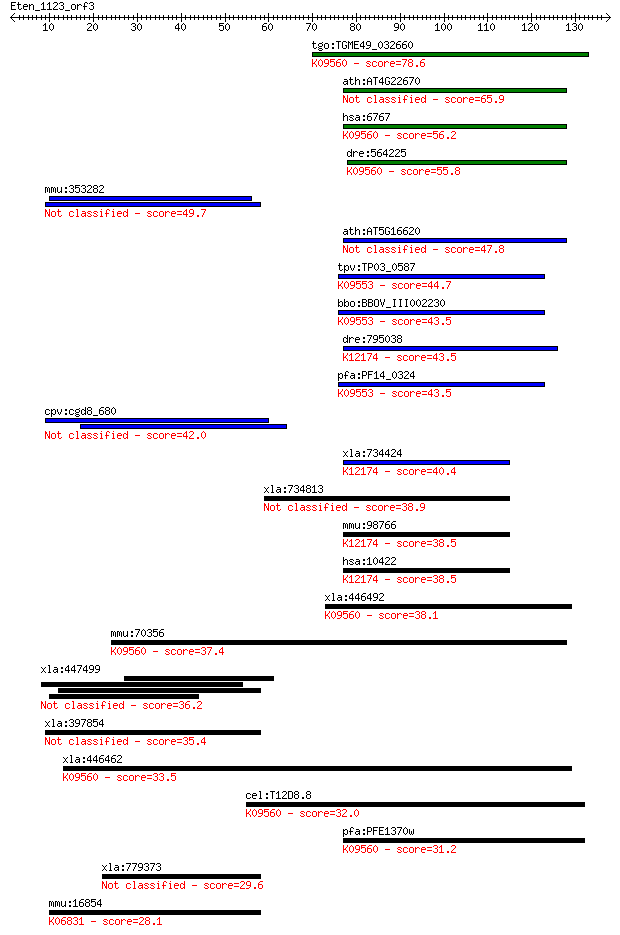
tgo:TGME49_032660 58 kDa phosphoprotein, putative ; K09560 sup... 78.6 6e-15
ath:AT4G22670 AtHip1; AtHip1 (Arabidopsis thaliana Hsp70-inter... 65.9 3e-11
hsa:6767 ST13, AAG2, FAM10A1, FAM10A4, FLJ27260, HIP, HOP, HSP... 56.2 3e-08
dre:564225 st13, MGC73267, MGC77089, wu:fd15g02, zgc:73267; su... 55.8 4e-08
mmu:353282 Sfmbt2, D2Wsu23e, D330030P06Rik; Scm-like with four... 49.7 2e-06
ath:AT5G16620 TIC40; TIC40 47.8 1e-05
tpv:TP03_0587 hypothetical protein; K09553 stress-induced-phos... 44.7 9e-05
bbo:BBOV_III002230 17.m07215; tetratricopeptide repeat domain ... 43.5 2e-04
dre:795038 fj67a11, wu:fj67a11; si:dkey-1o2.1; K12174 Kip1 ubi... 43.5 2e-04
pfa:PF14_0324 Hsp70/Hsp90 organizing protein, putative; K09553... 43.5 2e-04
cpv:cgd8_680 hypothetical protein 42.0 5e-04
xla:734424 ubac1, MGC115132; UBA domain containing 1; K12174 K... 40.4 0.002
xla:734813 hypothetical protein MGC131182 38.9 0.004
mmu:98766 Ubac1, 1110033G07Rik, AA407978, GBDR1, Gdbr1, Ubadc1... 38.5 0.006
hsa:10422 UBAC1, GBDR1, RP11-432J22.3, UBADC1; UBA domain cont... 38.5 0.007
xla:446492 MGC79131 protein; K09560 suppressor of tumorigenici... 38.1 0.008
mmu:70356 St13, 1110007I03Rik, 3110002K08Rik, AW555194, HIP, H... 37.4 0.014
xla:447499 anxa7, MGC82023, anx7, snx, synexin; annexin A7 36.2 0.031
xla:397854 anxa7; annexin VII 35.4 0.048
xla:446462 st13, MGC78939; suppression of tumorigenicity 13 (c... 33.5 0.21
cel:T12D8.8 hypothetical protein; K09560 suppressor of tumorig... 32.0 0.62
pfa:PFE1370w hsp70 interacting protein, putative; K09560 suppr... 31.2 0.92
xla:779373 anxa7, MGC83033; annexin A7 29.6 2.9
mmu:16854 Lgals3, GBP, L-34, Mac-2, gal3; lectin, galactose bi... 28.1 8.1
> tgo:TGME49_032660 58 kDa phosphoprotein, putative ; K09560 suppressor
of tumorigenicity protein 13
Length=425
Score = 78.6 bits (192), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 35/63 (55%), Positives = 49/63 (77%), Gaps = 0/63 (0%)
Query 70 AGGLLGSVNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLMRKFGGGM 129
AG L G++NDP+++++F NPKMMAAFQDI++NP +++KY DPEV A+ L KFGGG+
Sbjct 355 AGNLFGALNDPELKKLFENPKMMAAFQDIMSNPSSISKYASDPEVMAAMGSLTSKFGGGV 414
Query 130 AGG 132
G
Sbjct 415 PGA 417
> ath:AT4G22670 AtHip1; AtHip1 (Arabidopsis thaliana Hsp70-interacting
protein 1); binding
Length=441
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 25/51 (49%), Positives = 40/51 (78%), Gaps = 0/51 (0%)
Query 77 VNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLMRKFGG 127
+NDP++ FS+P++MAA QD++ NP N+AK++ +P+VA I+K+M KF G
Sbjct 389 LNDPELMTAFSDPEVMAALQDVMKNPANLAKHQANPKVAPVIAKMMGKFAG 439
> hsa:6767 ST13, AAG2, FAM10A1, FAM10A4, FLJ27260, HIP, HOP, HSPABP,
HSPABP1, MGC129952, P48, PRO0786, SNC6; suppression of
tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein);
K09560 suppressor of tumorigenicity protein 13
Length=369
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 25/51 (49%), Positives = 37/51 (72%), Gaps = 0/51 (0%)
Query 77 VNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLMRKFGG 127
++DP+V +P++M AFQD+ NP NM+KY+ +P+V + ISKL KFGG
Sbjct 317 LSDPEVLAAMQDPEVMVAFQDVAQNPANMSKYQSNPKVMNLISKLSAKFGG 367
> dre:564225 st13, MGC73267, MGC77089, wu:fd15g02, zgc:73267;
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting
protein); K09560 suppressor of tumorigenicity protein
13
Length=362
Score = 55.8 bits (133), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 24/50 (48%), Positives = 35/50 (70%), Gaps = 0/50 (0%)
Query 78 NDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLMRKFGG 127
NDP+V +P++MAAFQD+ NP N+AKY+ +P++ I+KL KF G
Sbjct 310 NDPEVLMAMQDPEVMAAFQDVAQNPANIAKYQSNPKIMALINKLSSKFAG 359
> mmu:353282 Sfmbt2, D2Wsu23e, D330030P06Rik; Scm-like with four
mbt domains 2
Length=975
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 19/46 (41%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 10 VPGGFPGGMPGGFPGEMSGGYPGGAPGGMSGSMPGGIPGGVPGGMP 55
VP G P G+P G P ++ G P G P + S+P GIP +P +P
Sbjct 742 VPAGVPAGVPAGVPEDIPAGIPEGIPASIPESIPEGIPESLPESLP 787
Score = 43.1 bits (100), Expect = 3e-04, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 24/49 (48%), Gaps = 0/49 (0%)
Query 9 GVPGGFPGGMPGGFPGEMSGGYPGGAPGGMSGSMPGGIPGGVPGGMPGG 57
GVP G P +P G P + P P G+ S+P +P +P +P G
Sbjct 749 GVPAGVPEDIPAGIPEGIPASIPESIPEGIPESLPESLPEAIPESIPKG 797
> ath:AT5G16620 TIC40; TIC40
Length=447
Score = 47.8 bits (112), Expect = 1e-05, Method: Composition-based stats.
Identities = 20/51 (39%), Positives = 31/51 (60%), Gaps = 0/51 (0%)
Query 77 VNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLMRKFGG 127
+ +PDV F NP++ AA + NP N+ KY++D EV D +K+ + F G
Sbjct 394 MENPDVAMAFQNPRVQAALMECSENPMNIMKYQNDKEVMDVFNKISQLFPG 444
> tpv:TP03_0587 hypothetical protein; K09553 stress-induced-phosphoprotein
1
Length=540
Score = 44.7 bits (104), Expect = 9e-05, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 76 SVNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLM 122
++ DP+VQQ+ +P+ + I NP MA+Y DP++++ I KL+
Sbjct 486 AMADPEVQQIICDPQFQLILKKISENPMTMAEYLKDPKISNGIQKLI 532
> bbo:BBOV_III002230 17.m07215; tetratricopeptide repeat domain
containing protein; K09553 stress-induced-phosphoprotein 1
Length=546
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 28/47 (59%), Gaps = 0/47 (0%)
Query 76 SVNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLM 122
++ DP+VQQ+ +P+ + + NP M +Y DP++A I KLM
Sbjct 492 AMADPEVQQMLGDPQFQIILKRLSENPAAMNEYLSDPKIAKGIQKLM 538
> dre:795038 fj67a11, wu:fj67a11; si:dkey-1o2.1; K12174 Kip1 ubiquitination-promoting
complex protein 2
Length=415
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 30/49 (61%), Gaps = 0/49 (0%)
Query 77 VNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLMRKF 125
+ +P VQ +NPK + AF+D+L NP N ++ +DPE + ++ R F
Sbjct 361 LENPVVQLGLTNPKTLLAFEDMLENPLNSTQWMNDPETGPVMLQISRIF 409
> pfa:PF14_0324 Hsp70/Hsp90 organizing protein, putative; K09553
stress-induced-phosphoprotein 1
Length=564
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 31/47 (65%), Gaps = 0/47 (0%)
Query 76 SVNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLM 122
S+ DP++QQ+ S+P+ Q + NP ++++Y DP++ + + KL+
Sbjct 510 SMADPEIQQIISDPQFQIILQKLNENPNSISEYIKDPKIFNGLQKLI 556
> cpv:cgd8_680 hypothetical protein
Length=1146
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 25/52 (48%), Positives = 28/52 (53%), Gaps = 6/52 (11%)
Query 9 GVPGGFPG-GMPGGFPGEMSGGYPGGAPGGMSGSMPGGIPGGVPGGMPGGMG 59
G+PGG G MPGG G P G PGG S P +PGGVP +PG G
Sbjct 148 GMPGGTSGPAMPGG---TTKPGVPSG-PGGQEASKPIAVPGGVP-VLPGAQG 194
Score = 32.0 bits (71), Expect = 0.57, Method: Composition-based stats.
Identities = 22/49 (44%), Positives = 24/49 (48%), Gaps = 7/49 (14%)
Query 17 GMPGGFPGEMSGGYPGGA--PGGMSGSMPGGIPGGVPGGMPGGMGGLEG 63
GMPGG G PGG PG SG PGG P +PGG+ L G
Sbjct 148 GMPGGTSGP---AMPGGTTKPGVPSG--PGGQEASKPIAVPGGVPVLPG 191
> xla:734424 ubac1, MGC115132; UBA domain containing 1; K12174
Kip1 ubiquitination-promoting complex protein 2
Length=406
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 26/38 (68%), Gaps = 0/38 (0%)
Query 77 VNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEV 114
V++P VQ +NPK + AF+D+L NP N ++ +DPE
Sbjct 352 VDNPVVQLGLTNPKTLLAFEDMLENPLNSTQWMNDPET 389
> xla:734813 hypothetical protein MGC131182
Length=401
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 33/56 (58%), Gaps = 5/56 (8%)
Query 59 GGLEGMSVVFFAGGLLGSVNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEV 114
G+E S +F A +++P VQ +NPK + AF+D+L NP N ++ +DPE
Sbjct 334 KGIETTSPLFQAI-----LDNPVVQLGLTNPKTLLAFEDMLENPLNSTQWMNDPET 384
> mmu:98766 Ubac1, 1110033G07Rik, AA407978, GBDR1, Gdbr1, Ubadc1;
ubiquitin associated domain containing 1; K12174 Kip1 ubiquitination-promoting
complex protein 2
Length=409
Score = 38.5 bits (88), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 26/38 (68%), Gaps = 0/38 (0%)
Query 77 VNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEV 114
+++P VQ +NPK + AF+D+L NP N ++ +DPE
Sbjct 355 LDNPVVQLGLTNPKTLLAFEDMLENPLNSTQWMNDPET 392
> hsa:10422 UBAC1, GBDR1, RP11-432J22.3, UBADC1; UBA domain containing
1; K12174 Kip1 ubiquitination-promoting complex protein
2
Length=405
Score = 38.5 bits (88), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 26/38 (68%), Gaps = 0/38 (0%)
Query 77 VNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEV 114
+++P VQ +NPK + AF+D+L NP N ++ +DPE
Sbjct 351 LDNPVVQLGLTNPKTLLAFEDMLENPLNSTQWMNDPET 388
> xla:446492 MGC79131 protein; K09560 suppressor of tumorigenicity
protein 13
Length=376
Score = 38.1 bits (87), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 30/56 (53%), Gaps = 18/56 (32%)
Query 73 LLGSVNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLMRKFGGG 128
++ AAFQD+ NP NM+KY+++P+V + I+KL KFGG
Sbjct 338 VM------------------AAFQDVAQNPANMSKYQNNPKVMNLITKLSSKFGGS 375
> mmu:70356 St13, 1110007I03Rik, 3110002K08Rik, AW555194, HIP,
HOP, HSPABP, HSPABP1, PRO0786, SNC6, p48; suppression of tumorigenicity
13; K09560 suppressor of tumorigenicity protein
13
Length=371
Score = 37.4 bits (85), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 41/104 (39%), Positives = 56/104 (53%), Gaps = 8/104 (7%)
Query 24 GEMSGGYPGGAPGGMSGSMPGGIPGGVPGGMPGGMGGLEGMSVVFFAGGLLGSVNDPDVQ 83
G Y G G G PGGMPG G + GM+ + GL ++DP+V
Sbjct 274 G---SQYGSFPGGFPGGMP-----GNFPGGMPGMGGAMPGMAGMAGMPGLNEILSDPEVL 325
Query 84 QVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLMRKFGG 127
+P++M AFQD+ NP NM+KY+ +P+V + ISKL KFGG
Sbjct 326 AAMQDPEVMVAFQDVAQNPSNMSKYQSNPKVMNLISKLSAKFGG 369
> xla:447499 anxa7, MGC82023, anx7, snx, synexin; annexin A7
Length=528
Score = 36.2 bits (82), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 20/34 (58%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 27 SGGYPGGAPGGMSGSMPGGIPGGVPGGMPGGMGG 60
S Y GAPG M G MPG +PG +PG MPG M G
Sbjct 157 SQNYGAGAPGQMPGQMPGQMPGQMPGQMPGQMPG 190
Score = 35.8 bits (81), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 22/46 (47%), Positives = 24/46 (52%), Gaps = 8/46 (17%)
Query 12 GGFPGGMPGGFPGEMSGGYPGGAPGGMSGSMPGGIPGGVPGGMPGG 57
G PG MPG PG+M PG M G MPG +PG P G P G
Sbjct 162 AGAPGQMPGQMPGQM--------PGQMPGQMPGQMPGQAPCGYPSG 199
Score = 35.8 bits (81), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 22/46 (47%), Positives = 23/46 (50%), Gaps = 8/46 (17%)
Query 8 CGVPGGFPGGMPGGFPGEMSGGYPGGAPGGMSGSMPGGIPGGVPGG 53
G PG PG MPG PG+M PG M G MPG P G P G
Sbjct 162 AGAPGQMPGQMPGQMPGQM--------PGQMPGQMPGQAPCGYPSG 199
Score = 29.6 bits (65), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 10 VPGGFPGGMPGGFPGEMSGGYPGGAPGGMSGSMP 43
+PG PG MPG PG+M G P G P G + + P
Sbjct 172 MPGQMPGQMPGQMPGQMPGQAPCGYPSGPAPAQP 205
> xla:397854 anxa7; annexin VII
Length=512
Score = 35.4 bits (80), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 25/49 (51%), Positives = 26/49 (53%), Gaps = 2/49 (4%)
Query 9 GVPGGFPGGMPGGFPGEMSGGYPGGAPGGMSGSMPGGIPGGVPGGMPGG 57
G PGG PG GGF S Y G PG M G MPG +PG P G P G
Sbjct 137 GSPGGAPGY--GGFSQPSSQSYGAGGPGQMPGQMPGQMPGQAPSGYPSG 183
> xla:446462 st13, MGC78939; suppression of tumorigenicity 13
(colon carcinoma) (Hsp70 interacting protein); K09560 suppressor
of tumorigenicity protein 13
Length=379
Score = 33.5 bits (75), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 43/116 (37%), Positives = 61/116 (52%), Gaps = 15/116 (12%)
Query 13 GFPGGMPGGFPGEMSGGYPGGAPGGMSGSMPGGIPGGVPGGMPGGMGGLEGMSVVFFAGG 72
GFPGG PGG PG GG PG GG+PG G+ +
Sbjct 278 GFPGGFPGGMPGMGGMPGMGGMPGMGGMPGMGGMPGMGGMAGMPGVSDI----------- 326
Query 73 LLGSVNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEVADAISKLMRKFGGG 128
++DP+V +P++MAAFQD+ NP N++KY+++P+V + I+KL KFGG
Sbjct 327 ----LSDPEVLAAMQDPEVMAAFQDVAQNPANISKYQNNPKVMNLITKLSSKFGGS 378
> cel:T12D8.8 hypothetical protein; K09560 suppressor of tumorigenicity
protein 13
Length=422
Score = 32.0 bits (71), Expect = 0.62, Method: Composition-based stats.
Identities = 23/77 (29%), Positives = 33/77 (42%), Gaps = 26/77 (33%)
Query 55 PGGMGGLEGMSVVFFAGGLLGSVNDPDVQQVFSNPKMMAAFQDILTNPGNMAKYKDDPEV 114
P ++ +P+++ A DI+ NP NM KY ++P+V
Sbjct 327 PEIAAAIQ-------------------------DPEVLPALMDIMQNPANMMKYINNPKV 361
Query 115 ADAISKLMRKFGGGMAG 131
A ISKL K G GM G
Sbjct 362 AKLISKLQSK-GAGMPG 377
> pfa:PFE1370w hsp70 interacting protein, putative; K09560 suppressor
of tumorigenicity protein 13
Length=458
Score = 31.2 bits (69), Expect = 0.92, Method: Composition-based stats.
Identities = 16/59 (27%), Positives = 32/59 (54%), Gaps = 8/59 (13%)
Query 77 VNDPDVQQVFSNPKMMAAFQDIL----TNPGNMAKYKDDPEVADAISKLMRKFGGGMAG 131
+N P+++++F+NP+ F ++ +NP ++KY +DP+ + L G M G
Sbjct 398 LNSPEMKELFNNPQ----FFQMMQNMMSNPQLISKYANDPKYKNLFENLKNSDFGNMMG 452
> xla:779373 anxa7, MGC83033; annexin A7
Length=520
Score = 29.6 bits (65), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 17/36 (47%), Positives = 18/36 (50%), Gaps = 0/36 (0%)
Query 22 FPGEMSGGYPGGAPGGMSGSMPGGIPGGVPGGMPGG 57
F S Y G PG M G MPG +PG P G P G
Sbjct 156 FSQPSSQSYGAGGPGQMPGQMPGQMPGQAPSGYPSG 191
> mmu:16854 Lgals3, GBP, L-34, Mac-2, gal3; lectin, galactose
binding, soluble 3; K06831 galectin-3
Length=264
Score = 28.1 bits (61), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 24/52 (46%), Positives = 29/52 (55%), Gaps = 5/52 (9%)
Query 10 VPGGFPG-GMPGGFPGEMS-GGYPG--GAPGGMSGSMPGGIPGGVPGGMPGG 57
PG +PG PG +PG + G +PG GAPG S PGG P P G+P G
Sbjct 76 APGAYPGPTAPGAYPGSTAPGAFPGQPGAPGAYP-SAPGGYPAAGPYGVPAG 126
Lambda K H
0.316 0.144 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2428006156
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40