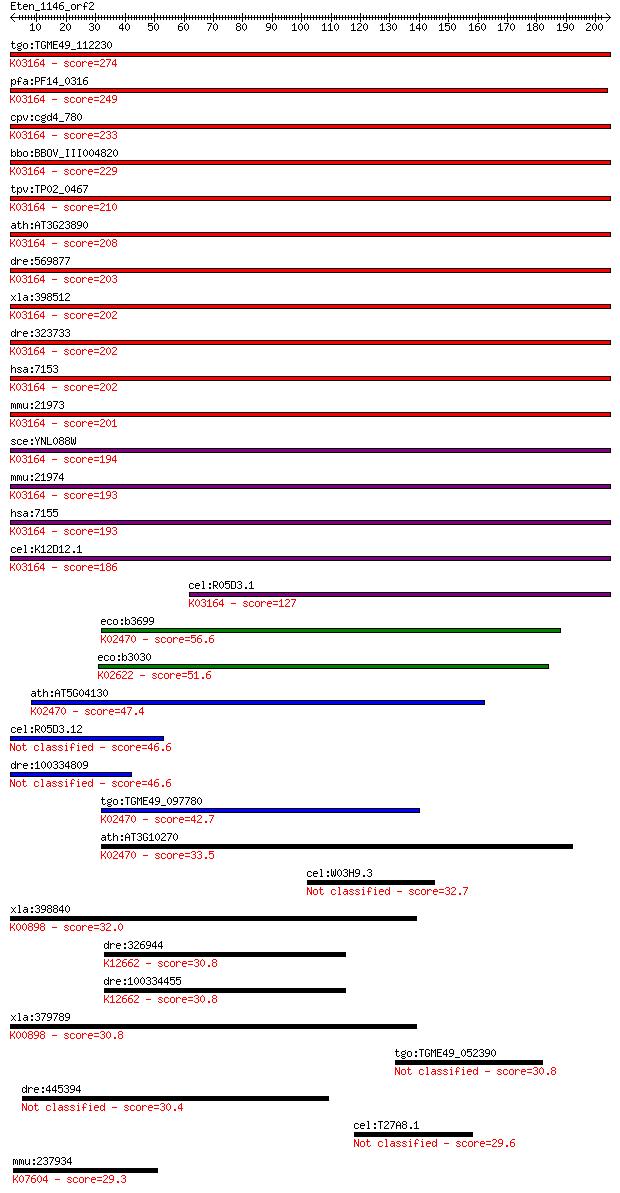
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1146_orf2
Length=204
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_112230 DNA topoisomerase II, putative ; K03164 DNA ... 274 2e-73
pfa:PF14_0316 DNA topoisomerase II, putative (EC:3.6.1.-); K03... 249 3e-66
cpv:cgd4_780 DNA topoisomerase II ; K03164 DNA topoisomerase I... 233 3e-61
bbo:BBOV_III004820 17.m07431; DNA topoisomerase II; K03164 DNA... 229 7e-60
tpv:TP02_0467 DNA topoisomerase II; K03164 DNA topoisomerase I... 210 3e-54
ath:AT3G23890 TOPII; TOPII (TOPOISOMERASE II); ATP binding / D... 208 1e-53
dre:569877 top2b, im:7138121, si:dkey-98n4.1, wu:fc38e02; topo... 203 4e-52
xla:398512 top2a, top2, tp2a; topoisomerase (DNA) II alpha 170... 202 6e-52
dre:323733 top2a, MGC92574, fc08c07, wu:fc08c07; topoisomerase... 202 7e-52
hsa:7153 TOP2A, TOP2, TP2A; topoisomerase (DNA) II alpha 170kD... 202 8e-52
mmu:21973 Top2a, Top-2; topoisomerase (DNA) II alpha (EC:5.99.... 201 2e-51
sce:YNL088W TOP2, TOR3, TRF3; Essential type II topoisomerase,... 194 2e-49
mmu:21974 Top2b, D230016L12Rik, Top-2; topoisomerase (DNA) II ... 193 3e-49
hsa:7155 TOP2B, TOPIIB, top2beta; topoisomerase (DNA) II beta ... 193 4e-49
cel:K12D12.1 top-2; TOPoisomerase family member (top-2); K0316... 186 4e-47
cel:R05D3.1 hypothetical protein; K03164 DNA topoisomerase II ... 127 2e-29
eco:b3699 gyrB, acrB, cou, ECK3691, himB, hisU, hopA, JW5625, ... 56.6 6e-08
eco:b3030 parE, ECK3021, JW2998, nfxD; DNA topoisomerase IV, s... 51.6 2e-06
ath:AT5G04130 DNA topoisomerase, ATP-hydrolyzing, putative / D... 47.4 4e-05
cel:R05D3.12 hypothetical protein 46.6 6e-05
dre:100334809 Top2a protein-like 46.6 6e-05
tgo:TGME49_097780 DNA gyrase subunit B, putative (EC:5.99.1.3)... 42.7 0.001
ath:AT3G10270 ATP binding / DNA binding / DNA topoisomerase (A... 33.5 0.51
cel:W03H9.3 hypothetical protein 32.7 0.90
xla:398840 hypothetical protein MGC68579; K00898 pyruvate dehy... 32.0 1.4
dre:326944 prpf4, mg:ab03a02, zgc:65943; PRP4 pre-mRNA process... 30.8 3.4
dre:100334455 PRP4 pre-mRNA processing factor 4 homolog; K1266... 30.8 3.4
xla:379789 pdk4, 3j828, MGC132353, MGC52849; pyruvate dehydrog... 30.8 3.5
tgo:TGME49_052390 hypothetical protein 30.8 3.6
dre:445394 wdr55, flj20195l, zgc:111796; WD repeat domain 55 30.4
cel:T27A8.1 hypothetical protein 29.6 7.6
mmu:237934 Krt39, 4732494G06Rik, Ka35; keratin 39; K07604 type... 29.3 9.6
> tgo:TGME49_112230 DNA topoisomerase II, putative ; K03164 DNA
topoisomerase II [EC:5.99.1.3]
Length=1687
Score = 274 bits (700), Expect = 2e-73, Method: Compositional matrix adjust.
Identities = 133/209 (63%), Positives = 160/209 (76%), Gaps = 6/209 (2%)
Query 1 YIGSTEISSQQLWVMPEGEPRMQQRVISFAPGLYKIVDEILVNAADVKARETEGGAA--- 57
YIGSTE+ +Q LWVM EG+ M + I++ PGLYKIVDEILVNAADVKARE E
Sbjct 57 YIGSTEVHTQALWVMEEGQETMVYKTINYVPGLYKIVDEILVNAADVKAREGEEQKGKPL 116
Query 58 -PRAK-MTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDNYDDTE 115
P A+ M+AIK+++N+A G I +WNDG+GIPVAIH+EHKVYV E+IFGQLLTSDNYDD+E
Sbjct 117 KPGARRMSAIKIDINKASGRITIWNDGEGIPVAIHQEHKVYVPEMIFGQLLTSDNYDDSE 176
Query 116 ARVTGGRNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDNMSKQDPPVISPFNGTGDY 175
RVTGGRNG+GAKLTNIFS F VECAD +R KK K+ WT NM ++ IS +NGT DY
Sbjct 177 CRVTGGRNGYGAKLTNIFSREFTVECADGERKKKFKMTWTKNMEQKSESEISSYNGT-DY 235
Query 176 VAVKFKPDLRRFGITTLDDDLISLLRKRC 204
V + F PD +FG+T DDD +SLLRKR
Sbjct 236 VKISFIPDFAKFGMTGFDDDSLSLLRKRA 264
> pfa:PF14_0316 DNA topoisomerase II, putative (EC:3.6.1.-); K03164
DNA topoisomerase II [EC:5.99.1.3]
Length=1472
Score = 249 bits (637), Expect = 3e-66, Method: Compositional matrix adjust.
Identities = 122/203 (60%), Positives = 148/203 (72%), Gaps = 4/203 (1%)
Query 1 YIGSTEISSQQLWVMPEGEPRMQQRVISFAPGLYKIVDEILVNAADVKARETEGGAAPRA 60
YIGS E+ +Q LWV + + RM Q+ I++ PGLYKI DEI+VNAADVKARE E P
Sbjct 27 YIGSVEMHTQLLWVWNKEKNRMVQKNITYVPGLYKIFDEIIVNAADVKAREKEKSENP-- 84
Query 61 KMTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDNYDDTEARVTG 120
MT IK+ +N+ I V+NDG+GIPV IHKE +YV +IFG+LLTSDNYDD E R+TG
Sbjct 85 -MTCIKIEINKENKRISVYNDGEGIPVDIHKEMNIYVPHMIFGELLTSDNYDDAEDRITG 143
Query 121 GRNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDNMSKQDPPVISPFNGTGDYVAVKF 180
GRNGFGAKLTNIFS F V+C DS R K+ K+ W+DNMSK P I +NG DYV V F
Sbjct 144 GRNGFGAKLTNIFSKEFIVQCGDSSRKKEFKMTWSDNMSKFSEPHIKNYNGK-DYVKVTF 202
Query 181 KPDLRRFGITTLDDDLISLLRKR 203
KPDL +FG+T +DDD+ SLL KR
Sbjct 203 KPDLNKFGMTEMDDDIESLLFKR 225
> cpv:cgd4_780 DNA topoisomerase II ; K03164 DNA topoisomerase
II [EC:5.99.1.3]
Length=1797
Score = 233 bits (594), Expect = 3e-61, Method: Compositional matrix adjust.
Identities = 118/206 (57%), Positives = 140/206 (67%), Gaps = 2/206 (0%)
Query 1 YIGSTEISSQQLWVMPEGEPRMQQRVISFAPGLYKIVDEILVNAADVKARE-TEGGAAPR 59
YIGSTE Q LWV + +M + I++ PGLYKI DEILVNAADVKARE +
Sbjct 53 YIGSTEFHEQALWVWDDSNMKMVYKTINYVPGLYKIFDEILVNAADVKAREISTKNNHIF 112
Query 60 AKMTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDNYDDTEARVT 119
K T IKV +N+ + I V+NDG+GIP+ IH+EH+VYV ELIFG LLTSDNYDD E RVT
Sbjct 113 EKCTCIKVEINREKKEITVFNDGEGIPIEIHREHQVYVPELIFGHLLTSDNYDDNEDRVT 172
Query 120 GGRNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDNMSKQDPPVISPFNGTGDYVAVK 179
GGRNGFGAKLTNIFS F+VEC D+ + K+ W DNMSK+ I YV +
Sbjct 173 GGRNGFGAKLTNIFSKEFKVECGDNNTGRIFKMTWYDNMSKKSEENIKENYSGKSYVKIT 232
Query 180 FKPDLRRFG-ITTLDDDLISLLRKRC 204
FKPDL RFG + DDD+ISLL KR
Sbjct 233 FKPDLSRFGNMKDFDDDIISLLNKRV 258
> bbo:BBOV_III004820 17.m07431; DNA topoisomerase II; K03164 DNA
topoisomerase II [EC:5.99.1.3]
Length=1437
Score = 229 bits (583), Expect = 7e-60, Method: Compositional matrix adjust.
Identities = 114/205 (55%), Positives = 142/205 (69%), Gaps = 4/205 (1%)
Query 1 YIGSTEISSQQLWVMPEGEPRMQQRVISFAPGLYKIVDEILVNAADVKARETEGGAAPRA 60
YIG+TE+ +QQ+WV + RM ++F PGLYKI DEILVNAADV AR+ + R
Sbjct 28 YIGNTEMLTQQMWVYNQEAKRMLYEPVTFIPGLYKIFDEILVNAADVHARQQSDPSLTR- 86
Query 61 KMTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDNYDDTEARVTG 120
MT IKVN N G I V+NDG+ IPV IHKEH VYV E+IFG+LLTSDNYDD + R+TG
Sbjct 87 -MTCIKVNFNATTGAITVFNDGEPIPVEIHKEHNVYVPEMIFGELLTSDNYDDEDGRITG 145
Query 121 GRNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDNMSK-QDPPVISPFNGTGDYVAVK 179
GRNGFGAKLTNIFS F V CADSKR+K+ + W NM++ P + ++G D+V V
Sbjct 146 GRNGFGAKLTNIFSKSFSVTCADSKRHKQFAMKWESNMTRVASPAKVGSYHGK-DFVKVT 204
Query 180 FKPDLRRFGITTLDDDLISLLRKRC 204
F PD RFGI T+D+ + +L KR
Sbjct 205 FIPDYERFGIPTMDEGTLKVLLKRV 229
> tpv:TP02_0467 DNA topoisomerase II; K03164 DNA topoisomerase
II [EC:5.99.1.3]
Length=1378
Score = 210 bits (534), Expect = 3e-54, Method: Compositional matrix adjust.
Identities = 110/205 (53%), Positives = 139/205 (67%), Gaps = 4/205 (1%)
Query 1 YIGSTEISSQQLWVMPEGEPRMQQRVISFAPGLYKIVDEILVNAADVKARETEGGAAPRA 60
YIGSTE+++QQ+WV E + RM I + PGLYKI DE+LVNAADV AR+
Sbjct 27 YIGSTELNNQQMWVFDEEKNRMVYETIDYVPGLYKIFDELLVNAADVYARQLSDPTL--E 84
Query 61 KMTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDNYDDTEARVTG 120
KMT IKV +++ G I V+NDG+ IP+ IH EHK+YV +LIFG+LLTSDNYDD+E RVTG
Sbjct 85 KMTCIKVTIDRGTGAIVVYNDGEPIPIQIHNEHKMYVPQLIFGELLTSDNYDDSEERVTG 144
Query 121 GRNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDNMSK-QDPPVISPFNGTGDYVAVK 179
GRNGFGAKL NIFS F V + K KK K+ W DNM+K + P I + G D+V +
Sbjct 145 GRNGFGAKLANIFSKTFTVVVGNKKTKKKFKMTWCDNMTKTKSPAQIESYMGP-DFVKIT 203
Query 180 FKPDLRRFGITTLDDDLISLLRKRC 204
F PD RF +T LD+ +L+KR
Sbjct 204 FIPDYSRFNLTELDEGTYKVLKKRV 228
> ath:AT3G23890 TOPII; TOPII (TOPOISOMERASE II); ATP binding /
DNA binding / DNA topoisomerase (ATP-hydrolyzing)/ DNA topoisomerase/
DNA-dependent ATPase; K03164 DNA topoisomerase II
[EC:5.99.1.3]
Length=1438
Score = 208 bits (529), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 105/204 (51%), Positives = 139/204 (68%), Gaps = 10/204 (4%)
Query 1 YIGSTEISSQQLWVMPEGEPRMQQRVISFAPGLYKIVDEILVNAADVKARETEGGAAPRA 60
YIGS E +Q LWV + E M QR +++ PGLYKI DEILVNAAD K R+ A
Sbjct 51 YIGSIEKHTQTLWVYEKDE--MVQRPVTYVPGLYKIFDEILVNAADNKQRD--------A 100
Query 61 KMTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDNYDDTEARVTG 120
KM +++V ++ + I V N G G+PV IH+E +YV E+IFG LLTS NYDD + TG
Sbjct 101 KMDSVQVVIDVEQNLISVCNSGAGVPVEIHQEEGIYVPEMIFGHLLTSSNYDDNVKKTTG 160
Query 121 GRNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDNMSKQDPPVISPFNGTGDYVAVKF 180
GRNG+GAKLTNIFST F +E AD KR KK K V+ +NM K+ PVI+ N + ++ V F
Sbjct 161 GRNGYGAKLTNIFSTEFIIETADGKRLKKYKQVFENNMGKKSEPVITKCNKSENWTKVTF 220
Query 181 KPDLRRFGITTLDDDLISLLRKRC 204
KPDL++F +T L+DD+++L+ KR
Sbjct 221 KPDLKKFNMTELEDDVVALMSKRV 244
> dre:569877 top2b, im:7138121, si:dkey-98n4.1, wu:fc38e02; topoisomerase
(DNA) II beta (EC:5.99.1.3); K03164 DNA topoisomerase
II [EC:5.99.1.3]
Length=1618
Score = 203 bits (516), Expect = 4e-52, Method: Compositional matrix adjust.
Identities = 106/204 (51%), Positives = 130/204 (63%), Gaps = 10/204 (4%)
Query 1 YIGSTEISSQQLWVMPEGEPRMQQRVISFAPGLYKIVDEILVNAADVKARETEGGAAPRA 60
YIGS E +QQ+WV E E M R ISF PGLYKI DEILVNAAD K R+
Sbjct 70 YIGSVEPVTQQMWVFDE-EIGMNLREISFVPGLYKIFDEILVNAADNKQRD--------K 120
Query 61 KMTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDNYDDTEARVTG 120
MT IK+ ++ TI VWN+G GIPV HK+ K+YV LIFG LLTS NYDD E +VTG
Sbjct 121 NMTTIKITIDPESNTISVWNNGKGIPVVEHKDEKMYVPALIFGHLLTSSNYDDNEKKVTG 180
Query 121 GRNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDNMSKQDPPVISPFNGTGDYVAVKF 180
GRNG+GAKL NIFS F VE A + + K WT+NM K + I F+G D+ V F
Sbjct 181 GRNGYGAKLCNIFSLKFTVETACKEYKRSFKQTWTENMGKSNEAKIKYFDGE-DFTCVTF 239
Query 181 KPDLRRFGITTLDDDLISLLRKRC 204
+PDL +F + LD D+++LL +R
Sbjct 240 QPDLAKFNMEKLDKDIVALLTRRA 263
> xla:398512 top2a, top2, tp2a; topoisomerase (DNA) II alpha 170kDa;
K03164 DNA topoisomerase II [EC:5.99.1.3]
Length=1579
Score = 202 bits (514), Expect = 6e-52, Method: Compositional matrix adjust.
Identities = 105/204 (51%), Positives = 130/204 (63%), Gaps = 10/204 (4%)
Query 1 YIGSTEISSQQLWVMPEGEPRMQQRVISFAPGLYKIVDEILVNAADVKARETEGGAAPRA 60
YIGS E +QQ+WV E E + R I+F PGLYKI DEILVNAAD K R+
Sbjct 48 YIGSVEPVTQQMWVFDEDE-GLNCRDITFVPGLYKIFDEILVNAADNKQRD--------P 98
Query 61 KMTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDNYDDTEARVTG 120
M+ IK+ ++ TI VWN+G GIPV HK KVYV LIFGQLLTS NYDD + +VTG
Sbjct 99 TMSCIKITIDPDNNTISVWNNGKGIPVVEHKVEKVYVPALIFGQLLTSSNYDDDQKKVTG 158
Query 121 GRNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDNMSKQDPPVISPFNGTGDYVAVKF 180
GRNG+GAKL NIFS F VE A + K K WT+NMS P I PF G D+ + F
Sbjct 159 GRNGYGAKLCNIFSNKFTVETACREYKKIFKQTWTNNMSNAPDPKIKPFEGE-DFTCITF 217
Query 181 KPDLRRFGITTLDDDLISLLRKRC 204
+PDL +F + LD D+++L+ +R
Sbjct 218 QPDLSKFKMQILDKDIVALMSRRA 241
> dre:323733 top2a, MGC92574, fc08c07, wu:fc08c07; topoisomerase
(DNA) II alpha (EC:5.99.1.3); K03164 DNA topoisomerase II
[EC:5.99.1.3]
Length=1574
Score = 202 bits (514), Expect = 7e-52, Method: Compositional matrix adjust.
Identities = 107/204 (52%), Positives = 128/204 (62%), Gaps = 10/204 (4%)
Query 1 YIGSTEISSQQLWVMPEGEPRMQQRVISFAPGLYKIVDEILVNAADVKARETEGGAAPRA 60
YIGS E +QQ+WV E + M R I+F PGLYKI DEILVNAAD K R+
Sbjct 49 YIGSVEPVTQQMWVFDE-DTGMNCRDITFVPGLYKIFDEILVNAADNKQRD--------K 99
Query 61 KMTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDNYDDTEARVTG 120
M IKVN++ TI VWN+G GIPV HK KVYV LIFGQLLTS NYDD E +VTG
Sbjct 100 TMNCIKVNIDPENNTISVWNNGKGIPVVEHKVEKVYVPALIFGQLLTSSNYDDDEKKVTG 159
Query 121 GRNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDNMSKQDPPVISPFNGTGDYVAVKF 180
GRNG+GAKL NIFS F VE A + + K W DNM + I PF+G DY + F
Sbjct 160 GRNGYGAKLCNIFSLKFTVETACRESKRCFKQTWFDNMERAGESKIKPFDG-DDYTCITF 218
Query 181 KPDLRRFGITTLDDDLISLLRKRC 204
+PDL +F + LD D ++LL +R
Sbjct 219 QPDLAKFKMQALDKDTVALLTRRA 242
> hsa:7153 TOP2A, TOP2, TP2A; topoisomerase (DNA) II alpha 170kDa
(EC:5.99.1.3); K03164 DNA topoisomerase II [EC:5.99.1.3]
Length=1531
Score = 202 bits (513), Expect = 8e-52, Method: Compositional matrix adjust.
Identities = 102/204 (50%), Positives = 132/204 (64%), Gaps = 10/204 (4%)
Query 1 YIGSTEISSQQLWVMPEGEPRMQQRVISFAPGLYKIVDEILVNAADVKARETEGGAAPRA 60
YIGS E+ +QQ+WV E + + R ++F PGLYKI DEILVNAAD K R+
Sbjct 50 YIGSVELVTQQMWVYDE-DVGINYREVTFVPGLYKIFDEILVNAADNKQRD--------P 100
Query 61 KMTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDNYDDTEARVTG 120
KM+ I+V ++ I +WN+G GIPV HK K+YV LIFGQLLTS NYDD E +VTG
Sbjct 101 KMSCIRVTIDPENNLISIWNNGKGIPVVEHKVEKMYVPALIFGQLLTSSNYDDDEKKVTG 160
Query 121 GRNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDNMSKQDPPVISPFNGTGDYVAVKF 180
GRNG+GAKL NIFST F VE A + K K W DNM + + PFNG DY + F
Sbjct 161 GRNGYGAKLCNIFSTKFTVETASREYKKMFKQTWMDNMGRAGEMELKPFNGE-DYTCITF 219
Query 181 KPDLRRFGITTLDDDLISLLRKRC 204
+PDL +F + +LD D+++L+ +R
Sbjct 220 QPDLSKFKMQSLDKDIVALMVRRA 243
> mmu:21973 Top2a, Top-2; topoisomerase (DNA) II alpha (EC:5.99.1.3);
K03164 DNA topoisomerase II [EC:5.99.1.3]
Length=1528
Score = 201 bits (510), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 101/204 (49%), Positives = 132/204 (64%), Gaps = 10/204 (4%)
Query 1 YIGSTEISSQQLWVMPEGEPRMQQRVISFAPGLYKIVDEILVNAADVKARETEGGAAPRA 60
YIGS E+ +QQ+WV E + + R ++F PGLYKI DEILVNAAD K R+
Sbjct 49 YIGSVELVTQQMWVYDE-DVGINYREVTFVPGLYKIFDEILVNAADNKQRD--------P 99
Query 61 KMTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDNYDDTEARVTG 120
KM+ I+V ++ I +WN+G GIPV HK K+YV LIFGQLLTS NYDD E +VTG
Sbjct 100 KMSCIRVTIDPENNVISIWNNGKGIPVVEHKVEKIYVPALIFGQLLTSSNYDDDEKKVTG 159
Query 121 GRNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDNMSKQDPPVISPFNGTGDYVAVKF 180
GRNG+GAKL NIFST F VE A + K K W DNM + + PF+G DY + F
Sbjct 160 GRNGYGAKLCNIFSTKFTVETASREYKKMFKQTWMDNMGRAGDMELKPFSGE-DYTCITF 218
Query 181 KPDLRRFGITTLDDDLISLLRKRC 204
+PDL +F + +LD D+++L+ +R
Sbjct 219 QPDLSKFKMQSLDKDIVALMVRRA 242
> sce:YNL088W TOP2, TOR3, TRF3; Essential type II topoisomerase,
relieves torsional strain in DNA by cleaving and re-sealing
the phosphodiester backbone of both positively and negatively
supercoiled DNA; cleaves complementary strands; localizes
to axial cores in meiosis (EC:5.99.1.3); K03164 DNA topoisomerase
II [EC:5.99.1.3]
Length=1428
Score = 194 bits (493), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 98/204 (48%), Positives = 126/204 (61%), Gaps = 8/204 (3%)
Query 1 YIGSTEISSQQLWVMPEGEPRMQQRVISFAPGLYKIVDEILVNAADVKARETEGGAAPRA 60
YIGS E Q W+ E M ++ ++ PGL+KI DEILVNAAD K R+
Sbjct 28 YIGSVETQEQLQWIYDEETDCMIEKNVTIVPGLFKIFDEILVNAADNKVRD--------P 79
Query 61 KMTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDNYDDTEARVTG 120
M I VN++ E TI V NDG GIP+ IH + +Y+ E+IFG LLTS NYDD E +VTG
Sbjct 80 SMKRIDVNIHAEEHTIEVKNDGKGIPIEIHNKENIYIPEMIFGHLLTSSNYDDDEKKVTG 139
Query 121 GRNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDNMSKQDPPVISPFNGTGDYVAVKF 180
GRNG+GAKL NIFST F +E AD +K W +NMS PP I+ + Y V F
Sbjct 140 GRNGYGAKLCNIFSTEFILETADLNVGQKYVQKWENNMSICHPPKITSYKKGPSYTKVTF 199
Query 181 KPDLRRFGITTLDDDLISLLRKRC 204
KPDL RFG+ LD+D++ ++R+R
Sbjct 200 KPDLTRFGMKELDNDILGVMRRRV 223
> mmu:21974 Top2b, D230016L12Rik, Top-2; topoisomerase (DNA) II
beta (EC:5.99.1.3); K03164 DNA topoisomerase II [EC:5.99.1.3]
Length=1612
Score = 193 bits (491), Expect = 3e-49, Method: Compositional matrix adjust.
Identities = 102/204 (50%), Positives = 127/204 (62%), Gaps = 10/204 (4%)
Query 1 YIGSTEISSQQLWVMPEGEPRMQQRVISFAPGLYKIVDEILVNAADVKARETEGGAAPRA 60
YIGS E +Q +WV E + M R ++F PGLYKI DEILVNAAD K R+
Sbjct 59 YIGSVEPLTQLMWVYDE-DVGMNCREVTFVPGLYKIFDEILVNAADNKQRD--------K 109
Query 61 KMTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDNYDDTEARVTG 120
MT IKV+++ I +WN+G GIPV HK KVYV LIFGQLLTS NYDD E +VTG
Sbjct 110 NMTCIKVSIDPESNIISIWNNGKGIPVVEHKVEKVYVPALIFGQLLTSSNYDDDEKKVTG 169
Query 121 GRNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDNMSKQDPPVISPFNGTGDYVAVKF 180
GRNG+GAKL NIFST F VE A + K W +NM K I F+G DY + F
Sbjct 170 GRNGYGAKLCNIFSTKFTVETACKEYKHSFKQTWMNNMMKTSEAKIKHFDGE-DYTCITF 228
Query 181 KPDLRRFGITTLDDDLISLLRKRC 204
+PDL +F + LD D+++L+ +R
Sbjct 229 QPDLSKFKMEKLDKDIVALMTRRA 252
> hsa:7155 TOP2B, TOPIIB, top2beta; topoisomerase (DNA) II beta
180kDa (EC:5.99.1.3); K03164 DNA topoisomerase II [EC:5.99.1.3]
Length=1621
Score = 193 bits (490), Expect = 4e-49, Method: Compositional matrix adjust.
Identities = 102/204 (50%), Positives = 127/204 (62%), Gaps = 10/204 (4%)
Query 1 YIGSTEISSQQLWVMPEGEPRMQQRVISFAPGLYKIVDEILVNAADVKARETEGGAAPRA 60
YIGS E +Q +WV E + M R ++F PGLYKI DEILVNAAD K R+
Sbjct 66 YIGSVEPLTQFMWVYDE-DVGMNCREVTFVPGLYKIFDEILVNAADNKQRD--------K 116
Query 61 KMTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDNYDDTEARVTG 120
MT IKV+++ I +WN+G GIPV HK KVYV LIFGQLLTS NYDD E +VTG
Sbjct 117 NMTCIKVSIDPESNIISIWNNGKGIPVVEHKVEKVYVPALIFGQLLTSSNYDDDEKKVTG 176
Query 121 GRNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDNMSKQDPPVISPFNGTGDYVAVKF 180
GRNG+GAKL NIFST F VE A + K W +NM K I F+G DY + F
Sbjct 177 GRNGYGAKLCNIFSTKFTVETACKEYKHSFKQTWMNNMMKTSEAKIKHFDGE-DYTCITF 235
Query 181 KPDLRRFGITTLDDDLISLLRKRC 204
+PDL +F + LD D+++L+ +R
Sbjct 236 QPDLSKFKMEKLDKDIVALMTRRA 259
> cel:K12D12.1 top-2; TOPoisomerase family member (top-2); K03164
DNA topoisomerase II [EC:5.99.1.3]
Length=1520
Score = 186 bits (473), Expect = 4e-47, Method: Compositional matrix adjust.
Identities = 97/205 (47%), Positives = 127/205 (61%), Gaps = 10/205 (4%)
Query 1 YIGSTEISSQQ-LWVMPEGEPRMQQRVISFAPGLYKIVDEILVNAADVKARETEGGAAPR 59
YIGS E + + +WV E +++QR IS+ PGLYKI DEILVNAAD K R+
Sbjct 83 YIGSVEHTEKTPMWVYNMEESKLEQRDISYVPGLYKIYDEILVNAADNKQRD-------- 134
Query 60 AKMTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDNYDDTEARVT 119
KM IK+ +N+ + I V+N+G GIPV HK KVYV ELIFG LLTS NY+D E +VT
Sbjct 135 PKMNTIKITINKEKNEISVYNNGKGIPVTQHKVEKVYVPELIFGTLLTSSNYNDDEKKVT 194
Query 120 GGRNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDNMSKQDPPVISPFNGTGDYVAVK 179
GGRNG+GAKL NIFST F +E + K W NM++ + P I + D+ +
Sbjct 195 GGRNGYGAKLCNIFSTKFTLETSSRDYKSAFKQTWIKNMTRDEEPKIVK-STDEDFTKIT 253
Query 180 FKPDLRRFGITTLDDDLISLLRKRC 204
F PDL +F + LDDD+ L+ +R
Sbjct 254 FSPDLAKFKMKELDDDICHLMARRA 278
> cel:R05D3.1 hypothetical protein; K03164 DNA topoisomerase II
[EC:5.99.1.3]
Length=1054
Score = 127 bits (320), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 65/145 (44%), Positives = 86/145 (59%), Gaps = 2/145 (1%)
Query 62 MTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDNYDDTEARVTGG 121
M +K + I VWN+G G+PV IH +YV L+FG L TS NYDD+E + GG
Sbjct 1 MYILKNKFLRETARISVWNNGSGLPVEIHPTEGIYVPTLVFGNLFTSSNYDDSEIKTVGG 60
Query 122 RNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDNMSK-QDPPVISPFNGT-GDYVAVK 179
RNG+GAKL NIFS F VE D++ ++ + W DNM K + V+ + T DY V+
Sbjct 61 RNGYGAKLCNIFSKEFIVETVDTRIKRRFRQKWYDNMKKCNEAEVVEILDETVKDYTKVE 120
Query 180 FKPDLRRFGITTLDDDLISLLRKRC 204
F PDL RF I L DD+I L+ +R
Sbjct 121 FVPDLERFQIDKLSDDVIDLIGRRV 145
> eco:b3699 gyrB, acrB, cou, ECK3691, himB, hisU, hopA, JW5625,
nalC, parA, pcbA, pcpA; DNA gyrase, subunit B (EC:5.99.1.3);
K02470 DNA gyrase subunit B [EC:5.99.1.3]
Length=804
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 48/157 (30%), Positives = 69/157 (43%), Gaps = 16/157 (10%)
Query 32 GLYKIVDEILVNAADVKARETEGGAAPRAKMTAIKVNVNQAEGTIRVWNDGDGIPVAIHK 91
GL+ +V E++ NA D A I V ++ A+ ++ V +DG GIP IH
Sbjct 35 GLHHMVFEVVDNAID---------EALAGHCKEIIVTIH-ADNSVSVQDDGRGIPTGIHP 84
Query 92 EHKVYVAELIFGQLLTSDNYDDTEARVTGGRNGFGAKLTNIFSTYFEVECADSKRNKKIK 151
E V AE+I L +DD +V+GG +G G + N S E+ +R KI
Sbjct 85 EEGVSAAEVIMTVLHAGGKFDDNSYKVSGGLHGVGVSVVNALSQKLELVI---QREGKIH 141
Query 152 VVWTDNMSKQDP-PVISPFNGTGDYVAVKFKPDLRRF 187
++ Q P V TG V+F P L F
Sbjct 142 RQIYEHGVPQAPLAVTGETEKTG--TMVRFWPSLETF 176
> eco:b3030 parE, ECK3021, JW2998, nfxD; DNA topoisomerase IV,
subunit B (EC:5.99.1.-); K02622 topoisomerase IV subunit B
[EC:5.99.1.-]
Length=630
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 43/161 (26%), Positives = 71/161 (44%), Gaps = 21/161 (13%)
Query 31 PGLY-------KIVDEILVNAADVKARETEGGAAPRAKMTAIKVNVNQAEGTIRVWNDGD 83
PG+Y + E++ N+ D E G A R + + A+ ++ V +DG
Sbjct 23 PGMYTDTTRPNHLGQEVIDNSVD----EALAGHAKRVDV------ILHADQSLEVIDDGR 72
Query 84 GIPVAIHKEHKVYVAELIFGQLLTSDNYDDTEARVTGGRNGFGAKLTNIFSTYFEVECAD 143
G+PV IH E V ELI +L + + + +GG +G G + N S EV +
Sbjct 73 GMPVDIHPEEGVPAVELILCRLHAGGKFSNKNYQFSGGLHGVGISVVNALSKRVEV---N 129
Query 144 SKRNKKIKVVWTDNMSK-QDPPVISPFNGTGDYVAVKFKPD 183
+R+ ++ + +N K QD V+ +V F PD
Sbjct 130 VRRDGQVYNIAFENGEKVQDLQVVGTCGKRNTGTSVHFWPD 170
> ath:AT5G04130 DNA topoisomerase, ATP-hydrolyzing, putative /
DNA topoisomerase II, putative / DNA gyrase, putative; K02470
DNA gyrase subunit B [EC:5.99.1.3]
Length=732
Score = 47.4 bits (111), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 45/161 (27%), Positives = 71/161 (44%), Gaps = 30/161 (18%)
Query 8 SSQQLWVMPEGEPRMQQRVISF-----APGLYKIVDEILVNAADVKARETEGGAAPRAKM 62
SS+Q+ V+ EG +++R + + GL+ +V EIL NA D E + G A + +
Sbjct 92 SSEQIQVL-EGLDPVRKRPGMYIGSTGSRGLHHLVYEILDNAID----EAQAGYASKVDV 146
Query 63 TAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDNYDDTEA--RVTG 120
V A+G++ V ++G GIP +H K E + L + T + V+G
Sbjct 147 ------VLHADGSVSVVDNGRGIPTDLHPATKKSSLETVLTVLHAGGKFGGTSSGYSVSG 200
Query 121 GRNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDNMSKQ 161
G +G G + N S EV VW D M +
Sbjct 201 GLHGVGLSVVNALSEALEVS------------VWRDGMEHK 229
> cel:R05D3.12 hypothetical protein
Length=144
Score = 46.6 bits (109), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 23/53 (43%), Positives = 33/53 (62%), Gaps = 1/53 (1%)
Query 1 YIGSTEISSQQL-WVMPEGEPRMQQRVISFAPGLYKIVDEILVNAADVKARET 52
YIG + Q+ W+ +M + +++ PGL KI DEILVNAAD KAR++
Sbjct 63 YIGGVAMREDQIIWLRDSENRKMIAKEVTYPPGLLKIFDEILVNAADNKARDS 115
> dre:100334809 Top2a protein-like
Length=95
Score = 46.6 bits (109), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 24/41 (58%), Positives = 28/41 (68%), Gaps = 1/41 (2%)
Query 1 YIGSTEISSQQLWVMPEGEPRMQQRVISFAPGLYKIVDEIL 41
YIGS E +QQ+WV E + M R I+F PGLYKI DEIL
Sbjct 49 YIGSVEPVTQQMWVFDE-DTGMNCRDITFVPGLYKIFDEIL 88
> tgo:TGME49_097780 DNA gyrase subunit B, putative (EC:5.99.1.3);
K02470 DNA gyrase subunit B [EC:5.99.1.3]
Length=1257
Score = 42.7 bits (99), Expect = 0.001, Method: Composition-based stats.
Identities = 31/108 (28%), Positives = 47/108 (43%), Gaps = 10/108 (9%)
Query 32 GLYKIVDEILVNAADVKARETEGGAAPRAKMTAIKVNVNQAEGTIRVWNDGDGIPVAIHK 91
GL+ +V E + NA D E E G R V V A+ + V +DG GIP +H
Sbjct 286 GLHHLVWEAIDNAVD----EVENGDCRRI------VVVLHADRRVTVADDGRGIPCDVHP 335
Query 92 EHKVYVAELIFGQLLTSDNYDDTEARVTGGRNGFGAKLTNIFSTYFEV 139
+ + E +F +L + GG +G G+ + N S+ V
Sbjct 336 QTGLSALETVFTKLHAGGKFSPASYAFAGGLHGVGSSVVNALSSQLRV 383
> ath:AT3G10270 ATP binding / DNA binding / DNA topoisomerase
(ATP-hydrolyzing); K02470 DNA gyrase subunit B [EC:5.99.1.3]
Length=693
Score = 33.5 bits (75), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 37/162 (22%), Positives = 64/162 (39%), Gaps = 12/162 (7%)
Query 32 GLYKIVDEILVNAADVKARETEGGAAPRAKMTAIKVNVNQAEGTIRVWNDGDGIPVAIHK 91
GL+ +V EIL NA D E + G A + + V ++ ++ + ++G GIP +H
Sbjct 46 GLHHLVYEILDNAID----EAQAGFASKIDV------VLHSDDSVSISDNGRGIPTDLHP 95
Query 92 EHKVYVAELIFGQLLTSDNYDDTEA--RVTGGRNGFGAKLTNIFSTYFEVECADSKRNKK 149
E + L + + V+GG +G G + N S EV +
Sbjct 96 ATGKSSLETVLTVLHAGGKFGGKSSGYSVSGGLHGVGLSVVNALSEALEVIVRRDGMEFQ 155
Query 150 IKVVWTDNMSKQDPPVISPFNGTGDYVAVKFKPDLRRFGITT 191
K ++ V+ P + ++F PD F + T
Sbjct 156 QKYSRGKPVTTLTCHVLPPESRGTQGTCIRFWPDKEGFALFT 197
> cel:W03H9.3 hypothetical protein
Length=96
Score = 32.7 bits (73), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 24/43 (55%), Gaps = 5/43 (11%)
Query 102 FGQLLTSDNYDDTEARVTGGRNGFGAKLTNIFSTYFEVECADS 144
+G T D++DD E R+ +N F AK+ N F V+ A+S
Sbjct 4 YGFFETVDDFDDVEYRIMPLQNSFSAKIVN-----FSVKSAES 41
> xla:398840 hypothetical protein MGC68579; K00898 pyruvate dehydrogenase
kinase [EC:2.7.11.2]
Length=404
Score = 32.0 bits (71), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 34/144 (23%), Positives = 67/144 (46%), Gaps = 23/144 (15%)
Query 1 YIGSTEISSQQLWVMPEGEPRMQQRVISFAPG-LYKIVDEILVNA--ADVKARETEGGAA 57
Y+ S E+ +Q G Q I + P LY ++ E+ NA A V++ ET +
Sbjct 216 YLASPELRIKQT----NGNDPSQPIHIVYVPSHLYHMLFELFKNAMRATVESHET----S 267
Query 58 PRAKMTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDN---YDDT 114
PR + V + + TI++ ++G G+P+ K+ E +F + ++ D++
Sbjct 268 PRLPPIKVNVVLGNEDVTIKISDNGGGVPL-----RKI---ERLFSYMYSTAPRPLMDNS 319
Query 115 EARVTGGRNGFGAKLTNIFSTYFE 138
G G+G ++ +++ YF+
Sbjct 320 RNAPLAGF-GYGLPISRLYARYFQ 342
> dre:326944 prpf4, mg:ab03a02, zgc:65943; PRP4 pre-mRNA processing
factor 4 homolog (yeast); K12662 U4/U6 small nuclear ribonucleoprotein
PRP4
Length=507
Score = 30.8 bits (68), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 24/92 (26%), Positives = 47/92 (51%), Gaps = 12/92 (13%)
Query 33 LYKIVDEILVNAADVKARETEGGA---APRAKMTAIKVNVNQA----EGTIRVWNDGDGI 85
L+K+ D L+ ++ T GA P+A +T + +VN A +G++++W+
Sbjct 242 LWKVPDCTLIRT--LRGHNTNVGAISFHPQATLTLEESDVNMASCAADGSVKLWSLDSDE 299
Query 86 PVAIHKEHKVYVAELIF---GQLLTSDNYDDT 114
P+A + H + VA + + G+ L + YD +
Sbjct 300 PIADIEGHSMRVARVTWHPSGRFLGTTCYDHS 331
> dre:100334455 PRP4 pre-mRNA processing factor 4 homolog; K12662
U4/U6 small nuclear ribonucleoprotein PRP4
Length=507
Score = 30.8 bits (68), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 24/92 (26%), Positives = 47/92 (51%), Gaps = 12/92 (13%)
Query 33 LYKIVDEILVNAADVKARETEGGA---APRAKMTAIKVNVNQA----EGTIRVWNDGDGI 85
L+K+ D L+ ++ T GA P+A +T + +VN A +G++++W+
Sbjct 242 LWKVPDCTLIRT--LRGHNTNVGAISFHPQATLTLEESDVNMASCAADGSVKLWSLDSDE 299
Query 86 PVAIHKEHKVYVAELIF---GQLLTSDNYDDT 114
P+A + H + VA + + G+ L + YD +
Sbjct 300 PIADIEGHSMRVARVTWHPSGRFLGTTCYDHS 331
> xla:379789 pdk4, 3j828, MGC132353, MGC52849; pyruvate dehydrogenase
kinase, isozyme 4 (EC:2.7.11.2); K00898 pyruvate dehydrogenase
kinase [EC:2.7.11.2]
Length=404
Score = 30.8 bits (68), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 33/144 (22%), Positives = 67/144 (46%), Gaps = 23/144 (15%)
Query 1 YIGSTEISSQQLWVMPEGEPRMQQRVISFAPG-LYKIVDEILVNA--ADVKARETEGGAA 57
Y+ S E+ +Q G+ Q I + P LY ++ E+ NA A ++ ET +
Sbjct 216 YLASPELRIKQA----NGKDPSQPIHIVYVPSHLYHMLFELFKNAMRATIENHET----S 267
Query 58 PRAKMTAIKVNVNQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQLLTSDN---YDDT 114
PR + V + + TI++ ++G G+P+ K+ E +F + ++ D++
Sbjct 268 PRLPPIKVNVVLGSEDLTIKISDNGGGVPL-----RKI---ERLFSYMYSTAPRPLMDNS 319
Query 115 EARVTGGRNGFGAKLTNIFSTYFE 138
G G+G ++ +++ YF+
Sbjct 320 RNAPLAGF-GYGLPISRLYARYFQ 342
> tgo:TGME49_052390 hypothetical protein
Length=103
Score = 30.8 bits (68), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 28/51 (54%), Gaps = 5/51 (9%)
Query 132 IFSTYFEVECADSK-RNKKIKVVWTDNMSKQDPPVISPFNGTGDYVAVKFK 181
+F F V C ++ +N KI VV D ++K D V P G GD + VKFK
Sbjct 40 VFLWCFSVSCIRTEAKNLKIDVV--DGITKTDAHV--PHLGAGDTITVKFK 86
> dre:445394 wdr55, flj20195l, zgc:111796; WD repeat domain 55
Length=386
Score = 30.4 bits (67), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 30/119 (25%), Positives = 51/119 (42%), Gaps = 17/119 (14%)
Query 5 TEISSQQLWVMPEGEPRMQQRVISFAPG---LYKIVDEILVNAADVKARETE-----GGA 56
TE +++LW G R + F+ L+ + + ++ DV+A + E
Sbjct 67 TEGENKELW--SSGHHLKSCRKVLFSSDGQKLFSVSKDKAIHIMDVEAGKLETRIPKAHK 124
Query 57 APRAKMTAIKVNV---NQAEGTIRVWNDGDGIPVAIHKEHKVYVAELIFGQ----LLTS 108
P M I N+ EGT++VW+ G K H+ Y++++ Q LLTS
Sbjct 125 VPINAMLLIDENIFATGDDEGTLKVWDMRKGTSFMDLKHHEDYISDITIDQAKRTLLTS 183
> cel:T27A8.1 hypothetical protein
Length=492
Score = 29.6 bits (65), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 12/40 (30%), Positives = 20/40 (50%), Gaps = 0/40 (0%)
Query 118 VTGGRNGFGAKLTNIFSTYFEVECADSKRNKKIKVVWTDN 157
V+GG + TN F E+ C + KK++ +W +N
Sbjct 304 VSGGMQDWNYLNTNCFEVTVEMNCEKFPQTKKLRYLWEEN 343
> mmu:237934 Krt39, 4732494G06Rik, Ka35; keratin 39; K07604 type
I keratin, acidic
Length=482
Score = 29.3 bits (64), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 31/49 (63%), Gaps = 0/49 (0%)
Query 2 IGSTEISSQQLWVMPEGEPRMQQRVISFAPGLYKIVDEILVNAADVKAR 50
I +T++++ L E E ++Q V + A GL +I+D + ++ AD++AR
Sbjct 166 IDNTKLAADDLRANYEAELSLRQLVEADANGLKQILDALTLSKADLEAR 214
Lambda K H
0.318 0.136 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6253620652
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40