bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1174_orf1
Length=100
Score E
Sequences producing significant alignments: (Bits) Value
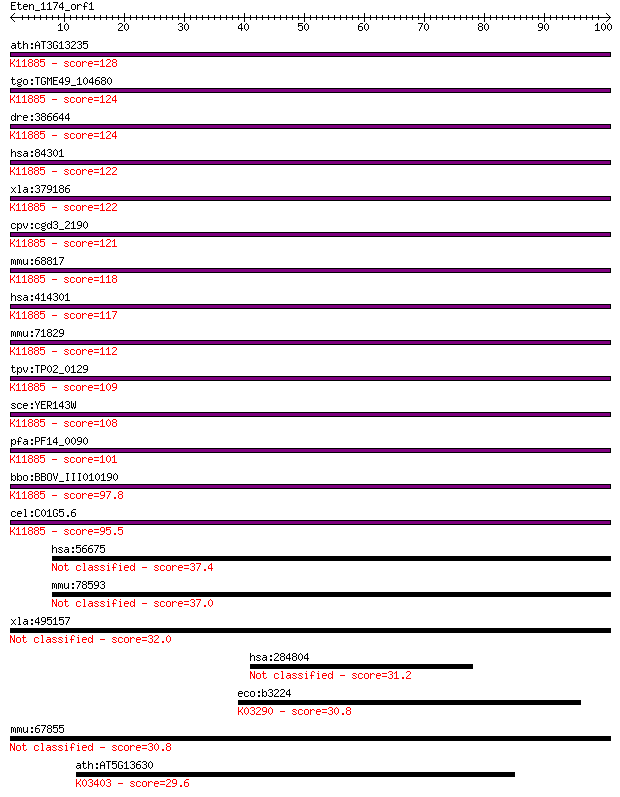
ath:AT3G13235 ubiquitin family protein; K11885 DNA damage-indu... 128 4e-30
tgo:TGME49_104680 DNA-damage inducible protein, putative ; K11... 124 6e-29
dre:386644 ddi2, zgc:63515; DNA-damage inducible protein 2; K1... 124 8e-29
hsa:84301 DDI2, MGC14844, RP4-680D5.5; DNA-damage inducible 1 ... 122 2e-28
xla:379186 ddi2, MGC53726; protein DDI1 homolog 2; K11885 DNA ... 122 2e-28
cpv:cgd3_2190 ubiquitin domain containing protein with a UBA d... 121 6e-28
mmu:68817 Ddi2, 1110056G13Rik, 1700027M01Rik, 9130022E05Rik, A... 118 4e-27
hsa:414301 DDI1, FLJ36017; DNA-damage inducible 1 homolog 1 (S... 117 6e-27
mmu:71829 Ddi1, 1700011N24Rik; DDI1, DNA-damage inducible 1, h... 112 2e-25
tpv:TP02_0129 hypothetical protein; K11885 DNA damage-inducibl... 109 2e-24
sce:YER143W DDI1, VSM1; DNA damage-inducible V-SNARE binding p... 108 3e-24
pfa:PF14_0090 DNA-damage inducible protein, putative; K11885 D... 101 6e-22
bbo:BBOV_III010190 17.m07883; hypothetical protein; K11885 DNA... 97.8 7e-21
cel:C01G5.6 hypothetical protein; K11885 DNA damage-inducible ... 95.5 4e-20
hsa:56675 NRIP3, C11orf14, NY-SAR-105; nuclear receptor intera... 37.4 0.013
mmu:78593 Nrip3, A330103B05Rik, D7H11orf14, ICRFP703B1614Q5.2;... 37.0 0.014
xla:495157 nrip3, ny-sar-105; nuclear receptor interacting pro... 32.0 0.49
hsa:284804 hypothetical protein LOC284804 31.2 0.92
eco:b3224 nanT, ECK3213, JW3193; sialic acid transporter; K032... 30.8 1.0
mmu:67855 Asprv1, 2300003P22Rik, AA986851, SASP, SASPase, Taps... 30.8 1.1
ath:AT5G13630 GUN5; GUN5 (GENOMES UNCOUPLED 5); magnesium chel... 29.6 2.7
> ath:AT3G13235 ubiquitin family protein; K11885 DNA damage-inducible
protein 1
Length=414
Score = 128 bits (322), Expect = 4e-30, Method: Composition-based stats.
Identities = 55/100 (55%), Positives = 78/100 (78%), Gaps = 0/100 (0%)
Query 1 YVHLEINGVACKAFVDSGAQQSILSLAFAEKCSLSSLIDKRFAGLALGVGRAPIIGRVHL 60
YV +E+NGV KAFVDSGAQ +I+S + AE+C L L+D+R+ G+A GVG+ I+GR+H+
Sbjct 203 YVDMEVNGVPLKAFVDSGAQSTIISKSCAERCGLLRLMDQRYKGIAHGVGQTEILGRIHV 262
Query 61 APLKLGSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
AP+K+G+ F PCSF+VL+ M+ L GLDML+++Q IDL
Sbjct 263 APIKIGNNFYPCSFVVLDSPNMEFLFGLDMLRKHQCTIDL 302
> tgo:TGME49_104680 DNA-damage inducible protein, putative ; K11885
DNA damage-inducible protein 1
Length=527
Score = 124 bits (312), Expect = 6e-29, Method: Composition-based stats.
Identities = 55/100 (55%), Positives = 77/100 (77%), Gaps = 0/100 (0%)
Query 1 YVHLEINGVACKAFVDSGAQQSILSLAFAEKCSLSSLIDKRFAGLALGVGRAPIIGRVHL 60
++ +E+NGV KAFVDSGAQ + +S A A+KCSL L+D R+ G+A GVG+ I+G++HL
Sbjct 300 FIDIEVNGVPIKAFVDSGAQSTFMSYACAQKCSLLRLMDTRYRGVAQGVGKTEIVGKIHL 359
Query 61 APLKLGSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
A LK+G +F P SF VL+D+K++ L GLD+L+RYQ IDL
Sbjct 360 ATLKIGQRFFPSSFTVLQDNKVEFLFGLDLLRRYQCCIDL 399
> dre:386644 ddi2, zgc:63515; DNA-damage inducible protein 2;
K11885 DNA damage-inducible protein 1
Length=409
Score = 124 bits (311), Expect = 8e-29, Method: Composition-based stats.
Identities = 58/100 (58%), Positives = 76/100 (76%), Gaps = 0/100 (0%)
Query 1 YVHLEINGVACKAFVDSGAQQSILSLAFAEKCSLSSLIDKRFAGLALGVGRAPIIGRVHL 60
Y++ ++NG KAFVDSGAQ +I+S A AE+C++ L+D+R+AG+A GVG IIGRVHL
Sbjct 247 YINCKVNGHPVKAFVDSGAQMTIMSQACAERCNIMRLVDRRWAGIAKGVGTQKIIGRVHL 306
Query 61 APLKLGSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
A +++ F PCSF +LED M MLLGLDMLKR+Q IDL
Sbjct 307 AQVQIEGDFLPCSFSILEDQPMDMLLGLDMLKRHQCSIDL 346
> hsa:84301 DDI2, MGC14844, RP4-680D5.5; DNA-damage inducible
1 homolog 2 (S. cerevisiae); K11885 DNA damage-inducible protein
1
Length=399
Score = 122 bits (307), Expect = 2e-28, Method: Composition-based stats.
Identities = 57/100 (57%), Positives = 76/100 (76%), Gaps = 0/100 (0%)
Query 1 YVHLEINGVACKAFVDSGAQQSILSLAFAEKCSLSSLIDKRFAGLALGVGRAPIIGRVHL 60
Y++ ++NG KAFVDSGAQ +I+S A AE+C++ L+D+R+AG+A GVG IIGRVHL
Sbjct 237 YINCKVNGHPVKAFVDSGAQMTIMSQACAERCNIMRLVDRRWAGIAKGVGTQKIIGRVHL 296
Query 61 APLKLGSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
A +++ F PCSF +LE+ M MLLGLDMLKR+Q IDL
Sbjct 297 AQVQIEGDFLPCSFSILEEQPMDMLLGLDMLKRHQCSIDL 336
> xla:379186 ddi2, MGC53726; protein DDI1 homolog 2; K11885 DNA
damage-inducible protein 1
Length=393
Score = 122 bits (307), Expect = 2e-28, Method: Composition-based stats.
Identities = 57/100 (57%), Positives = 75/100 (75%), Gaps = 0/100 (0%)
Query 1 YVHLEINGVACKAFVDSGAQQSILSLAFAEKCSLSSLIDKRFAGLALGVGRAPIIGRVHL 60
Y++ ++NG KAFVDSGAQ +I+S A AE+C + L+D+R+AG+A GVG IIGRVHL
Sbjct 231 YINCKVNGYPVKAFVDSGAQMTIMSQACAERCHIMRLVDRRWAGIAKGVGTQKIIGRVHL 290
Query 61 APLKLGSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
A +++ F PCSF +LE+ M MLLGLDMLKR+Q IDL
Sbjct 291 AQVQIEGDFLPCSFSILEEQPMDMLLGLDMLKRHQCSIDL 330
> cpv:cgd3_2190 ubiquitin domain containing protein with a UBA
domain at the C-terminus ; K11885 DNA damage-inducible protein
1
Length=384
Score = 121 bits (303), Expect = 6e-28, Method: Composition-based stats.
Identities = 55/100 (55%), Positives = 76/100 (76%), Gaps = 0/100 (0%)
Query 1 YVHLEINGVACKAFVDSGAQQSILSLAFAEKCSLSSLIDKRFAGLALGVGRAPIIGRVHL 60
Y++ E+NG++ KAFVDSGAQ +I+S AEKC+L LID RF+G+A GVG + I+G++H+
Sbjct 205 YINAEVNGISIKAFVDSGAQTTIMSKKCAEKCNLVRLIDYRFSGIAQGVGTSKIVGKIHV 264
Query 61 APLKLGSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
A +K+G+ F P S VLE+S + L GLD+LKRYQ IDL
Sbjct 265 AQMKIGNSFFPFSITVLEESHVDFLFGLDLLKRYQCCIDL 304
> mmu:68817 Ddi2, 1110056G13Rik, 1700027M01Rik, 9130022E05Rik,
AI604911, AU040698; DNA-damage inducible protein 2; K11885
DNA damage-inducible protein 1
Length=399
Score = 118 bits (296), Expect = 4e-27, Method: Composition-based stats.
Identities = 56/100 (56%), Positives = 74/100 (74%), Gaps = 0/100 (0%)
Query 1 YVHLEINGVACKAFVDSGAQQSILSLAFAEKCSLSSLIDKRFAGLALGVGRAPIIGRVHL 60
Y++ +NG KAFVDSGAQ +I+S A AE+C++ L+D+R+AG+A GVG IIGRVHL
Sbjct 237 YINCRVNGHPVKAFVDSGAQMTIMSQACAERCNIMRLVDRRWAGIAKGVGTQKIIGRVHL 296
Query 61 APLKLGSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
A +++ F CSF +LE+ M MLLGLDMLKR+Q IDL
Sbjct 297 AQVQIEGDFLACSFSILEEQPMDMLLGLDMLKRHQCSIDL 336
> hsa:414301 DDI1, FLJ36017; DNA-damage inducible 1 homolog 1
(S. cerevisiae); K11885 DNA damage-inducible protein 1
Length=396
Score = 117 bits (294), Expect = 6e-27, Method: Composition-based stats.
Identities = 56/100 (56%), Positives = 75/100 (75%), Gaps = 0/100 (0%)
Query 1 YVHLEINGVACKAFVDSGAQQSILSLAFAEKCSLSSLIDKRFAGLALGVGRAPIIGRVHL 60
Y++ ++NG KAFVDSGAQ +I+S A AE+C++ L+D+R+AG+A GVG IIGRVHL
Sbjct 245 YINCKVNGHPLKAFVDSGAQMTIMSQACAERCNIMRLVDRRWAGVAKGVGTQRIIGRVHL 304
Query 61 APLKLGSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
A +++ F CSF +LED M MLLGLDML+R+Q IDL
Sbjct 305 AQIQIEGDFLQCSFSILEDQPMDMLLGLDMLRRHQCSIDL 344
> mmu:71829 Ddi1, 1700011N24Rik; DDI1, DNA-damage inducible 1,
homolog 1 (S. cerevisiae); K11885 DNA damage-inducible protein
1
Length=408
Score = 112 bits (281), Expect = 2e-25, Method: Composition-based stats.
Identities = 52/100 (52%), Positives = 74/100 (74%), Gaps = 0/100 (0%)
Query 1 YVHLEINGVACKAFVDSGAQQSILSLAFAEKCSLSSLIDKRFAGLALGVGRAPIIGRVHL 60
Y++ ++NG KAFVDSGAQ +I+S A AE+C++ L+D+R+ G+A GVG I+GRVHL
Sbjct 251 YINCKVNGHPLKAFVDSGAQMTIMSQACAERCNIMRLVDRRWGGVAKGVGTQRIMGRVHL 310
Query 61 APLKLGSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
A +++ F CSF +LE+ M +LLGLDML+R+Q IDL
Sbjct 311 AQIQIEGDFLQCSFSILEEQPMDILLGLDMLRRHQCSIDL 350
> tpv:TP02_0129 hypothetical protein; K11885 DNA damage-inducible
protein 1
Length=359
Score = 109 bits (272), Expect = 2e-24, Method: Composition-based stats.
Identities = 49/100 (49%), Positives = 74/100 (74%), Gaps = 0/100 (0%)
Query 1 YVHLEINGVACKAFVDSGAQQSILSLAFAEKCSLSSLIDKRFAGLALGVGRAPIIGRVHL 60
Y+ +EIN V KA VD+GAQ +I+S A +C+L SL+D+RF G+A+GVG +G++HL
Sbjct 200 YIKVEINNVVVKALVDTGAQNTIMSKECALQCNLLSLVDERFKGVAVGVGSTKTLGKIHL 259
Query 61 APLKLGSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
A +K+GS F P SFIV++ + ++ +LGLD+L+RY I+L
Sbjct 260 ADMKIGSIFIPVSFIVIDGANLEFILGLDILRRYTCDINL 299
> sce:YER143W DDI1, VSM1; DNA damage-inducible V-SNARE binding
protein, contains a ubiquitin-associated (UBA) domain, may
act as a negative regulator of constitutive exocytosis, may
play a role in S-phase checkpoint control; K11885 DNA damage-inducible
protein 1
Length=428
Score = 108 bits (271), Expect = 3e-24, Method: Composition-based stats.
Identities = 52/100 (52%), Positives = 72/100 (72%), Gaps = 1/100 (1%)
Query 1 YVHLEINGVACKAFVDSGAQQSILSLAFAEKCSLSSLIDKRFAGLALGVGRAPIIGRVHL 60
Y+++EIN KAFVD+GAQ +I+S A+K LS +IDKRF G A GVG IIGR+H
Sbjct 205 YINIEINNYPVKAFVDTGAQTTIMSTRLAKKTGLSRMIDKRFIGEARGVGTGKIIGRIHQ 264
Query 61 APLKLGSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
A +K+ +++ PCSF VL D+ + +L+GLDMLKR+ +DL
Sbjct 265 AQVKIETQYIPCSFTVL-DTDIDVLIGLDMLKRHLACVDL 303
> pfa:PF14_0090 DNA-damage inducible protein, putative; K11885
DNA damage-inducible protein 1
Length=382
Score = 101 bits (251), Expect = 6e-22, Method: Composition-based stats.
Identities = 46/100 (46%), Positives = 68/100 (68%), Gaps = 0/100 (0%)
Query 1 YVHLEINGVACKAFVDSGAQQSILSLAFAEKCSLSSLIDKRFAGLALGVGRAPIIGRVHL 60
Y+ +EIN AFVDSGAQ SI+S A+KC++ L+DKRF G+A GVG I+G++H+
Sbjct 247 YIPVEINKNTVHAFVDSGAQSSIMSKKCAQKCNILRLMDKRFTGIAKGVGTKTILGKIHM 306
Query 61 APLKLGSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
+K+G+ F S ++ED + + GLD+LKR+Q +ID
Sbjct 307 IDIKIGNYFYAVSLTIIEDYDIDFIFGLDLLKRHQCLIDF 346
> bbo:BBOV_III010190 17.m07883; hypothetical protein; K11885 DNA
damage-inducible protein 1
Length=500
Score = 97.8 bits (242), Expect = 7e-21, Method: Composition-based stats.
Identities = 41/100 (41%), Positives = 74/100 (74%), Gaps = 0/100 (0%)
Query 1 YVHLEINGVACKAFVDSGAQQSILSLAFAEKCSLSSLIDKRFAGLALGVGRAPIIGRVHL 60
++ ++INGV +A VD+GAQ S++ + +AEKC+L ++ID+RF G+A+G+ + IIG++H+
Sbjct 326 FIPVQINGVNLEALVDTGAQNSVMRIDYAEKCNLLNIIDRRFQGVAVGISKERIIGKIHM 385
Query 61 APLKLGSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
A +K+G+ F S V+E + ++GLD++++YQ V+ L
Sbjct 386 AQMKIGNLFLLFSSSVIEQLNVGFIIGLDIMRQYQCVVSL 425
> cel:C01G5.6 hypothetical protein; K11885 DNA damage-inducible
protein 1
Length=389
Score = 95.5 bits (236), Expect = 4e-20, Method: Composition-based stats.
Identities = 46/101 (45%), Positives = 70/101 (69%), Gaps = 1/101 (0%)
Query 1 YVHLEINGVACKAFVDSGAQQSILSLAFAEKCSLSSLIDKRFAGLALGVGRAPII-GRVH 59
Y++L INGV KAF+DSGAQ+SI+S+A AE+C L+ LID+RF +A GVG I G++H
Sbjct 246 YINLTINGVPVKAFIDSGAQKSIMSMACAERCGLNGLIDRRFQSMARGVGGTEKIEGKIH 305
Query 60 LAPLKLGSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
L +K+ C F V+ +M +L+GL++L+++ I+L
Sbjct 306 LCDVKVEDAHFSCPFEVMARREMDLLIGLNVLRKHGCCINL 346
> hsa:56675 NRIP3, C11orf14, NY-SAR-105; nuclear receptor interacting
protein 3
Length=241
Score = 37.4 bits (85), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 26/95 (27%), Positives = 49/95 (51%), Gaps = 2/95 (2%)
Query 8 GVACKAFVDSGAQQSILSLAFAEKCSLSSLI-DKRFAGLALGVGRA-PIIGRVHLAPLKL 65
G KA VD+G +++SLA ++ L + + G L + R ++G++ + L
Sbjct 116 GKDVKALVDTGCLYNLISLACVDRLGLKEHVKSHKHEGEKLSLPRHLKVVGQIEHLVITL 175
Query 66 GSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
GS C V++D++ + LGL L+ + +I+L
Sbjct 176 GSLRLDCPAAVVDDNEKNLSLGLQTLRSLKCIINL 210
> mmu:78593 Nrip3, A330103B05Rik, D7H11orf14, ICRFP703B1614Q5.2;
nuclear receptor interacting protein 3
Length=240
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 25/95 (26%), Positives = 49/95 (51%), Gaps = 2/95 (2%)
Query 8 GVACKAFVDSGAQQSILSLAFAEKCSLSSLI-DKRFAGLALGVGRA-PIIGRVHLAPLKL 65
G KA VD+G Q +++S A ++ L + + G L + R ++G++ + +
Sbjct 115 GKDVKALVDTGCQYNLISSACVDRLGLKDHVKSHKHEGEKLSLPRHLKVVGQIEHLMITV 174
Query 66 GSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
GS C V++D++ + LGL L+ + +I+L
Sbjct 175 GSLRLDCQAAVVDDNEKSLSLGLQTLRSLKCIINL 209
> xla:495157 nrip3, ny-sar-105; nuclear receptor interacting protein
3
Length=247
Score = 32.0 bits (71), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 27/103 (26%), Positives = 47/103 (45%), Gaps = 4/103 (3%)
Query 1 YVHLEINGVACKAFVDSGAQQSILSLAFAEKCSLSS---LIDKRFAGLALGVGRAPIIGR 57
YV + G+ +A +D+ Q S++ ++ L L K +L IG+
Sbjct 114 YVCCQCAGMEVRALIDTSCQYSLIPAPCLDRLGLKEHFRLYKKEEEPPSLPYS-VKAIGQ 172
Query 58 VHLAPLKLGSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
V + LG C+ IV++++ + LGL LK + VI+L
Sbjct 173 VERVAVMLGGATVECTAIVIDNNDRNLSLGLQTLKSLKCVINL 215
> hsa:284804 hypothetical protein LOC284804
Length=346
Score = 31.2 bits (69), Expect = 0.92, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 22/41 (53%), Gaps = 4/41 (9%)
Query 41 RFAGLALGVGRAPIIGRVHLAPLKLGS----KFCPCSFIVL 77
R G LG GR P +G+V P+K GS +F C+ I L
Sbjct 282 RSKGFRLGSGREPKMGQVVYVPVKTGSLCSIQFYGCTIIYL 322
> eco:b3224 nanT, ECK3213, JW3193; sialic acid transporter; K03290
MFS transporter, SHS family, sialic acid transporter
Length=496
Score = 30.8 bits (68), Expect = 1.0, Method: Composition-based stats.
Identities = 25/64 (39%), Positives = 32/64 (50%), Gaps = 9/64 (14%)
Query 39 DKRFAGL-------ALGVGRAPIIGRVHLAPLKLGSKFCPCSFIVLEDSKMQMLLGLDML 91
D+R AGL ALG APIIG + L LG+ SF + + +L+GLDM
Sbjct 398 DQRAAGLGFTYNVGALGGALAPIIGALIAQRLDLGTALASLSFSL--TFVVILLIGLDMP 455
Query 92 KRYQ 95
R Q
Sbjct 456 SRVQ 459
> mmu:67855 Asprv1, 2300003P22Rik, AA986851, SASP, SASPase, Taps;
aspartic peptidase, retroviral-like 1
Length=339
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 27/103 (26%), Positives = 46/103 (44%), Gaps = 4/103 (3%)
Query 1 YVHLEINGVACKAFVDSGAQQSILSLAFAEKCSLSSLIDKRFAGLALGVGRAP---IIGR 57
Y+ ++ V + VDSGAQ S++ A E+ + L R + V I+G
Sbjct 195 YLKGKVGHVPVRFLVDSGAQVSVVHPALWEEVTDGDLDTLRPFNNVVKVANGAEMKILG- 253
Query 58 VHLAPLKLGSKFCPCSFIVLEDSKMQMLLGLDMLKRYQMVIDL 100
V + LG F+V S + ++G D+L+ + V+D
Sbjct 254 VWDTEISLGKTKLKAEFLVANASAEEAIIGTDVLQDHNAVLDF 296
> ath:AT5G13630 GUN5; GUN5 (GENOMES UNCOUPLED 5); magnesium chelatase;
K03403 magnesium chelatase subunit H [EC:6.6.1.1]
Length=1263
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 19/76 (25%), Positives = 39/76 (51%), Gaps = 3/76 (3%)
Query 12 KAFVDSGAQQSILS--LAFAEKCSLSSLIDKRFAGLALG-VGRAPIIGRVHLAPLKLGSK 68
++ D+G I+S ++ A++C+L +D GL L R ++G+V+ +++ S+
Sbjct 747 QSLKDTGRGPQIVSSIISTAKQCNLDKDVDLPDEGLELSPKDRDSVVGKVYSKIMEIESR 806
Query 69 FCPCSFIVLEDSKMQM 84
PC V+ + M
Sbjct 807 LLPCGLHVIGEPPSAM 822
Lambda K H
0.328 0.142 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2041372988
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40