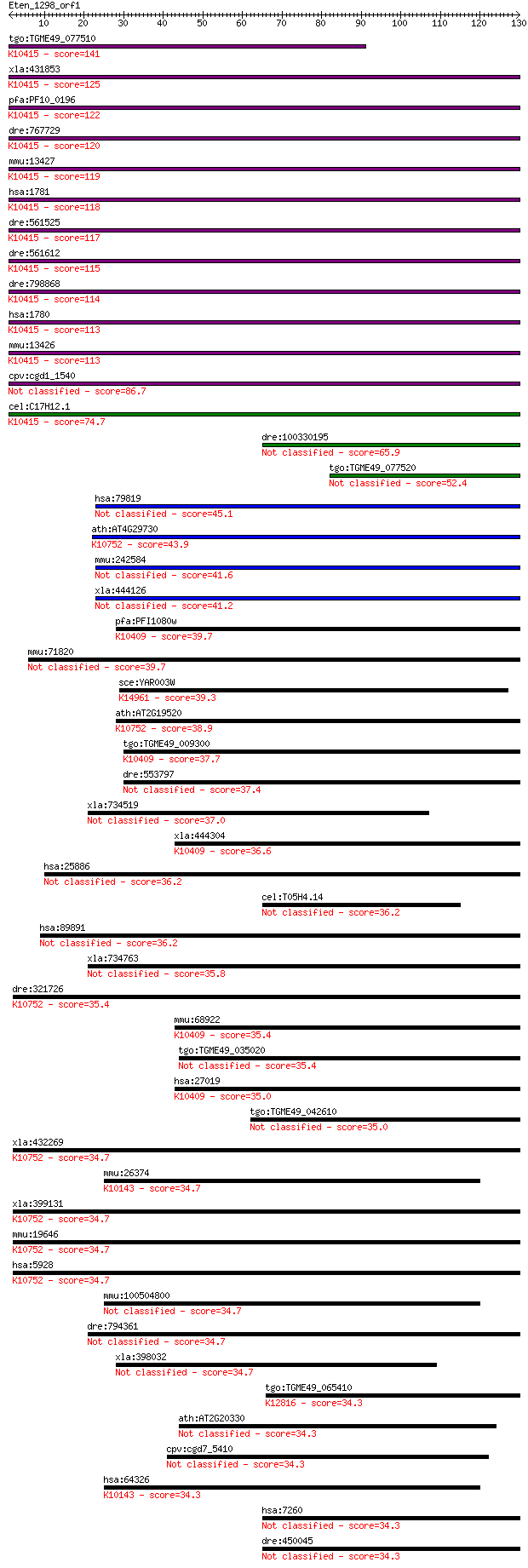
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1298_orf1
Length=129
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_077510 dynein intermediate chain, putative ; K10415... 141 5e-34
xla:431853 dync1i2, MGC83936, dnci2, ic2; dynein, cytoplasmic ... 125 4e-29
pfa:PF10_0196 cytoplasmic dynein intermediate chain, putative;... 122 2e-28
dre:767729 dync1i2a, MGC153318, dync1i2, zgc:153318; dynein, c... 120 1e-27
mmu:13427 Dync1i2, 3110079H08Rik, AW554389, Dncic2; dynein cyt... 119 3e-27
hsa:1781 DYNC1I2, DNCI2, FLJ21089, FLJ90842, IC2, MGC104199, M... 118 5e-27
dre:561525 dync1i1, MGC158394, sb:eu1013, zgc:158394; dynein, ... 117 9e-27
dre:561612 dync1i2b; dynein, cytoplasmic 1, intermediate chain... 115 4e-26
dre:798868 dynein cytoplasmic 1 intermediate chain 1-like; K10... 114 6e-26
hsa:1780 DYNC1I1, DNCI1, DNCIC1; dynein, cytoplasmic 1, interm... 113 1e-25
mmu:13426 Dync1i1, DH_IC-1, DHIC-1, DIC, Dncic1, IC74; dynein ... 113 1e-25
cpv:cgd1_1540 dynein intermediate chain 86.7 2e-17
cel:C17H12.1 dyci-1; DYnein Chain, light Intermediate family m... 74.7 6e-14
dre:100330195 dynein, cytoplasmic 1, intermediate chain 2a-like 65.9 3e-11
tgo:TGME49_077520 hypothetical protein 52.4 4e-07
hsa:79819 WDR78, FLJ23129; WD repeat domain 78 45.1
ath:AT4G29730 NFC5; NFC5 (Nucleosome/chromatin assembly factor... 43.9 1e-04
mmu:242584 Wdr78, BC028975, MGC31722, MGC38950; WD repeat doma... 41.6 7e-04
xla:444126 wdr78, MGC80539; WD repeat domain 78 41.2
pfa:PFI1080w dynein intermediate chain 2, ciliary; K10409 dyne... 39.7 0.002
mmu:71820 Wdr34, 3200002I06Rik, MGC103259; WD repeat domain 34 39.7
sce:YAR003W SWD1, CPS50, FUN16, SAF49; Subunit of the COMPASS ... 39.3 0.003
ath:AT2G19520 FVE; FVE; metal ion binding; K10752 histone-bind... 38.9 0.005
tgo:TGME49_009300 dynein intermediate chain, putative ; K10409... 37.7 0.010
dre:553797 wdr78, MGC111979, zgc:111979; WD repeat domain 78 37.4
xla:734519 poc1b, MGC114911, pix1, tuwd12, wdr51b; POC1 centri... 37.0 0.016
xla:444304 MGC80975 protein; K10409 dynein intermediate chain ... 36.6 0.018
hsa:25886 POC1A, DKFZp434C245, MGC131902, PIX2, WDR51A; POC1 c... 36.2 0.023
cel:T05H4.14 gad-1; GAstrulation Defective family member (gad-1) 36.2 0.026
hsa:89891 WDR34, MGC20486, bA216B9.3; WD repeat domain 34 36.2
xla:734763 wdr34, MGC130908; WD repeat domain 34 35.8
dre:321726 rbb4, RBBP4, rbb4-2, wu:fb33a09, wu:fb40e10, zgc:55... 35.4 0.043
mmu:68922 Dnaic1, 1110066F04Rik, BB124644, Dnai1; dynein, axon... 35.4 0.045
tgo:TGME49_035020 coatomer protein complex subunit beta, putative 35.4 0.046
hsa:27019 DNAI1, CILD1, ICS, ICS1, MGC26204, PCD; dynein, axon... 35.0 0.054
tgo:TGME49_042610 WD domain, G-beta repeat-containing protein 35.0 0.056
xla:432269 rbbp4-b, MGC82618, nurf55, rbap48, xrbbp4; retinobl... 34.7 0.074
mmu:26374 Rfwd2, AI316802, C80879, Cop1; ring finger and WD re... 34.7 0.074
xla:399131 rbbp4-a, MGC79922, nurf55, rbap48, rbbp4, xrbbp4; r... 34.7 0.076
mmu:19646 Rbbp4, mRbAp48; retinoblastoma binding protein 4; K1... 34.7 0.078
hsa:5928 RBBP4, NURF55, RBAP48; retinoblastoma binding protein... 34.7 0.078
mmu:100504800 e3 ubiquitin-protein ligase RFWD2-like 34.7 0.079
dre:794361 dync2i1, MGC171284, wdr34, zgc:171284; dynein, cyto... 34.7 0.083
xla:398032 katnb1; katanin p80 (WD repeat containing) subunit B 1 34.7
tgo:TGME49_065410 pre-mRNA splicing factor, putative ; K12816 ... 34.3 0.089
ath:AT2G20330 transducin family protein / WD-40 repeat family ... 34.3 0.094
cpv:cgd7_5410 WD40 protein 34.3 0.094
hsa:64326 RFWD2, COP1, FLJ10416, RNF200; ring finger and WD re... 34.3 0.100
hsa:7260 TSSC1; tumor suppressing subtransferable candidate 1 34.3
dre:450045 tssc1, zgc:101720; tumor suppressing subtransferabl... 34.3 0.11
> tgo:TGME49_077510 dynein intermediate chain, putative ; K10415
dynein intermediate chain, cytosolic
Length=539
Score = 141 bits (355), Expect = 5e-34, Method: Composition-based stats.
Identities = 60/91 (65%), Positives = 73/91 (80%), Gaps = 1/91 (1%)
Query 1 RLCQWSLQMMGQPAETLDLKRGSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNKA 60
RLC WSL M+G P ETLD+KRG+++VCVEC++FAE EVN L LG+EDG ++ ANIHGNK
Sbjct 434 RLCVWSLHMLGAPTETLDMKRGNKEVCVECIAFAENEVNSLVLGTEDGAMLKANIHGNKP 493
Query 61 GVMDVYEAHGGALTSLHFHPGMTE-GSRDYS 90
GV D Y++H GALTSLHFHP E G RDY+
Sbjct 494 GVTDAYDSHNGALTSLHFHPAAAEPGQRDYT 524
> xla:431853 dync1i2, MGC83936, dnci2, ic2; dynein, cytoplasmic
1, intermediate chain 2; K10415 dynein intermediate chain,
cytosolic
Length=637
Score = 125 bits (313), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 57/130 (43%), Positives = 91/130 (70%), Gaps = 4/130 (3%)
Query 1 RLCQWSLQMMGQPAETLDL-KRGSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNK 59
++C WSL M+ QP ++++L + S+ V V C+SF +VN+ +GSE+G + TA HG+K
Sbjct 405 KICSWSLDMLSQPQDSMELVHKQSKAVAVTCMSFPVGDVNNFVVGSEEGSVYTACRHGSK 464
Query 60 AGVMDVYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEG 119
AG+ +++E H G +T +H H + G+ D+SHL ++SS DW++KLW++KN ++PL FE
Sbjct 465 AGISEMFEGHQGPITGIHCHSAV--GAIDFSHLFVTSSFDWTVKLWTTKN-SKPLYSFED 521
Query 120 SDAYVYDVKW 129
+ YVYDV W
Sbjct 522 NSDYVYDVMW 531
> pfa:PF10_0196 cytoplasmic dynein intermediate chain, putative;
K10415 dynein intermediate chain, cytosolic
Length=769
Score = 122 bits (306), Expect = 2e-28, Method: Composition-based stats.
Identities = 56/129 (43%), Positives = 81/129 (62%), Gaps = 1/129 (0%)
Query 1 RLCQWSLQMMGQPAETLDLKRGSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNKA 60
RLC WSL M P+ T+DLK+G+R++ +F E ++N L G+EDG L A IHG K
Sbjct 540 RLCNWSLSMFTYPSNTIDLKKGNREISCTTFAFPEGDINTLYGGTEDGTLFQAQIHGMKV 599
Query 61 GVMDVYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEGS 120
G+++ H G +TS FHP + E D + L+L+SSVDW+ +L S KN+ PL F+
Sbjct 600 GIINSDIIHDGPITSAQFHPSLDE-MNDNNDLILTSSVDWTCQLLSIKNIKNPLIVFDSF 658
Query 121 DAYVYDVKW 129
+ Y+ D+KW
Sbjct 659 EDYIMDIKW 667
> dre:767729 dync1i2a, MGC153318, dync1i2, zgc:153318; dynein,
cytoplasmic 1, intermediate chain 2a; K10415 dynein intermediate
chain, cytosolic
Length=471
Score = 120 bits (300), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 56/130 (43%), Positives = 87/130 (66%), Gaps = 4/130 (3%)
Query 1 RLCQWSLQMMGQPAETLDLK-RGSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNK 59
++C WSL M+ QP ++++L + S+ V V +SF +VN+ +GSEDG + A+ HG++
Sbjct 239 KMCSWSLDMLSQPQDSMELVFKQSKSVAVTSMSFPLGDVNNFVVGSEDGSVYAASRHGSR 298
Query 60 AGVMDVYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEG 119
AG+ +++E H G +T +H H G D+SHL L++S DW++KLWS+KN +PL FE
Sbjct 299 AGISEMFEGHHGPITGIHCH--TAAGPVDFSHLFLTASFDWTVKLWSNKN-NKPLYSFED 355
Query 120 SDAYVYDVKW 129
+ YVYDV W
Sbjct 356 NSDYVYDVMW 365
> mmu:13427 Dync1i2, 3110079H08Rik, AW554389, Dncic2; dynein cytoplasmic
1 intermediate chain 2; K10415 dynein intermediate
chain, cytosolic
Length=655
Score = 119 bits (297), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 55/130 (42%), Positives = 88/130 (67%), Gaps = 4/130 (3%)
Query 1 RLCQWSLQMMGQPAETLDL-KRGSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNK 59
++C WSL M+ P ++++L + S+ V V +SF +VN+ +GSE+G + TA HG+K
Sbjct 423 KICSWSLDMLSHPQDSMELVHKQSKAVAVTSMSFPVGDVNNFVVGSEEGSVYTACRHGSK 482
Query 60 AGVMDVYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEG 119
AG+ +++E H G +T +H H + G+ D+SHL ++SS DW++KLW++KN +PL FE
Sbjct 483 AGISEMFEGHQGPITGIHCHAAV--GAVDFSHLFVTSSFDWTVKLWTTKN-NKPLYSFED 539
Query 120 SDAYVYDVKW 129
+ YVYDV W
Sbjct 540 NSDYVYDVMW 549
> hsa:1781 DYNC1I2, DNCI2, FLJ21089, FLJ90842, IC2, MGC104199,
MGC111094, MGC9324; dynein, cytoplasmic 1, intermediate chain
2; K10415 dynein intermediate chain, cytosolic
Length=638
Score = 118 bits (296), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 55/130 (42%), Positives = 88/130 (67%), Gaps = 4/130 (3%)
Query 1 RLCQWSLQMMGQPAETLDL-KRGSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNK 59
++C WSL M+ P ++++L + S+ V V +SF +VN+ +GSE+G + TA HG+K
Sbjct 406 KICSWSLDMLSHPQDSMELVHKQSKAVAVTSMSFPVGDVNNFVVGSEEGSVYTACRHGSK 465
Query 60 AGVMDVYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEG 119
AG+ +++E H G +T +H H + G+ D+SHL ++SS DW++KLW++KN +PL FE
Sbjct 466 AGISEMFEGHQGPITGIHCHAAV--GAVDFSHLFVTSSFDWTVKLWTTKN-NKPLYSFED 522
Query 120 SDAYVYDVKW 129
+ YVYDV W
Sbjct 523 NADYVYDVMW 532
> dre:561525 dync1i1, MGC158394, sb:eu1013, zgc:158394; dynein,
cytoplasmic 1, intermediate chain 1; K10415 dynein intermediate
chain, cytosolic
Length=627
Score = 117 bits (293), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 55/130 (42%), Positives = 88/130 (67%), Gaps = 4/130 (3%)
Query 1 RLCQWSLQMMGQPAETLDL-KRGSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNK 59
++C WSL M+ QP E+++L S+ V V C++F ++VN+ +GSE+G + TA+ HG+K
Sbjct 396 KMCSWSLDMLSQPQESMELIYNKSKPVAVTCMAFPASDVNNYVVGSEEGTVYTASRHGSK 455
Query 60 AGVMDVYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEG 119
AG+ ++++ H G +T + H + G D+SHL +SS DW++KLWS+K+ +PL FE
Sbjct 456 AGIGEIFDGHQGPVTGISCHNAV--GPVDFSHLFTTSSFDWTVKLWSTKH-NKPLYSFED 512
Query 120 SDAYVYDVKW 129
+ YVYDV W
Sbjct 513 NADYVYDVMW 522
> dre:561612 dync1i2b; dynein, cytoplasmic 1, intermediate chain
2b; K10415 dynein intermediate chain, cytosolic
Length=608
Score = 115 bits (288), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 57/130 (43%), Positives = 84/130 (64%), Gaps = 4/130 (3%)
Query 1 RLCQWSLQMMGQPAETLDLK-RGSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNK 59
++C WSL M+ P ++L+L + S+ V V +SF +VN+ +GSEDG + TA HG+K
Sbjct 376 KMCSWSLDMLSTPQDSLELVFKQSKSVAVTSMSFPLGDVNNFVVGSEDGSVYTACRHGSK 435
Query 60 AGVMDVYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEG 119
AG+ +++E H G +T L H G D+SHL ++SS DW++KLW +KN +PL FE
Sbjct 436 AGISEMFEGHHGPITGLDCH--TAAGPVDFSHLFVTSSFDWTVKLWGTKN-NKPLYSFED 492
Query 120 SDAYVYDVKW 129
+ YVYDV W
Sbjct 493 NSDYVYDVMW 502
> dre:798868 dynein cytoplasmic 1 intermediate chain 1-like; K10415
dynein intermediate chain, cytosolic
Length=591
Score = 114 bits (286), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 56/130 (43%), Positives = 86/130 (66%), Gaps = 4/130 (3%)
Query 1 RLCQWSLQMMGQPAETLDL-KRGSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNK 59
R+C WSL M+ QP E+++L S+ V V ++F +VN+ +GSE+G + TA+ HG+K
Sbjct 396 RMCSWSLDMLSQPQESMELVYNKSKPVAVTGMAFPTGDVNNYIVGSEEGTVYTASRHGSK 455
Query 60 AGVMDVYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEG 119
AG+ +++E H G +T + H + G D+SHL ++SS DW++KLWS+K +PL FE
Sbjct 456 AGICEMFEGHQGPVTGISCHSAV--GPVDFSHLFVTSSFDWTVKLWSTKQ-NKPLYSFED 512
Query 120 SDAYVYDVKW 129
+ YVYDV W
Sbjct 513 NADYVYDVMW 522
> hsa:1780 DYNC1I1, DNCI1, DNCIC1; dynein, cytoplasmic 1, intermediate
chain 1; K10415 dynein intermediate chain, cytosolic
Length=628
Score = 113 bits (282), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 55/130 (42%), Positives = 86/130 (66%), Gaps = 4/130 (3%)
Query 1 RLCQWSLQMMGQPAETLDL-KRGSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNK 59
++C WSL M+ P E+++L S+ V V ++F +VN+ +GSE+G + TA HG+K
Sbjct 397 KMCSWSLDMLSTPQESMELVYNKSKPVAVTGMAFPTGDVNNFVVGSEEGTVYTACRHGSK 456
Query 60 AGVMDVYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEG 119
AG+ +V+E H G +T ++ H M G D+SHL ++SS DW++KLW++K+ +PL FE
Sbjct 457 AGIGEVFEGHQGPVTGINCH--MAVGPIDFSHLFVTSSFDWTVKLWTTKH-NKPLYSFED 513
Query 120 SDAYVYDVKW 129
+ YVYDV W
Sbjct 514 NADYVYDVMW 523
> mmu:13426 Dync1i1, DH_IC-1, DHIC-1, DIC, Dncic1, IC74; dynein
cytoplasmic 1 intermediate chain 1; K10415 dynein intermediate
chain, cytosolic
Length=625
Score = 113 bits (282), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 55/130 (42%), Positives = 86/130 (66%), Gaps = 4/130 (3%)
Query 1 RLCQWSLQMMGQPAETLDL-KRGSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNK 59
++C WSL M+ P E+++L S+ V V ++F +VN+ +GSE+G + TA HG+K
Sbjct 394 KMCSWSLDMLSTPQESMELVYNKSKPVAVTGMAFPTGDVNNFVVGSEEGTVYTACRHGSK 453
Query 60 AGVMDVYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEG 119
AG+ +V+E H G +T ++ H M G D+SHL ++SS DW++KLW++K+ +PL FE
Sbjct 454 AGIGEVFEGHQGPVTGINCH--MAVGPIDFSHLFVTSSFDWTVKLWTTKH-NKPLYSFED 510
Query 120 SDAYVYDVKW 129
+ YVYDV W
Sbjct 511 NADYVYDVMW 520
> cpv:cgd1_1540 dynein intermediate chain
Length=677
Score = 86.7 bits (213), Expect = 2e-17, Method: Composition-based stats.
Identities = 51/136 (37%), Positives = 72/136 (52%), Gaps = 15/136 (11%)
Query 1 RLCQWSLQMMGQPAETLDLKR-GSRDVCVECVSFAE-AEVNDLCLGSEDGCLMTANIHGN 58
+LC W+L + +P E+ L++ S+DV ++C++ ++ N + GSEDG L I N
Sbjct 428 KLCNWNLTNLSEPVESFQLRKTNSKDVSIQCMTLSKLINPNAIICGSEDGSLYQTMIKTN 487
Query 59 KAGVMD--VYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEP--- 113
K GV+ AH G +TSL +HP + LSS DW+IKLW LA
Sbjct 488 KPGVISSTFQNAHNGYITSLDYHP--------INDCFLSSGADWTIKLWIPNPLASSFTL 539
Query 114 LGCFEGSDAYVYDVKW 129
L FE S+ YV DV W
Sbjct 540 LYTFESSENYVIDVAW 555
> cel:C17H12.1 dyci-1; DYnein Chain, light Intermediate family
member (dyci-1); K10415 dynein intermediate chain, cytosolic
Length=643
Score = 74.7 bits (182), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 43/133 (32%), Positives = 69/133 (51%), Gaps = 7/133 (5%)
Query 1 RLCQWSLQMMGQPAETLDL-KRGSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNK 59
R+C W++ + QP + +L + + +SF E+ +G++ G ++ A+ +G
Sbjct 410 RICSWNVDNLTQPVDGKELMTKEGKAASATRMSFPIGEMQTFVVGTDTGEILAASRNGAD 469
Query 60 AGVMDV---YEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGC 116
V+ Y+ H G LT + FH G D+SHL +SSS DW++KLWS+K+ A
Sbjct 470 PAVIKTDVCYKGHSGTLTGIEFH--RASGVVDFSHLFISSSHDWTVKLWSTKD-ANLKFS 526
Query 117 FEGSDAYVYDVKW 129
FE V DV W
Sbjct 527 FESHPDLVLDVAW 539
> dre:100330195 dynein, cytoplasmic 1, intermediate chain 2a-like
Length=168
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 31/65 (47%), Positives = 43/65 (66%), Gaps = 3/65 (4%)
Query 65 VYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEGSDAYV 124
++E H G +T +H H G D+SHL L++S DW++KLWS+KN +PL FE + YV
Sbjct 1 MFEGHHGPITGIHCH--TAAGPVDFSHLFLTASFDWTVKLWSNKN-NKPLYSFEDNSDYV 57
Query 125 YDVKW 129
YDV W
Sbjct 58 YDVMW 62
> tgo:TGME49_077520 hypothetical protein
Length=167
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 23/49 (46%), Positives = 35/49 (71%), Gaps = 1/49 (2%)
Query 82 MTEGSRDYSHLLLSSSVDWSIKLWSSK-NLAEPLGCFEGSDAYVYDVKW 129
+T+ R +LLLS+SVDW++KLWS+K + A+PL F+ + Y+YD W
Sbjct 5 VTKFRRPTINLLLSTSVDWTVKLWSAKRSFAQPLKTFDDFEVYIYDAAW 53
> hsa:79819 WDR78, FLJ23129; WD repeat domain 78
Length=848
Score = 45.1 bits (105), Expect = 5e-05, Method: Composition-based stats.
Identities = 29/108 (26%), Positives = 50/108 (46%), Gaps = 11/108 (10%)
Query 23 SRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNKAGVMDVYEAHGGALTSLHFHPGM 82
SR C +F + N G+E+G + + N+ +D Y H G + + ++P
Sbjct 658 SRQAPGMCFAFHPKDTNIYLAGTEEGHIHKCSCSYNEQ-YLDTYRGHKGPVYKVTWNP-- 714
Query 83 TEGSRDYSH-LLLSSSVDWSIKLWSSKNLAEPLGCFEGSDAYVYDVKW 129
+ H + LS S DW + +W +N+ +P F + + VYDV W
Sbjct 715 ------FCHDVFLSCSADWGVIIWQQENV-KPSLSFYPATSVVYDVAW 755
> ath:AT4G29730 NFC5; NFC5 (Nucleosome/chromatin assembly factor
group C 5); K10752 histone-binding protein RBBP4
Length=487
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 31/113 (27%), Positives = 54/113 (47%), Gaps = 13/113 (11%)
Query 22 GSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNKAGVMDVYEAHGGALTSLHFHPG 81
G +D VE V+F + + C +D CLM + + M V +AH L + ++P
Sbjct 270 GHKDT-VEDVAFCPSSAQEFCSVGDDSCLMLWDARTGTSPAMKVEKAHDADLHCVDWNP- 327
Query 82 MTEGSRDYSHLLLSSSVDWSIKLWSSKNL-----AEPLGCFEGSDAYVYDVKW 129
++L+L+ S D +++++ +NL P+ FEG A V V+W
Sbjct 328 ------HDNNLILTGSADNTVRVFDRRNLTSNGVGSPVYKFEGHRAAVLCVQW 374
> mmu:242584 Wdr78, BC028975, MGC31722, MGC38950; WD repeat domain
78
Length=807
Score = 41.6 bits (96), Expect = 7e-04, Method: Composition-based stats.
Identities = 26/107 (24%), Positives = 47/107 (43%), Gaps = 9/107 (8%)
Query 23 SRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNKAGVMDVYEAHGGALTSLHFHPGM 82
SR C +F + N G+E+G + + N+ ++ Y H G + + ++P
Sbjct 617 SRQAPGMCFAFHPKDTNIYLAGTEEGLIHKCSCSYNEQ-YLETYRGHKGPVYKVTWNPFC 675
Query 83 TEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEGSDAYVYDVKW 129
+ + LS S DW + +W ++ +P F + VYDV W
Sbjct 676 PD-------VFLSCSADWGVMIW-HQDTVKPFLSFYPTTYVVYDVSW 714
> xla:444126 wdr78, MGC80539; WD repeat domain 78
Length=742
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 28/107 (26%), Positives = 46/107 (42%), Gaps = 9/107 (8%)
Query 23 SRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNKAGVMDVYEAHGGALTSLHFHPGM 82
SR C F + N G+E+G + + N+ +D Y H G + + + P
Sbjct 587 SRQAPGMCFDFLPKDSNIYLAGTEEGHIHKCSCSYNEQ-FLDTYRGHKGPVYKIVWSPFC 645
Query 83 TEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEGSDAYVYDVKW 129
+ + LS S DW I LW +++ P+ F + VYD+ W
Sbjct 646 PD-------VFLSCSADWCIHLW-QQDVLLPILTFSNTTNAVYDIMW 684
> pfa:PFI1080w dynein intermediate chain 2, ciliary; K10409 dynein
intermediate chain 1, axonemal
Length=820
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 26/103 (25%), Positives = 56/103 (54%), Gaps = 12/103 (11%)
Query 28 VECVSFAEAEVNDLCLGSEDGCL-MTANIHGNKAGVMDVYEAHGGALTSLHFHPGMTEGS 86
+ C+ F ++N +G+ G + + ++ + + +++Y H ++ SL ++ +
Sbjct 627 ITCIDFNPFKINLFLIGTNKGKIYLYSSTYSDH--YLNIYNEHTMSINSLSYN----KFK 680
Query 87 RDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEGSDAYVYDVKW 129
RD + +SSS DW+I++W +++ L F+ + VYDVKW
Sbjct 681 RD---IFISSSYDWTIRIW-TQSRKTSLIIFDIKEC-VYDVKW 718
> mmu:71820 Wdr34, 3200002I06Rik, MGC103259; WD repeat domain
34
Length=537
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 31/137 (22%), Positives = 59/137 (43%), Gaps = 21/137 (15%)
Query 6 SLQMMGQPAETLDLKRGSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGN------- 58
++Q + + + RG +V V V+F+ + + LG+E G + ++
Sbjct 316 AVQQLPRSTKLKKPPRGETEVGVTSVAFSSFDSSLFVLGTEGGFPLKCSLASEVAALTRM 375
Query 59 ------KAGVMDVYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAE 112
+A V + HGG + S+ P + +L LS+ D + L+S A+
Sbjct 376 PSSVPLRAPVQFTFSPHGGPVYSVSCSPF-------HRNLFLSAGTDGHVHLYSMLQ-AQ 427
Query 113 PLGCFEGSDAYVYDVKW 129
PL + S Y++ V+W
Sbjct 428 PLTSLQLSHKYLFAVRW 444
> sce:YAR003W SWD1, CPS50, FUN16, SAF49; Subunit of the COMPASS
(Set1C) complex, which methylates histone H3 on lysine 4 and
is required in transcriptional silencing near telomeres;
WD40 beta propeller superfamily member with similarity to mammalian
Rbbp7; K14961 COMPASS component SWD1
Length=426
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 40/130 (30%), Positives = 53/130 (40%), Gaps = 47/130 (36%)
Query 29 ECVSFAEAEVNDLCLGSEDGCLMTAN--------IHGNKAGVMDVYEAHGGALTSLHFHP 80
EC+ F+ + L LG +G L+ + + GN G AH +TS+ + P
Sbjct 30 ECLQFSPCG-DYLALGCANGALVIYDMDTFRPICVPGNMLG------AHVRPITSIAWSP 82
Query 81 GMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPL----------GC-------------- 116
LLL+SS DWSIKLW ++PL GC
Sbjct 83 D--------GRLLLTSSRDWSIKLWDLSKPSKPLKEIRFDSPIWGCQWLDAKRRLCVATI 134
Query 117 FEGSDAYVYD 126
FE SDAYV D
Sbjct 135 FEESDAYVID 144
> ath:AT2G19520 FVE; FVE; metal ion binding; K10752 histone-binding
protein RBBP4
Length=507
Score = 38.9 bits (89), Expect = 0.005, Method: Composition-based stats.
Identities = 28/107 (26%), Positives = 48/107 (44%), Gaps = 12/107 (11%)
Query 28 VECVSFAEAEVNDLCLGSEDGCLMTANIHGNKAGVMDVYEAHGGALTSLHFHPGMTEGSR 87
VE V+F+ + C +D CL+ + V V +AH L + ++P
Sbjct 295 VEDVAFSPTSAQEFCSVGDDSCLILWDARTGTNPVTKVEKAHDADLHCVDWNP------- 347
Query 88 DYSHLLLSSSVDWSIKLWSSKNL-----AEPLGCFEGSDAYVYDVKW 129
+L+L+ S D +++L+ + L P+ FEG A V V+W
Sbjct 348 HDDNLILTGSADNTVRLFDRRKLTANGVGSPIYKFEGHKAAVLCVQW 394
> tgo:TGME49_009300 dynein intermediate chain, putative ; K10409
dynein intermediate chain 1, axonemal
Length=619
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 25/102 (24%), Positives = 50/102 (49%), Gaps = 14/102 (13%)
Query 30 CVSFAEAEVNDLCLGSEDGCLM--TANIHGNKAGVMDVYEAHGGALTSLHFHPGMTEGSR 87
C F ++ + +G+ +G + + N G + Y H ++T + ++P
Sbjct 480 CFDFHPSQSHVFIVGTTEGVIFKCSKNYSGQ---YLQTYLGHDMSITGVEWNPF------ 530
Query 88 DYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEGSDAYVYDVKW 129
+ + +SSS DW++K+W ++L P+ F+ A V DV+W
Sbjct 531 -HPRVFISSSSDWTVKIW-EESLTSPVLTFDFEMA-VGDVRW 569
> dre:553797 wdr78, MGC111979, zgc:111979; WD repeat domain 78
Length=778
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 26/100 (26%), Positives = 46/100 (46%), Gaps = 9/100 (9%)
Query 30 CVSFAEAEVNDLCLGSEDGCLMTANIHGNKAGVMDVYEAHGGALTSLHFHPGMTEGSRDY 89
C F + +G+E+G + + N+ +D Y+AH + + + P
Sbjct 596 CFDFHPNDSKIYLVGTEEGHIHKCSSSYNEQ-YLDSYKAHKRPVYKVTWSPFC------- 647
Query 90 SHLLLSSSVDWSIKLWSSKNLAEPLGCFEGSDAYVYDVKW 129
S + LS S DW+I+LW ++L P+ F V+D+ W
Sbjct 648 SDVFLSCSSDWTIQLW-RQDLQIPVMGFTSGQRVVFDIMW 686
> xla:734519 poc1b, MGC114911, pix1, tuwd12, wdr51b; POC1 centriolar
protein homolog B (Chlamydomonas)
Length=468
Score = 37.0 bits (84), Expect = 0.016, Method: Composition-based stats.
Identities = 26/86 (30%), Positives = 38/86 (44%), Gaps = 12/86 (13%)
Query 21 RGSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNKAGVMDVYEAHGGALTSLHFHP 80
+G +D V CV F+ + L S D C+M N Y H A+T + F P
Sbjct 15 KGHKDA-VTCVDFS-PDSKQLASSSADACVMIWNFKPQSRAYK--YPGHKEAVTCVQFSP 70
Query 81 GMTEGSRDYSHLLLSSSVDWSIKLWS 106
HL+ SSS D +++LW+
Sbjct 71 S--------GHLVASSSKDRTVRLWA 88
> xla:444304 MGC80975 protein; K10409 dynein intermediate chain
1, axonemal
Length=702
Score = 36.6 bits (83), Expect = 0.018, Method: Composition-based stats.
Identities = 22/87 (25%), Positives = 46/87 (52%), Gaps = 10/87 (11%)
Query 43 LGSEDGCLMTANIHGNKAGVMDVYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSI 102
+G+E+G + + + +D Y+AH A+ ++ ++P + + +S S DW++
Sbjct 504 VGTEEGKIHKCS-KAYSSKFLDTYDAHNMAVDTVRWNPF-------HPKVFISCSSDWTV 555
Query 103 KLWSSKNLAEPLGCFEGSDAYVYDVKW 129
K+W ++ P+ F+ +A V DV W
Sbjct 556 KIW-QHDVKTPMFVFD-LNAPVADVAW 580
> hsa:25886 POC1A, DKFZp434C245, MGC131902, PIX2, WDR51A; POC1
centriolar protein homolog A (Chlamydomonas)
Length=359
Score = 36.2 bits (82), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 37/124 (29%), Positives = 51/124 (41%), Gaps = 17/124 (13%)
Query 10 MGQP-AETLDLKR---GSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNKAGVMDV 65
M P AE L+R G RD V CV F+ L GS D CLM ++
Sbjct 1 MAAPCAEDPSLERHFKGHRD-AVTCVDFS-INTKQLASGSMDSCLMVWHMKPQSRAYR-- 56
Query 66 YEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEGSDAYVY 125
+ H A+T ++F P HLL S S D ++++W N+ F A V
Sbjct 57 FTGHKDAVTCVNFSPS--------GHLLASGSRDKTVRIW-VPNVKGESTVFRAHTATVR 107
Query 126 DVKW 129
V +
Sbjct 108 SVHF 111
> cel:T05H4.14 gad-1; GAstrulation Defective family member (gad-1)
Length=620
Score = 36.2 bits (82), Expect = 0.026, Method: Composition-based stats.
Identities = 20/50 (40%), Positives = 27/50 (54%), Gaps = 8/50 (16%)
Query 65 VYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPL 114
V +AH G++TS+ F P S LLS D ++K+WS N EPL
Sbjct 320 VRKAHNGSITSIAFSPD--------SKRLLSRGFDDTLKMWSLDNPKEPL 361
> hsa:89891 WDR34, MGC20486, bA216B9.3; WD repeat domain 34
Length=536
Score = 36.2 bits (82), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 32/137 (23%), Positives = 57/137 (41%), Gaps = 24/137 (17%)
Query 9 MMGQPAETLDLK---RGSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNKAGVMDV 65
+M Q + LK RG +V V+F+ + LG+E G + ++ +A + +
Sbjct 315 VMQQLPRSTKLKKHPRGETEVGATAVAFSSFDPRLFILGTEGGFPLKCSLAAGEAALTRM 374
Query 66 -------------YEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAE 112
+ HGG + S+ P + +L LS+ D + L+S A
Sbjct 375 PSSVPLRAPAQFTFSPHGGPIYSVSCSPF-------HRNLFLSAGTDGHVHLYSMLQ-AP 426
Query 113 PLGCFEGSDAYVYDVKW 129
PL + S Y++ V+W
Sbjct 427 PLTSLQLSLKYLFAVRW 443
> xla:734763 wdr34, MGC130908; WD repeat domain 34
Length=503
Score = 35.8 bits (81), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 30/122 (24%), Positives = 51/122 (41%), Gaps = 21/122 (17%)
Query 21 RGSRDVCVECVSFAEAEVNDLCLGSEDGCLM-------------TANIHGNKAGVMDVYE 67
RG V V C+S++ + N +G E G L+ T + +A +
Sbjct 297 RGDTAVGVTCLSYSHFDPNLFIVGVEGGYLLKCSSSVQTSAAIGTGSSIPLRAPAQFTFS 356
Query 68 AHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEGSDAYVYDV 127
HGG + S+ P + +L LS+ D L+S A+PL + S Y++ +
Sbjct 357 PHGGPVYSVSCSPF-------HRNLFLSAGTDGHAHLYSMLQ-AKPLVSLQLSQQYLFSI 408
Query 128 KW 129
+W
Sbjct 409 RW 410
> dre:321726 rbb4, RBBP4, rbb4-2, wu:fb33a09, wu:fb40e10, zgc:55349,
zgc:77854; retinoblastoma binding protein 4; K10752 histone-binding
protein RBBP4
Length=424
Score = 35.4 bits (80), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 31/132 (23%), Positives = 54/132 (40%), Gaps = 12/132 (9%)
Query 2 LCQWSLQMMGQPAETLDLKR--GSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNK 59
+C W + + + + +D K VE VS+ + ++D LM + N
Sbjct 202 ICLWDISTVPKEGKIVDAKTIFTGHTAVVEDVSWHLLHESLFGSVADDQKLMIWDTRSNN 261
Query 60 AGVMD-VYEAHGGALTSLHFHPGMTEGSRDYSHLLLSS-SVDWSIKLWSSKNLAEPLGCF 117
+AH + L F+P YS +L++ S D ++ LW +NL L F
Sbjct 262 TSKPSHAVDAHTAEVNCLSFNP--------YSEFILATGSADKTVALWDLRNLKLKLHSF 313
Query 118 EGSDAYVYDVKW 129
E ++ V+W
Sbjct 314 ESHKDEIFQVQW 325
> mmu:68922 Dnaic1, 1110066F04Rik, BB124644, Dnai1; dynein, axonemal,
intermediate chain 1; K10409 dynein intermediate chain
1, axonemal
Length=701
Score = 35.4 bits (80), Expect = 0.045, Method: Composition-based stats.
Identities = 22/87 (25%), Positives = 46/87 (52%), Gaps = 10/87 (11%)
Query 43 LGSEDGCLMTANIHGNKAGVMDVYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSI 102
+G+E+G + + + +D Y+AH A+ ++ ++P T+ + +S S DW++
Sbjct 515 VGTEEGKIYKCS-KSYSSQFLDTYDAHNMAVDAVLWNPYHTK-------VFMSCSSDWTV 566
Query 103 KLWSSKNLAEPLGCFEGSDAYVYDVKW 129
K+W + P+ ++ + A V DV W
Sbjct 567 KIW-DHTIKTPMFIYDLNSA-VGDVAW 591
> tgo:TGME49_035020 coatomer protein complex subunit beta, putative
Length=1256
Score = 35.4 bits (80), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 24/86 (27%), Positives = 37/86 (43%), Gaps = 9/86 (10%)
Query 44 GSEDGCLMTANIHGNKAGVMDVYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIK 103
G +D L NIH V ++ AHG + + H L+LSSS D ++K
Sbjct 75 GGDDCALRVFNIH-TLEKVKEIPSAHGDYIRHISVHAAKP--------LVLSSSDDMTVK 125
Query 104 LWSSKNLAEPLGCFEGSDAYVYDVKW 129
LW + + + +E YV +W
Sbjct 126 LWHYEKNWQKVASYEQHTHYVMQTQW 151
> hsa:27019 DNAI1, CILD1, ICS, ICS1, MGC26204, PCD; dynein, axonemal,
intermediate chain 1; K10409 dynein intermediate chain
1, axonemal
Length=699
Score = 35.0 bits (79), Expect = 0.054, Method: Composition-based stats.
Identities = 21/87 (24%), Positives = 46/87 (52%), Gaps = 10/87 (11%)
Query 43 LGSEDGCLMTANIHGNKAGVMDVYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSI 102
+G+E+G + + + +D Y+AH ++ ++ ++P T+ + +S S DW++
Sbjct 514 VGTEEGKIYKCS-KSYSSQFLDTYDAHNMSVDTVSWNPYHTK-------VFMSCSSDWTV 565
Query 103 KLWSSKNLAEPLGCFEGSDAYVYDVKW 129
K+W + P+ ++ + A V DV W
Sbjct 566 KIW-DHTIKTPMFIYDLNSA-VGDVAW 590
> tgo:TGME49_042610 WD domain, G-beta repeat-containing protein
Length=554
Score = 35.0 bits (79), Expect = 0.056, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 37/68 (54%), Gaps = 8/68 (11%)
Query 62 VMDVYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEGSD 121
VM+ +H +TS+ F P D +LLL+ S D +++++ ++L PL ++ S
Sbjct 390 VMENKRSHSMGVTSITFFP-------DNPNLLLTGSYDEAVRIFDRRHLQSPLTSYKVSG 442
Query 122 AYVYDVKW 129
V+ +KW
Sbjct 443 G-VWRLKW 449
> xla:432269 rbbp4-b, MGC82618, nurf55, rbap48, xrbbp4; retinoblastoma
binding protein 4; K10752 histone-binding protein RBBP4
Length=425
Score = 34.7 bits (78), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 31/132 (23%), Positives = 54/132 (40%), Gaps = 12/132 (9%)
Query 2 LCQWSLQMMGQPAETLDLKR--GSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNK 59
+C W + + + + +D K VE VS+ + ++D LM + N
Sbjct 202 ICLWDISAVPKEGKVVDAKTIFTGHTAVVEDVSWHLLHESLFGSVADDQKLMIWDTRSNN 261
Query 60 AGVMD-VYEAHGGALTSLHFHPGMTEGSRDYSHLLLSS-SVDWSIKLWSSKNLAEPLGCF 117
+AH + L F+P YS +L++ S D ++ LW +NL L F
Sbjct 262 TSKPSHSVDAHTAEVNCLSFNP--------YSEFILATGSADKTVALWDLRNLKLKLHSF 313
Query 118 EGSDAYVYDVKW 129
E ++ V+W
Sbjct 314 ESHKDEIFQVQW 325
> mmu:26374 Rfwd2, AI316802, C80879, Cop1; ring finger and WD
repeat domain 2; K10143 E3 ubiquitin-protein ligase RFWD2 [EC:6.3.2.19]
Length=733
Score = 34.7 bits (78), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 45/97 (46%), Gaps = 17/97 (17%)
Query 25 DVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNKAGVMDVYEAHGGALTSLHFHPGMTE 84
+VC CV F+ + L G D C+ ++ K +M V++ H A++ F G
Sbjct 559 NVC--CVKFSPSSRYHLAFGCADHCVHYYDLRNTKQPIM-VFKGHRKAVSYAKFVSGEE- 614
Query 85 GSRDYSHLLLSSSVDWSIKLWSSKNLAEP--LGCFEG 119
++S+S D +KLW N+ +P L F+G
Sbjct 615 --------IVSASTDSQLKLW---NVGKPYCLRSFKG 640
> xla:399131 rbbp4-a, MGC79922, nurf55, rbap48, rbbp4, xrbbp4;
retinoblastoma binding protein 4; K10752 histone-binding protein
RBBP4
Length=425
Score = 34.7 bits (78), Expect = 0.076, Method: Compositional matrix adjust.
Identities = 31/132 (23%), Positives = 54/132 (40%), Gaps = 12/132 (9%)
Query 2 LCQWSLQMMGQPAETLDLKR--GSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNK 59
+C W + + + + +D K VE VS+ + ++D LM + N
Sbjct 202 ICLWDISAVPKEGKVVDAKTIFTGHTAVVEDVSWHLLHESLFGSVADDQKLMIWDTRSNN 261
Query 60 AGVMD-VYEAHGGALTSLHFHPGMTEGSRDYSHLLLSS-SVDWSIKLWSSKNLAEPLGCF 117
+AH + L F+P YS +L++ S D ++ LW +NL L F
Sbjct 262 TSKPSHSVDAHTAEVNCLSFNP--------YSEFILATGSADKTVALWDLRNLKLKLHSF 313
Query 118 EGSDAYVYDVKW 129
E ++ V+W
Sbjct 314 ESHKDEIFQVQW 325
> mmu:19646 Rbbp4, mRbAp48; retinoblastoma binding protein 4;
K10752 histone-binding protein RBBP4
Length=425
Score = 34.7 bits (78), Expect = 0.078, Method: Compositional matrix adjust.
Identities = 31/132 (23%), Positives = 54/132 (40%), Gaps = 12/132 (9%)
Query 2 LCQWSLQMMGQPAETLDLKR--GSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNK 59
+C W + + + + +D K VE VS+ + ++D LM + N
Sbjct 202 ICLWDISAVPKEGKVVDAKTIFTGHTAVVEDVSWHLLHESLFGSVADDQKLMIWDTRSNN 261
Query 60 AGVMD-VYEAHGGALTSLHFHPGMTEGSRDYSHLLLSS-SVDWSIKLWSSKNLAEPLGCF 117
+AH + L F+P YS +L++ S D ++ LW +NL L F
Sbjct 262 TSKPSHSVDAHTAEVNCLSFNP--------YSEFILATGSADKTVALWDLRNLKLKLHSF 313
Query 118 EGSDAYVYDVKW 129
E ++ V+W
Sbjct 314 ESHKDEIFQVQW 325
> hsa:5928 RBBP4, NURF55, RBAP48; retinoblastoma binding protein
4; K10752 histone-binding protein RBBP4
Length=424
Score = 34.7 bits (78), Expect = 0.078, Method: Compositional matrix adjust.
Identities = 31/132 (23%), Positives = 54/132 (40%), Gaps = 12/132 (9%)
Query 2 LCQWSLQMMGQPAETLDLKR--GSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNK 59
+C W + + + + +D K VE VS+ + ++D LM + N
Sbjct 201 ICLWDISAVPKEGKVVDAKTIFTGHTAVVEDVSWHLLHESLFGSVADDQKLMIWDTRSNN 260
Query 60 AGVMD-VYEAHGGALTSLHFHPGMTEGSRDYSHLLLSS-SVDWSIKLWSSKNLAEPLGCF 117
+AH + L F+P YS +L++ S D ++ LW +NL L F
Sbjct 261 TSKPSHSVDAHTAEVNCLSFNP--------YSEFILATGSADKTVALWDLRNLKLKLHSF 312
Query 118 EGSDAYVYDVKW 129
E ++ V+W
Sbjct 313 ESHKDEIFQVQW 324
> mmu:100504800 e3 ubiquitin-protein ligase RFWD2-like
Length=677
Score = 34.7 bits (78), Expect = 0.079, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 45/97 (46%), Gaps = 17/97 (17%)
Query 25 DVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNKAGVMDVYEAHGGALTSLHFHPGMTE 84
+VC CV F+ + L G D C+ ++ K +M V++ H A++ F G
Sbjct 503 NVC--CVKFSPSSRYHLAFGCADHCVHYYDLRNTKQPIM-VFKGHRKAVSYAKFVSGEE- 558
Query 85 GSRDYSHLLLSSSVDWSIKLWSSKNLAEP--LGCFEG 119
++S+S D +KLW N+ +P L F+G
Sbjct 559 --------IVSASTDSQLKLW---NVGKPYCLRSFKG 584
> dre:794361 dync2i1, MGC171284, wdr34, zgc:171284; dynein, cytoplasmic
2, intermediate chain 1
Length=501
Score = 34.7 bits (78), Expect = 0.083, Method: Compositional matrix adjust.
Identities = 29/123 (23%), Positives = 49/123 (39%), Gaps = 22/123 (17%)
Query 21 RGSRDVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNKAGVMD--------------VY 66
RGS + V V+ + +++ +GSE G ++ + A + +
Sbjct 294 RGSLTIGVTAVALSPWDLDTFLVGSEGGLVLKCSFSSETAAAVPSDGESVILRAPMQFSF 353
Query 67 EAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEGSDAYVYD 126
GG + S+HF P + +L +S D L S PL SD+YV+
Sbjct 354 SPRGGPIHSVHFSPF-------HRNLFVSVGTDGLAHLHSVLQPC-PLLALRVSDSYVFG 405
Query 127 VKW 129
V+W
Sbjct 406 VRW 408
> xla:398032 katnb1; katanin p80 (WD repeat containing) subunit
B 1
Length=655
Score = 34.7 bits (78), Expect = 0.084, Method: Composition-based stats.
Identities = 24/81 (29%), Positives = 37/81 (45%), Gaps = 11/81 (13%)
Query 28 VECVSFAEAEVNDLCLGSEDGCLMTANIHGNKAGVMDVYEAHGGALTSLHFHPGMTEGSR 87
VE V F +E + GS+ G L ++ K ++ H ++SL FHP
Sbjct 66 VESVRFNNSE-ELIVAGSQSGSLRIWDLEAAK--ILRTLMGHKANVSSLDFHP------- 115
Query 88 DYSHLLLSSSVDWSIKLWSSK 108
Y + S S+D +IKLW +
Sbjct 116 -YGEFVASGSLDTNIKLWDVR 135
> tgo:TGME49_065410 pre-mRNA splicing factor, putative ; K12816
pre-mRNA-processing factor 17
Length=607
Score = 34.3 bits (77), Expect = 0.089, Method: Composition-based stats.
Identities = 19/64 (29%), Positives = 30/64 (46%), Gaps = 7/64 (10%)
Query 66 YEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEGSDAYVY 125
Y H G + ++ F P HLLLS+S+D ++K+W N + + V
Sbjct 314 YVGHTGGVQAIRFFP-------RSGHLLLSASMDSTLKIWDVLNQRKLQCTYTAHKQAVR 366
Query 126 DVKW 129
DV+W
Sbjct 367 DVQW 370
> ath:AT2G20330 transducin family protein / WD-40 repeat family
protein
Length=648
Score = 34.3 bits (77), Expect = 0.094, Method: Composition-based stats.
Identities = 26/82 (31%), Positives = 38/82 (46%), Gaps = 10/82 (12%)
Query 44 GSEDGCLMTANIHGNKAGVMDVY--EAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWS 101
G DG + ++ D+Y +AH +TS+ F S D +LLS S D S
Sbjct 345 GVGDGSIQIWSLKPGWGSRPDIYVGKAHTDDITSVKF-------SSD-GRILLSRSFDGS 396
Query 102 IKLWSSKNLAEPLGCFEGSDAY 123
+K+W + + E L FEG Y
Sbjct 397 LKVWDLRQMKEALKVFEGLPNY 418
> cpv:cgd7_5410 WD40 protein
Length=890
Score = 34.3 bits (77), Expect = 0.094, Method: Composition-based stats.
Identities = 28/100 (28%), Positives = 43/100 (43%), Gaps = 26/100 (26%)
Query 41 LCLGSEDGCLMTANIHGNKAGVMDVY-------------------EAHGGALTSLHFHPG 81
L LGS+DG + G++ G + +AH A+ SLH+
Sbjct 505 LILGSQDGSIRQWKFKGDEFGDSGMISSSGHQIEPFFNEKEDFNIQAHSNAIISLHWE-- 562
Query 82 MTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEGSD 121
+ + SH LSSS+D ++KLW + + EP SD
Sbjct 563 ----NDEGSHRFLSSSMDRTVKLWEAGS-TEPNAVINCSD 597
> hsa:64326 RFWD2, COP1, FLJ10416, RNF200; ring finger and WD
repeat domain 2; K10143 E3 ubiquitin-protein ligase RFWD2 [EC:6.3.2.19]
Length=707
Score = 34.3 bits (77), Expect = 0.100, Method: Compositional matrix adjust.
Identities = 27/97 (27%), Positives = 45/97 (46%), Gaps = 17/97 (17%)
Query 25 DVCVECVSFAEAEVNDLCLGSEDGCLMTANIHGNKAGVMDVYEAHGGALTSLHFHPGMTE 84
+VC CV F+ + L G D C+ ++ K +M V++ H A++ F G
Sbjct 533 NVC--CVKFSPSSRYHLAFGCADHCVHYYDLRNTKQPIM-VFKGHRKAVSYAKFVSGEE- 588
Query 85 GSRDYSHLLLSSSVDWSIKLWSSKNLAEP--LGCFEG 119
++S+S D +KLW N+ +P L F+G
Sbjct 589 --------IVSASTDSQLKLW---NVGKPYCLRSFKG 614
> hsa:7260 TSSC1; tumor suppressing subtransferable candidate
1
Length=387
Score = 34.3 bits (77), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 17/65 (26%), Positives = 31/65 (47%), Gaps = 7/65 (10%)
Query 65 VYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEGSDAYV 124
+ AHG + L F+P + L S D +K W ++N+ EP+ E +V
Sbjct 223 IENAHGQLVRDLDFNPNK-------QYYLASCGDDCKVKFWDTRNVTEPVKTLEEHSHWV 275
Query 125 YDVKW 129
++V++
Sbjct 276 WNVRY 280
> dre:450045 tssc1, zgc:101720; tumor suppressing subtransferable
candidate 1
Length=387
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 17/65 (26%), Positives = 31/65 (47%), Gaps = 7/65 (10%)
Query 65 VYEAHGGALTSLHFHPGMTEGSRDYSHLLLSSSVDWSIKLWSSKNLAEPLGCFEGSDAYV 124
+ AHG + L F+P + L S D +K W ++++EP+ C E +V
Sbjct 223 IENAHGQLVRDLDFNPNK-------QYYLASCGDDCKVKFWDVRHISEPVKCLEEHSHWV 275
Query 125 YDVKW 129
+ V++
Sbjct 276 WSVRY 280
Lambda K H
0.318 0.133 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2044474180
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40