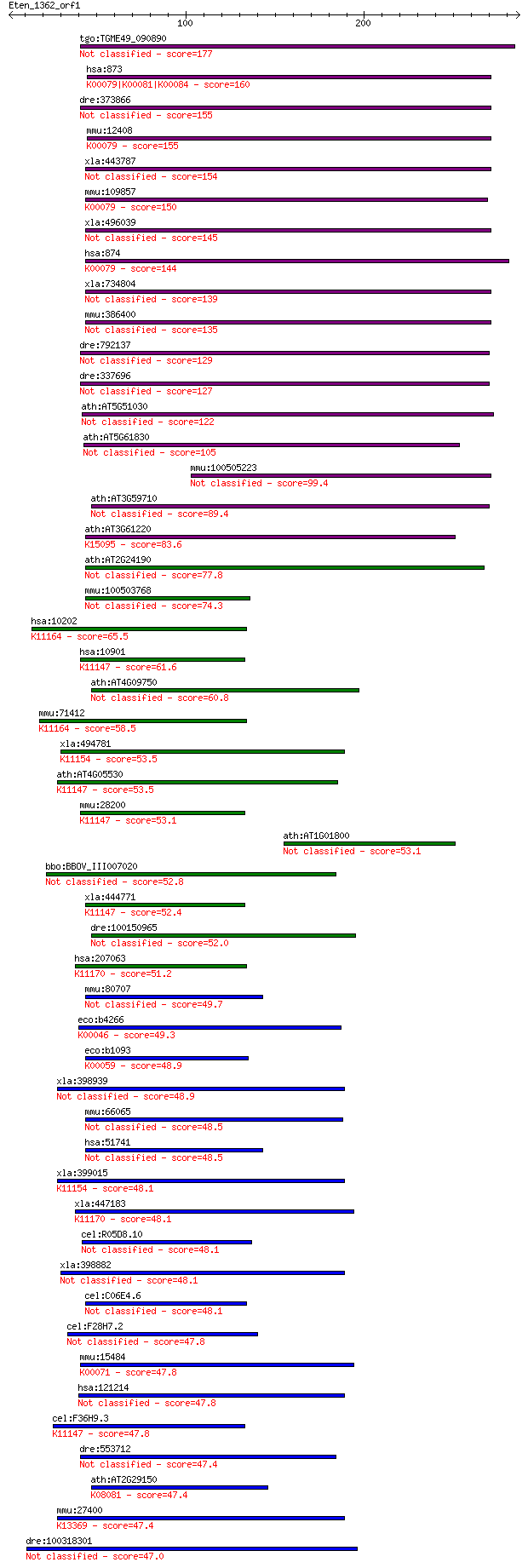
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1362_orf1
Length=286
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_090890 carbonyl reductase, putative (EC:1.1.1.189) 177 4e-44
hsa:873 CBR1, CBR, SDR21C1, hCBR1; carbonyl reductase 1 (EC:1.... 160 7e-39
dre:373866 cbr1; carbonyl reductase 1 155 1e-37
mmu:12408 Cbr1, AW261796, CR, Cbr; carbonyl reductase 1 (EC:1.... 155 2e-37
xla:443787 cbr3, MGC81473; carbonyl reductase 3 (EC:1.1.1.184) 154 2e-37
mmu:109857 Cbr3, 1110001J05Rik, C81353, MGC99956; carbonyl red... 150 6e-36
xla:496039 hypothetical LOC496039 145 2e-34
hsa:874 CBR3, SDR21C2, hCBR3; carbonyl reductase 3 (EC:1.1.1.1... 144 3e-34
xla:734804 cbr1, MGC131152; carbonyl reductase 1 139 2e-32
mmu:386400 carbonyl reductase [NADPH] 1-like 135 2e-31
dre:792137 hypothetical protein LOC792137 129 1e-29
dre:337696 cbr1l, fp17a06, wu:fa92b03, wu:fp17a06; carbonyl re... 127 4e-29
ath:AT5G51030 short-chain dehydrogenase/reductase (SDR) family... 122 2e-27
ath:AT5G61830 short-chain dehydrogenase/reductase (SDR) family... 105 2e-22
mmu:100505223 carbonyl reductase [NADPH] 1-like 99.4 1e-20
ath:AT3G59710 short-chain dehydrogenase/reductase (SDR) family... 89.4 1e-17
ath:AT3G61220 short-chain dehydrogenase/reductase (SDR) family... 83.6 8e-16
ath:AT2G24190 short-chain dehydrogenase/reductase (SDR) family... 77.8 4e-14
mmu:100503768 carbonyl reductase [NADPH] 1-like 74.3 5e-13
hsa:10202 DHRS2, HEP27, SDR25C1; dehydrogenase/reductase (SDR ... 65.5 2e-10
hsa:10901 DHRS4, CR, FLJ11008, NRDR, PHCR, PSCD, SCAD-SRL, SDR... 61.6 3e-09
ath:AT4G09750 short-chain dehydrogenase/reductase (SDR) family... 60.8 6e-09
mmu:71412 Dhrs2, 5430405K24Rik, Hep27, MGC143467; dehydrogenas... 58.5 2e-08
xla:494781 rdh7.2; retinol dehydrogenase 7, gene 2; K11154 ret... 53.5 8e-07
ath:AT4G05530 IBR1; IBR1 (INDOLE-3-BUTYRIC ACID RESPONSE 1); b... 53.5 9e-07
mmu:28200 Dhrs4, AI043103, AI790593, CR, D14Ucla2, NDRD, PHCR,... 53.1 1e-06
ath:AT1G01800 short-chain dehydrogenase/reductase (SDR) family... 53.1 1e-06
bbo:BBOV_III007020 17.m07620; hypothetical protein 52.8 1e-06
xla:444771 dhrs4, MGC81922; dehydrogenase/reductase (SDR famil... 52.4 2e-06
dre:100150965 retinol dehydrogenase 12-like 52.0 3e-06
hsa:207063 DHRSX, CXorf11, DHRS5X, DHRS5Y, DHRSXY, DHRSY, SDR4... 51.2 4e-06
mmu:80707 Wwox, 5330426P09Rik, 9030416C10Rik, WOX1; WW domain-... 49.7 1e-05
eco:b4266 idnO, ECK4259, JW4223, yjgU; 5-keto-D-gluconate-5-re... 49.3 2e-05
eco:b1093 fabG, ECK1079, JW1079; 3-oxoacyl-[acyl-carrier-prote... 48.9 2e-05
xla:398939 hsd17b6, MGC68605; hydroxysteroid (17-beta) dehydro... 48.9 2e-05
mmu:66065 Hsd17b14, 0610039E24Rik, Dhrs10, retSDR3; hydroxyste... 48.5 3e-05
hsa:51741 WWOX, D16S432E, FOR, FRA16D, HHCMA56, PRO0128, SDR41... 48.5 3e-05
xla:399015 hypothetical protein MGC68519; K11154 retinol dehyd... 48.1 3e-05
xla:447183 dhrsx, MGC85576; dehydrogenase/reductase (SDR famil... 48.1 4e-05
cel:R05D8.10 dhs-15; DeHydrogenases, Short chain family member... 48.1 4e-05
xla:398882 rdh16, MGC68820; retinol dehydrogenase 16 (all-trans) 48.1 4e-05
cel:C06E4.6 hypothetical protein 48.1 4e-05
cel:F28H7.2 hypothetical protein 47.8 4e-05
mmu:15484 Hsd11b2, 11HSD2; hydroxysteroid 11-beta dehydrogenas... 47.8 4e-05
hsa:121214 SDR9C7, FLJ16333, MGC126600, MGC126602, RDHS, SDR-O... 47.8 4e-05
cel:F36H9.3 dhs-13; DeHydrogenases, Short chain family member ... 47.8 5e-05
dre:553712 MGC112332; zgc:112332 47.4 6e-05
ath:AT2G29150 tropinone reductase, putative / tropine dehydrog... 47.4 6e-05
mmu:27400 Hsd17b6, 17betaHSD9, Hsd17b9, Rdh8; hydroxysteroid (... 47.4 7e-05
dre:100318301 dhrsx; dehydrogenase/reductase (SDR family) X-li... 47.0 8e-05
> tgo:TGME49_090890 carbonyl reductase, putative (EC:1.1.1.189)
Length=305
Score = 177 bits (449), Expect = 4e-44, Method: Compositional matrix adjust.
Identities = 104/250 (41%), Positives = 148/250 (59%), Gaps = 8/250 (3%)
Query 41 MAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLK--A 98
MA++V +VTGGNKGIGF +KLC L ++ VV++ +RD NG +AL L GL
Sbjct 1 MAKKVALVTGGNKGIGFGVTRKLCERLPKDSWVVLLGTRDVSNGERALTNLKECGLPMLP 60
Query 99 EMELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYA 158
+ LDIT S + I KYG +D LVNN+GFAFK+ ATE QAK T G+NY+
Sbjct 61 VVHQLDITDSTSCKQMKDFIHQKYGGLDLLVNNSGFAFKRNATESKYEQAKHTIGVNYFG 120
Query 159 TRDITLDMMGLFKPGSRIVNVASAAGEMALQEMSAELRHRLMSKSARQEDIDKVVDDFIV 218
T+ IT ++ + + G+RI++VAS G+M L+ MS E R ++S E +D ++ +I
Sbjct 121 TKQITETVLPIMRDGARIISVASMCGKMGLEHMSEEHRRAILSPDLSFEKLDDMMKQYIE 180
Query 219 ACEKGQ--QEGWPSSTYGLSKAAVIALTAAWARKAD-HCPSMEACRDMVITCCC--CCCD 273
A + + + GWP STY +SKA VIA T WA+ AD + + + + M + CCC C
Sbjct 181 AAKTDELTKLGWPESTYEMSKAGVIAATELWAQAADKNALTPQGTKGMFVACCCPGWCRT 240
Query 274 DGEGKDAHKP 283
D G + H P
Sbjct 241 DMAGYE-HPP 249
> hsa:873 CBR1, CBR, SDR21C1, hCBR1; carbonyl reductase 1 (EC:1.1.1.189
1.1.1.197 1.1.1.184); K00079 carbonyl reductase (NADPH)
[EC:1.1.1.184]; K00081 prostaglandin-E2 9-reductase [EC:1.1.1.189];
K00084 15-hydroxyprostaglandin dehydrogenase (NADP)
[EC:1.1.1.197]
Length=277
Score = 160 bits (404), Expect = 7e-39, Method: Compositional matrix adjust.
Identities = 89/228 (39%), Positives = 132/228 (57%), Gaps = 9/228 (3%)
Query 45 VFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELLD 104
V +VTGGNKGIG + LCR G+ VV+T+RD G A+ +L AEGL LD
Sbjct 7 VALVTGGNKGIGLAIVRDLCRLFSGD---VVLTARDVTRGQAAVQQLQAEGLSPRFHQLD 63
Query 105 ITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDITL 164
I +S + ++ +YG +D LVNNAG AFK A P +QA+VT N++ TRD+
Sbjct 64 IDDLQSIRALRDFLRKEYGGLDVLVNNAGIAFKVADPTPFHIQAEVTMKTNFFGTRDVCT 123
Query 165 DMMGLFKPGSRIVNVASAAGEMALQEMSAELRHRLMSKSARQEDIDKVVDDFIVACEKG- 223
+++ L KP R+VNV+S AL+ S EL+ + S++ +E++ +++ F+ +KG
Sbjct 124 ELLPLIKPQGRVVNVSSIMSVRALKSCSPELQQKFRSETITEEELVGLMNKFVEDTKKGV 183
Query 224 -QQEGWPSSTYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCCCC 270
Q+EGWPSS YG++K V L+ ARK S + D ++ CC
Sbjct 184 HQKEGWPSSAYGVTKIGVTVLSRIHARKL----SEQRKGDKILLNACC 227
> dre:373866 cbr1; carbonyl reductase 1
Length=276
Score = 155 bits (393), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 89/233 (38%), Positives = 128/233 (54%), Gaps = 10/233 (4%)
Query 41 MAQ-RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAE 99
M+Q +V +VTG NKGIGF + LC++ G+ V ++SRD G A+ L EGL
Sbjct 1 MSQCKVALVTGANKGIGFAIVRALCKEYTGD---VYLSSRDVGRGTAAVDSLKKEGLHPL 57
Query 100 MELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYAT 159
LDI S + + KYG +D L+NNAG AFK A T P QA VT N++AT
Sbjct 58 FHQLDINDPNSVRTARDFFQEKYGGLDVLINNAGIAFKMADTTPFGTQADVTLKTNFFAT 117
Query 160 RDITLDMMGLFKPGSRIVNVASAAGEMALQEMSAELRHRLMSKSARQEDIDKVVDDFIVA 219
RD+ + + KPG R+VNV+S G MAL S EL+ R S +E+++ +++ F+
Sbjct 118 RDMCNVFLPIIKPGGRLVNVSSGMGSMALGRCSPELQARFRSDDITEEELNGLMERFVRE 177
Query 220 CEKG--QQEGWPSSTYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCCCC 270
++G + GWPS+ YG+SK + LT AR + E D ++ CC
Sbjct 178 AQEGVHSERGWPSTAYGISKTGLTTLTRIQARNL----TKERPGDGILCNACC 226
> mmu:12408 Cbr1, AW261796, CR, Cbr; carbonyl reductase 1 (EC:1.1.1.189
1.1.1.197 1.1.1.184); K00079 carbonyl reductase (NADPH)
[EC:1.1.1.184]
Length=277
Score = 155 bits (391), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 85/228 (37%), Positives = 129/228 (56%), Gaps = 9/228 (3%)
Query 45 VFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELLD 104
V +VTG NKGIGF + LCR G+ VV+ +RD+E G A+ KL AEGL LD
Sbjct 7 VALVTGANKGIGFAITRDLCRKFSGD---VVLAARDEERGQTAVQKLQAEGLSPRFHQLD 63
Query 105 ITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDITL 164
I +S + + +YG +D LVNNAG AFK P +QA+VT N++ TRD+
Sbjct 64 IDNPQSIRALRDFLLKEYGGLDVLVNNAGIAFKVNDDTPFHIQAEVTMKTNFFGTRDVCK 123
Query 165 DMMGLFKPGSRIVNVASAAGEMALQEMSAELRHRLMSKSARQEDIDKVVDDFIVACEKG- 223
+++ L KP R+VNV+S AL+ EL+ + S++ +E++ +++ F+ +KG
Sbjct 124 ELLPLIKPQGRVVNVSSMVSLRALKNCRLELQQKFRSETITEEELVGLMNKFVEDTKKGV 183
Query 224 -QQEGWPSSTYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCCCC 270
+EGWP+S YG++K V L+ ARK + + D ++ CC
Sbjct 184 HAEEGWPNSAYGVTKIGVTVLSRILARKLNE----QRRGDKILLNACC 227
> xla:443787 cbr3, MGC81473; carbonyl reductase 3 (EC:1.1.1.184)
Length=277
Score = 154 bits (390), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 89/230 (38%), Positives = 130/230 (56%), Gaps = 10/230 (4%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAA-EGLKAEMEL 102
+V +VTGGNKGIG + LC+ KG+ V +T+RD + G +A+ L EGL
Sbjct 5 KVAVVTGGNKGIGLAIVRALCKQFKGD---VYLTARDPKLGEEAVRALKEKEGLSPLFHQ 61
Query 103 LDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDI 162
LDI +S + + +K KYG +D L+NNAG AFK A T P QA+VT N++ATRDI
Sbjct 62 LDINNLQSIRTLGSFLKEKYGGIDVLINNAGIAFKVADTTPFGTQAEVTLKTNFFATRDI 121
Query 163 TLDMMGLFKPGSRIVNVASAAGEMALQEMSAELRHRLMSKSARQEDIDKVVDDFIVACEK 222
+++ L KP R+VNV+S A MAL+ S EL+ S + +E++ ++ F+ +K
Sbjct 122 CNELLPLIKPHGRVVNVSSMASYMALERCSPELQKVFRSDTITEEELVTFMEKFVEDAKK 181
Query 223 GQQE--GWPSSTYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCCCC 270
G E GWP+ YG SK V L+ AR+ + + D ++ CC
Sbjct 182 GVHEAQGWPNMAYGTSKVGVTVLSRIQARELNE----KRKNDGILLNACC 227
> mmu:109857 Cbr3, 1110001J05Rik, C81353, MGC99956; carbonyl reductase
3 (EC:1.1.1.184); K00079 carbonyl reductase (NADPH)
[EC:1.1.1.184]
Length=277
Score = 150 bits (378), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 82/227 (36%), Positives = 128/227 (56%), Gaps = 7/227 (3%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELL 103
RV +VTG NKGIGF + LCR G+ VV+T+RD+ G A+ +L AEGL L
Sbjct 6 RVALVTGANKGIGFAITRDLCRKFSGD---VVLTARDEARGRAAVQQLQAEGLSPRFHQL 62
Query 104 DITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDIT 163
DI +S + ++ +YG ++ LVNNAG AF+ P +QA+VT N++ATR++
Sbjct 63 DIDDPQSIRALRDFLRKEYGGLNVLVNNAGIAFRMDDPTPFDIQAEVTLKTNFFATRNVC 122
Query 164 LDMMGLFKPGSRIVNVASAAGEMALQEMSAELRHRLMSKSARQEDIDKVVDDFI--VACE 221
+++ + KP R+VN++S G AL+ +L+ + + + D+ ++ F+ E
Sbjct 123 TELLPIMKPHGRVVNISSLQGLKALENCREDLQEKFRCDTLTEVDLVDLMKKFVEDTKNE 182
Query 222 KGQQEGWPSSTYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCC 268
++EGWP S YG+SK V LT AR+ D +A R ++ CC
Sbjct 183 VHEREGWPDSAYGVSKLGVTVLTRILARQLDE--KRKADRILLNACC 227
> xla:496039 hypothetical LOC496039
Length=277
Score = 145 bits (365), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 86/230 (37%), Positives = 129/230 (56%), Gaps = 10/230 (4%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAA-EGLKAEMEL 102
+V +VTGGNKGIG + LC+ KG+ V +T+R+ + G +A+ L EGL
Sbjct 5 KVAVVTGGNKGIGLAIVRALCKHFKGD---VYLTARNTKLGEEAVKGLKEKEGLSPLFHQ 61
Query 103 LDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDI 162
LDI +S + + +K KYG +D LVNNAG AFK A T P QA+VT N++ATRDI
Sbjct 62 LDINDLQSIRTLGSFLKEKYGGIDVLVNNAGIAFKVADTTPFGTQAEVTLKTNFFATRDI 121
Query 163 TLDMMGLFKPGSRIVNVASAAGEMALQEMSAELRHRLMSKSARQEDIDKVVDDFIVACEK 222
+ + L K R+VNV+S A AL S EL+ + +E++ ++ F+ +K
Sbjct 122 CNEFLPLIKSHGRVVNVSSMASYGALGRCSPELQKVFRRDNITEEELVTFMEKFVEDAKK 181
Query 223 G--QQEGWPSSTYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCCCC 270
G Q+EGWP++ YG+SK + L+ AR+ + + D ++ CC
Sbjct 182 GIHQKEGWPNTAYGVSKVGLTVLSRIQARELNE----KRKSDGILLNACC 227
> hsa:874 CBR3, SDR21C2, hCBR3; carbonyl reductase 3 (EC:1.1.1.184);
K00079 carbonyl reductase (NADPH) [EC:1.1.1.184]
Length=277
Score = 144 bits (364), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 84/240 (35%), Positives = 135/240 (56%), Gaps = 8/240 (3%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELL 103
RV +VTG N+GIG A++LCR G+ VV+T+RD G A+ +L AEGL L
Sbjct 6 RVALVTGANRGIGLAIARELCRQFSGD---VVLTARDVARGQAAVQQLQAEGLSPRFHQL 62
Query 104 DITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDIT 163
DI +S + ++ +YG ++ LVNNA AFK P ++A++T N++ATR++
Sbjct 63 DIDDLQSIRALRDFLRKEYGGLNVLVNNAAVAFKSDDPMPFDIKAEMTLKTNFFATRNMC 122
Query 164 LDMMGLFKPGSRIVNVASAAGEMALQEMSAELRHRLMSKSARQEDIDKVVDDFI--VACE 221
+++ + KP R+VN++S A + S +L+ R S++ + D+ ++ F+ E
Sbjct 123 NELLPIMKPHGRVVNISSLQCLRAFENCSEDLQERFHSETLTEGDLVDLMKKFVEDTKNE 182
Query 222 KGQQEGWPSSTYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCCCCCCD-DGEGKDA 280
++EGWP+S YG+SK V L+ AR+ D +A R +V CC D +GKD+
Sbjct 183 VHEREGWPNSPYGVSKLGVTVLSRILARRLDE--KRKADRILVNACCPGPVKTDMDGKDS 240
> xla:734804 cbr1, MGC131152; carbonyl reductase 1
Length=277
Score = 139 bits (349), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 86/230 (37%), Positives = 129/230 (56%), Gaps = 10/230 (4%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAA-EGLKAEMEL 102
+V IVTGGNKGIG + LC+ G+ V +T+R+ + G +A+ L EGL
Sbjct 5 KVAIVTGGNKGIGLAIVRALCKHFMGD---VYLTARNTKLGEEAVKALKEKEGLSPLFHQ 61
Query 103 LDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDI 162
LDI +S + + +K KYG +D L+NNAG AFK A P +QA VT N++ATRDI
Sbjct 62 LDINDLQSIRTLGSFLKEKYGGIDVLINNAGIAFKGADPTPFGIQAHVTLKTNFFATRDI 121
Query 163 TLDMMGLFKPGSRIVNVASAAGEMALQEMSAELRHRLMSKSARQEDIDKVVDDFIVACEK 222
+++ KP R+VNV+S ALQ S ELR + +E++ +++ F+ +K
Sbjct 122 CNELLPQIKPQGRVVNVSSVLSSSALQGCSPELRKVFRRDNITEEELVTLMEKFVEDAKK 181
Query 223 G--QQEGWPSSTYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCCCC 270
G Q+EGWP++ YG+SK V L+ AR+ + + D ++ CC
Sbjct 182 GIHQKEGWPNTAYGVSKVGVTVLSRIQARELNE----KRKSDGILLNACC 227
> mmu:386400 carbonyl reductase [NADPH] 1-like
Length=277
Score = 135 bits (339), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 76/229 (33%), Positives = 125/229 (54%), Gaps = 9/229 (3%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELL 103
R+ +VTG NKGIGF + LC+ G+ VV+T+RD+ G+ A+ KL AEGL L
Sbjct 6 RIALVTGANKGIGFAITRDLCQQFSGD---VVLTARDEARGLAAVQKLQAEGLIPRFHQL 62
Query 104 DITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDIT 163
DI +S + + +YG +D LVNNAG A+K + + N++ T+ +
Sbjct 63 DINDPQSIHALRNFLLKEYGGLDVLVNNAGIAYKGTDLTHFHILREAAMKTNFFGTQAVC 122
Query 164 LDMMGLFKPGSRIVNVASAAGEMALQEMSAELRHRLMSKSARQEDIDKVVDDFIVACEKG 223
+++ L K R+VN++S AL+ S EL+ + S++ +E++ +++ F+ +KG
Sbjct 123 TELLPLIKTQGRVVNISSLISLEALKNCSLELQQKFRSETITEEELVGLMNKFVEDTKKG 182
Query 224 --QQEGWPSSTYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCCCC 270
+EGWP+S YG+SK V L+ ARK + + D ++ CC
Sbjct 183 VHAKEGWPNSAYGVSKIGVTVLSRILARKLNE----QRRGDKILLNACC 227
> dre:792137 hypothetical protein LOC792137
Length=277
Score = 129 bits (325), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 77/232 (33%), Positives = 130/232 (56%), Gaps = 8/232 (3%)
Query 41 MAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLK-AE 99
M+Q+V +VTG NKGIG K LC+ G +++T+R+++ G +A+A L +EG K
Sbjct 1 MSQKVAVVTGANKGIGLAIVKGLCK--AGFTGDILLTARNEKLGQEAIAGLQSEGFKNVV 58
Query 100 MELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYAT 159
LDI + S ++ KYG +D L+NNAG AFK AATEP QA+VT N++ T
Sbjct 59 FHQLDICDQGSCMKLKKFLEEKYGGLDVLINNAGIAFKNAATEPFGEQAEVTMRTNFWGT 118
Query 160 RDITLDMMGLFKPGSRIVNVASAAGEMALQEMSAELRHRLMSKSARQEDIDKVVDDFIVA 219
++ + + +R+VNV+S + +L + SAEL+ + +K +E++ ++ +F+
Sbjct 119 LWACHALLPILRANARVVNVSSFVSKKSLDQCSAELQAKFRNKDLSEEELCLLMGEFVQD 178
Query 220 CEKGQQ--EGWPSSTYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCCC 269
+ G +GWP++ YG +K V L+ AR + + +++ CC
Sbjct 179 AQAGDHSAKGWPNTAYGTTKIGVTVLSRIQARVLNETRPGDG---ILLNACC 227
> dre:337696 cbr1l, fp17a06, wu:fa92b03, wu:fp17a06; carbonyl
reductase 1-like
Length=277
Score = 127 bits (320), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 76/232 (32%), Positives = 130/232 (56%), Gaps = 8/232 (3%)
Query 41 MAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLK-AE 99
M+++V +VTG NKGIG K LC+ G +++T+R+++ G +A+A L +EG K
Sbjct 1 MSKKVAVVTGANKGIGLAIVKGLCK--AGFTGDILLTARNEKLGQEAIAGLQSEGFKNVV 58
Query 100 MELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYAT 159
LDI + S ++ KYG +D L+NNAG AFK AATEP QA+VT N++ T
Sbjct 59 FHQLDICDQGSCMKLKKFLEEKYGGLDVLINNAGIAFKNAATEPFGEQAEVTMRTNFWGT 118
Query 160 RDITLDMMGLFKPGSRIVNVASAAGEMALQEMSAELRHRLMSKSARQEDIDKVVDDFIVA 219
++ + + +R+VNV+S + +L + SAEL+ + +K +E++ ++ +F+
Sbjct 119 LWACHALLPILRANARVVNVSSFVSKKSLDQCSAELQAKFRNKDLSEEELCLLMGEFVQD 178
Query 220 CEKGQQ--EGWPSSTYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCCC 269
+ G +GWP++ YG +K V L+ AR + + +++ CC
Sbjct 179 AQAGDHSAKGWPNTAYGTTKIGVTVLSRIQARVLNETRPGDG---ILLNACC 227
> ath:AT5G51030 short-chain dehydrogenase/reductase (SDR) family
protein
Length=314
Score = 122 bits (306), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 85/241 (35%), Positives = 119/241 (49%), Gaps = 21/241 (8%)
Query 42 AQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEME 101
++ V +VTG N+GIGFE + R L G V++TSRD+ GV+A L G +
Sbjct 36 SETVAVVTGANRGIGFE----MVRQLAGHGLTVILTSRDENVGVEAAKILQEGGFNVDFH 91
Query 102 LLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRD 161
LDI S + F IK KYG +D L+NNAG + + V + + NYY T++
Sbjct 92 RLDILDSSSIQEFCEWIKEKYGFIDVLINNAGVNYNVGSDNSVEF-SHMVISTNYYGTKN 150
Query 162 ITLDMMGLFK---PGSRIVNVASAAGEMALQEMSAE---LRHRLMS-KSARQEDIDKVVD 214
I M+ L + G+RIVNV S G + + E +R +LM S +E +DK V
Sbjct 151 IINAMIPLMRHACQGARIVNVTSRLGRLKGRHSKLENEDVRAKLMDVDSLTEEIVDKTVS 210
Query 215 DFIVACEKGQQE--GWPSS--TYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCCCC 270
+F+ E+G E GWP S Y +SK AV A T A++ P E I C
Sbjct 211 EFLKQVEEGTWESGGWPHSFTDYSVSKMAVNAYTRVLAKELSERPEGEK-----IYANCF 265
Query 271 C 271
C
Sbjct 266 C 266
> ath:AT5G61830 short-chain dehydrogenase/reductase (SDR) family
protein
Length=316
Score = 105 bits (262), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 78/223 (34%), Positives = 117/223 (52%), Gaps = 18/223 (8%)
Query 43 QRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKL--AAEGLKAEM 100
+ V +VTG N+GIGFE A++L + G VV+T+R+ G++A+ L EGLK
Sbjct 36 ENVAVVTGSNRGIGFEIARQLA--VHG--LTVVLTARNVNAGLEAVKSLRHQEEGLKVYF 91
Query 101 ELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATR 160
LD+T S F +K +G +D LVNNAG + + V A+ NY T+
Sbjct 92 HQLDVTDSSSIREFGCWLKQTFGGLDILVNNAGVNYNLGSDNTVEF-AETVISTNYQGTK 150
Query 161 DITLDMMGLFKP---GSRIVNVASAAGEMALQE---MSAELRHRLMSKSARQED-IDKVV 213
++T M+ L +P G+R+VNV+S G + + + ELR +L S E+ ID+ V
Sbjct 151 NMTKAMIPLMRPSPHGARVVNVSSRLGRVNGRRNRLANVELRDQLSSPDLLTEELIDRTV 210
Query 214 DDFIVACEKGQQE--GWPS--STYGLSKAAVIALTAAWARKAD 252
FI + G E GWP + Y +SK AV A T A++ +
Sbjct 211 SKFINQVKDGTWESGGWPQTFTDYSMSKLAVNAYTRLMAKELE 253
> mmu:100505223 carbonyl reductase [NADPH] 1-like
Length=218
Score = 99.4 bits (246), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 55/170 (32%), Positives = 93/170 (54%), Gaps = 6/170 (3%)
Query 103 LDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDI 162
LDI +S + + +Y +D LVNNAG AFK P ++ +VT N++ TRD+
Sbjct 3 LDIDDPQSIHALRDFLLKEYKGLDVLVNNAGIAFKVDDPTPFHIKGEVTMKTNFFGTRDV 62
Query 163 TLDMMGLFKPGSRIVNVASAAGEMALQEMSAELRHRLMSKSARQEDIDKVVDDFIVACEK 222
+M+ L KP R+VNV+S AL++ S EL+ + S++ +E++ +++ ++ +K
Sbjct 63 CKEMLPLIKPQGRVVNVSSMVSLSALKDCSPELQQKFRSETITEEELVGLMNKYVEDAKK 122
Query 223 G--QQEGWPSSTYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCCCC 270
G +E WP+S Y +SK V L+ ARK + + D ++ CC
Sbjct 123 GVYVKEDWPNSAYEVSKIGVTVLSRILARKLNE----QRRGDKILLNACC 168
> ath:AT3G59710 short-chain dehydrogenase/reductase (SDR) family
protein
Length=302
Score = 89.4 bits (220), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 70/231 (30%), Positives = 110/231 (47%), Gaps = 21/231 (9%)
Query 47 IVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGL-KAEMELLDI 105
+VTG NKGIGF K+L VV+T+R+ ENG QA L G LDI
Sbjct 32 VVTGANKGIGFAVVKRLLE----LGLTVVLTARNAENGSQAAESLRRIGFGNVHFCCLDI 87
Query 106 TKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDITLD 165
+ S +F + G +D LVNNA +F A E + + + N+Y + +T
Sbjct 88 SDPSSIAAFASWFGRNLGILDILVNNAAVSF-NAVGENLIKEPETIIKTNFYGAKLLTEA 146
Query 166 MMGLFKPG---SRIVNVASAAGEMALQEMSAELRHRLMSKSARQEDIDKVVDDFIVACEK 222
++ LF+ SRI+N++S G + + S +R L S+ E ID + F+ +
Sbjct 147 LLPLFRRSVSVSRILNMSSRLGTLN-KLRSPSIRRILESEDLTNEQIDATLTQFLQDVKS 205
Query 223 G--QQEGWPSS--TYGLSKAAVIALTAAWARKADHCPSMEACRDMVITCCC 269
G +++GWP + Y +SK A+ A + AR+ D + + + C C
Sbjct 206 GTWEKQGWPENWPDYAISKLALNAYSRVLARRYDG-------KKLSVNCLC 249
> ath:AT3G61220 short-chain dehydrogenase/reductase (SDR) family
protein; K15095 (+)-neomenthol dehydrogenase [EC:1.1.1.208]
Length=296
Score = 83.6 bits (205), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 73/243 (30%), Positives = 119/243 (48%), Gaps = 48/243 (19%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMEL- 102
R +VTG N+GIGFE +CR L E VV+TSRD+ G++A+ E LK E+E+
Sbjct 7 RYAVVTGANRGIGFE----ICRQLASEGIRVVLTSRDENRGLEAV-----ETLKKELEIS 57
Query 103 --------LDITKKESRESFVAAIKSKYGHVDSLVNNA-------------------GFA 135
LD+ S S +K+++G +D LVNNA GF
Sbjct 58 DQSLLFHQLDVADPASITSLAEFVKTQFGKLDILVNNAGIGGIITDAEALRAGAGKEGFK 117
Query 136 FKKAATEPVAVQAKVTCGINYYATRDITLDMMGLFK--PGSRIVNVASAAGEMALQEMSA 193
+ + TE + + INYY + + + L K RIVNV+S+ G+ L+ +
Sbjct 118 WDEIITETYELTEEC-IKINYYGPKRMCEAFIPLLKLSDSPRIVNVSSSMGQ--LKNVLN 174
Query 194 ELRHRLMS--KSARQEDIDKVVDDFIVACEKG--QQEGWPS--STYGLSKAAVIALTAAW 247
E ++S ++ +E ID+V++ + ++G +++ W S Y +SKA++ T
Sbjct 175 EWAKGILSDAENLTEERIDQVINQLLNDFKEGTVKEKNWAKFMSAYVVSKASLNGYTRVL 234
Query 248 ARK 250
A+K
Sbjct 235 AKK 237
> ath:AT2G24190 short-chain dehydrogenase/reductase (SDR) family
protein
Length=301
Score = 77.8 bits (190), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 72/259 (27%), Positives = 121/259 (46%), Gaps = 48/259 (18%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMEL- 102
R IVTGGN+GIGFE +CR L + V++TSRD++ G++A+ E LK E+E+
Sbjct 12 RYAIVTGGNRGIGFE----ICRQLANKGIRVILTSRDEKQGLEAV-----ETLKKELEIS 62
Query 103 --------LDITKKESRESFVAAIKSKYGHVDSLVNNA-------------------GFA 135
LD++ S S +K+ +G +D L+NNA GF
Sbjct 63 DQSIVFHQLDVSDPVSVTSLAEFVKTHFGKLDILINNAGVGGVITDVDALRAGTGKEGFK 122
Query 136 FKKAATEPVAVQAKVTCGINYYATRDITLDMMGLFK--PGSRIVNVASAAGEMALQEMSA 193
+++ TE + A+ INYY + + + L + RI+NV+S G+ ++ +
Sbjct 123 WEETITETYEL-AEECIKINYYGPKRMCEAFIPLLQLSDSPRIINVSSFMGQ--VKNLVN 179
Query 194 ELRHRLMSKSARQED--IDKVVDDFI--VACEKGQQEGWPS--STYGLSKAAVIALTAAW 247
E ++S + + ID+V++ + + + + + W S Y +SKA + A T
Sbjct 180 EWAKGILSDAENLTEVRIDQVINQLLNDLKEDTAKTKYWAKVMSAYVVSKAGLNAYTRIL 239
Query 248 ARKADHCPSMEACRDMVIT 266
A+K C V T
Sbjct 240 AKKHPEIRVNSVCPGFVKT 258
> mmu:100503768 carbonyl reductase [NADPH] 1-like
Length=149
Score = 74.3 bits (181), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 39/92 (42%), Positives = 53/92 (57%), Gaps = 3/92 (3%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELL 103
R+ +VTG NKGIGF + LCR G+ VV+T+RD+ G+ A+ KL AEGL L
Sbjct 6 RIALVTGANKGIGFAITRDLCRQFSGD---VVLTARDEARGLAAVQKLQAEGLSPRFHQL 62
Query 104 DITKKESRESFVAAIKSKYGHVDSLVNNAGFA 135
DI + + + +YG +D LVNNAG
Sbjct 63 DINDPQRICALCDFLLKEYGGLDVLVNNAGIG 94
> hsa:10202 DHRS2, HEP27, SDR25C1; dehydrogenase/reductase (SDR
family) member 2; K11164 dehydrogenase/reductase SDR family
member 2 [EC:1.1.-.-]
Length=280
Score = 65.5 bits (158), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 48/125 (38%), Positives = 64/125 (51%), Gaps = 9/125 (7%)
Query 14 LYSIFRSFICRFSPLPRF-LRSKFTFC----TMAQRVFIVTGGNKGIGFETAKKLCRDLK 68
L ++ R + F P R +R T +A RV +VTG GIGF A++L RD
Sbjct 2 LSAVARGYQGWFHPCARLSVRMSSTGIDRKGVLANRVAVVTGSTSGIGFAIARRLARD-- 59
Query 69 GENAVVVITSRDKENGVQALAKLAAEGLKAEMELLDITKKESRESFVAAIKSKYGHVDSL 128
A VVI+SR ++N +A+AKL EGL + + K E RE VA G VD L
Sbjct 60 --GAHVVISSRKQQNVDRAMAKLQGEGLSVAGIVCHVGKAEDREQLVAKALEHCGGVDFL 117
Query 129 VNNAG 133
V +AG
Sbjct 118 VCSAG 122
> hsa:10901 DHRS4, CR, FLJ11008, NRDR, PHCR, PSCD, SCAD-SRL, SDR-SRL,
SDR25C1, SDR25C2; dehydrogenase/reductase (SDR family)
member 4 (EC:1.1.1.184); K11147 dehydrogenase/reductase SDR
family member 4 [EC:1.1.-.-]
Length=278
Score = 61.6 bits (148), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 52/92 (56%), Gaps = 4/92 (4%)
Query 41 MAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEM 100
+A +V +VT GIGF A++L +D A VV++SR ++N QA+A L EGL
Sbjct 30 LANKVALVTASTDGIGFAIARRLAQD----GAHVVVSSRKQQNVDQAVATLQGEGLSVTG 85
Query 101 ELLDITKKESRESFVAAIKSKYGHVDSLVNNA 132
+ + K E RE VA +G +D LV+NA
Sbjct 86 TVCHVGKAEDRERLVATAVKLHGGIDILVSNA 117
> ath:AT4G09750 short-chain dehydrogenase/reductase (SDR) family
protein
Length=322
Score = 60.8 bits (146), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 47/155 (30%), Positives = 76/155 (49%), Gaps = 13/155 (8%)
Query 47 IVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAE--GLKAEMELLD 104
+VTG N GIGF A+ L A V + R+KE G +AL+K+ +E+ D
Sbjct 47 VVTGANSGIGFAAAEGLAS----RGATVYMVCRNKERGQEALSKIQTSTGNQNVYLEVCD 102
Query 105 ITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDITL 164
++ +SF ++ SK V LVNNAG K T P + ++ +N T +T
Sbjct 103 LSSVNEIKSFASSFASKDVPVHVLVNNAGLLENKRTTTPEGFE--LSFAVNVLGTYTMTE 160
Query 165 DMMGLFK---PGSRIVNVASAAGEMALQEMSAELR 196
M+ L + P ++++ VAS G M ++ +L+
Sbjct 161 LMLPLLEKATPDAKVITVAS--GGMYTSPLTTDLQ 193
> mmu:71412 Dhrs2, 5430405K24Rik, Hep27, MGC143467; dehydrogenase/reductase
member 2; K11164 dehydrogenase/reductase SDR family
member 2 [EC:1.1.-.-]
Length=282
Score = 58.5 bits (140), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 39/116 (33%), Positives = 59/116 (50%), Gaps = 4/116 (3%)
Query 18 FRSFICRFSPLPRFLRSKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVIT 77
FR +C RF ++ ++A +V ++TG +GIGF A++L +D A VVI+
Sbjct 12 FRGSLCLPLSARRFSKTADENRSLAGKVAVITGSTRGIGFAIARRLAQD----GAHVVIS 67
Query 78 SRDKENGVQALAKLAAEGLKAEMELLDITKKESRESFVAAIKSKYGHVDSLVNNAG 133
SR +EN +A+ L EGL + + K E R+ V G +D LV AG
Sbjct 68 SRKQENVDEAVTILKEEGLSVTGTMCHVGKAEDRQHLVTTALKHSGGIDFLVCVAG 123
> xla:494781 rdh7.2; retinol dehydrogenase 7, gene 2; K11154 retinol
dehydrogenase 16 [EC:1.1.1.-]
Length=317
Score = 53.5 bits (127), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 45/164 (27%), Positives = 78/164 (47%), Gaps = 13/164 (7%)
Query 30 RFLRSKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALA 89
R+ + + + + ++TG + G G A+KL R + V+ + + G + L
Sbjct 16 RWYKDRHILDNLTDKYVLITGCDSGFGNLLARKLDR-----RGIHVLAACLTDRGAEELK 70
Query 90 KLAAEGLKAEMELLDITKK---ESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAV 146
K + GL+ LLD+T +S E + + I G + LVNNAG +F A E +
Sbjct 71 KQTSSGLQT--ILLDVTDSKSVKSAEKWASQIVGDKG-LWGLVNNAGISFPCAPNEWLTK 127
Query 147 QAKV-TCGINYYATRDITLDMMGLF-KPGSRIVNVASAAGEMAL 188
+ + +N D+T++M+ L K R+VN+AS AG +
Sbjct 128 EDFIKIMNVNLLGLVDVTINMLHLIRKARGRVVNIASVAGRITF 171
> ath:AT4G05530 IBR1; IBR1 (INDOLE-3-BUTYRIC ACID RESPONSE 1);
binding / catalytic/ oxidoreductase; K11147 dehydrogenase/reductase
SDR family member 4 [EC:1.1.-.-]
Length=254
Score = 53.5 bits (127), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 46/161 (28%), Positives = 75/161 (46%), Gaps = 20/161 (12%)
Query 28 LPRFLRSKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQA 87
LPR L K V IVT +GIGF ++ E A VV++SR + N +A
Sbjct 5 LPRRLEGK---------VAIVTASTQGIGFGITERFGL----EGASVVVSSRKQANVDEA 51
Query 88 LAKLAAEGLKAEMELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQ 147
+AKL ++G+ A + ++ + R + V KYG +D +V NA +T+P+
Sbjct 52 VAKLKSKGIDAYGIVCHVSNAQHRRNLVEKTVQKYGKIDIVVCNAA---ANPSTDPILSS 108
Query 148 AKVTCG----INYYATRDITLDMMGLFKPGSRIVNVASAAG 184
+ IN ++ + DM + GS ++ + S AG
Sbjct 109 KEAVLDKLWEINVKSSILLLQDMAPHLEKGSSVIFITSIAG 149
> mmu:28200 Dhrs4, AI043103, AI790593, CR, D14Ucla2, NDRD, PHCR,
PSCD, RRD, mouNRDR; dehydrogenase/reductase (SDR family)
member 4 (EC:1.1.1.184); K11147 dehydrogenase/reductase SDR
family member 4 [EC:1.1.-.-]
Length=279
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 50/92 (54%), Gaps = 4/92 (4%)
Query 41 MAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEM 100
++ +V +VT GIGF A++L D A VV++SR ++N +A+A L EGL
Sbjct 31 LSNKVALVTASTDGIGFAIARRLAED----GAHVVVSSRKQQNVDRAVATLQGEGLSVTG 86
Query 101 ELLDITKKESRESFVAAIKSKYGHVDSLVNNA 132
+ + K E RE + ++ +D LV+NA
Sbjct 87 IVCHVGKAEDREKLITTALKRHQGIDILVSNA 118
> ath:AT1G01800 short-chain dehydrogenase/reductase (SDR) family
protein
Length=260
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 37/104 (35%), Positives = 63/104 (60%), Gaps = 10/104 (9%)
Query 155 NYYATRDITLDMMGLFKPG--SRIVNVASAAGEMALQEMSAELRHRLMS--KSARQEDID 210
NYY + + M+ L + RIV++AS G+ L+ +S E ++S ++ +E ID
Sbjct 101 NYYGVKRMCEAMIPLLQSSDSPRIVSIASTMGK--LENVSNEWAKGVLSDAENLTEEKID 158
Query 211 KVVDDFIVACEKG--QQEGWPS--STYGLSKAAVIALTAAWARK 250
+V+++++ ++G Q +GWP+ S Y LSKAAVIALT A++
Sbjct 159 EVINEYLKDYKEGALQVKGWPTVMSGYILSKAAVIALTRVLAKR 202
> bbo:BBOV_III007020 17.m07620; hypothetical protein
Length=389
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 53/167 (31%), Positives = 82/167 (49%), Gaps = 15/167 (8%)
Query 22 ICRFSPLPRFLRSKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDK 81
I + + +P+ L ++ M ++V IVTGG+ G+G E A C+ LK N VVIT+RDK
Sbjct 93 IAQGTKVPQILINR---TDMHRKVAIVTGGSSGVGKEVA---CQLLKW-NCKVVITARDK 145
Query 82 E---NGVQALAKLAAEGLKA-EMELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFK 137
N V+ L K A K + +D+ +S + V I +D L+NNAG A
Sbjct 146 TKGANAVEYLRKQANVDFKMIQFVEMDLNDPKSIKLAVEEILKTNASIDFLINNAGIA-- 203
Query 138 KAATEPVAVQAKVTCGINYYATRDIT-LDMMGLFKPGSRIVNVASAA 183
A + + N+ +T L M L + +R++NV+S A
Sbjct 204 -TNVHLNAYKQEAMFATNFLGHFQLTKLLMPTLVRFKARVINVSSIA 249
> xla:444771 dhrs4, MGC81922; dehydrogenase/reductase (SDR family)
member 4; K11147 dehydrogenase/reductase SDR family member
4 [EC:1.1.-.-]
Length=261
Score = 52.4 bits (124), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 50/89 (56%), Gaps = 4/89 (4%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELL 103
+V +VT +GIG A++L D A V+++SR ++N +A+ L EGL+ E +
Sbjct 16 KVALVTASTEGIGLAIARRLGHD----GARVLLSSRKQQNVDRAVQDLRNEGLEVEGTVC 71
Query 104 DITKKESRESFVAAIKSKYGHVDSLVNNA 132
+ +E RE + ++G +D LV+NA
Sbjct 72 HVGNREDREKLIETAVQRFGGIDILVSNA 100
> dre:100150965 retinol dehydrogenase 12-like
Length=296
Score = 52.0 bits (123), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 45/155 (29%), Positives = 74/155 (47%), Gaps = 13/155 (8%)
Query 47 IVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLA--AEGLKAEMELLD 104
IVTG N GIG ETAK DL A V++ RD QA + ++ E + LD
Sbjct 24 IVTGANTGIGKETAK----DLANRGARVILACRDLVKAEQAASDISRDVENANVVVRKLD 79
Query 105 ITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYATRDITL 164
+ +S F I + + L+NNAG A +T + + G+N+ +T
Sbjct 80 LADTKSICEFAELIYNTEKSLHLLINNAGVAICPYSTTVDGFETQF--GVNHLGHFFLTF 137
Query 165 DMMGLFKPG--SRIVNVASAA---GEMALQEMSAE 194
++ L K SR++NV+S G++ +++++E
Sbjct 138 LLIDLLKHSAPSRVINVSSLVHPMGKIHFEDLNSE 172
> hsa:207063 DHRSX, CXorf11, DHRS5X, DHRS5Y, DHRSXY, DHRSY, SDR46C1;
dehydrogenase/reductase (SDR family) X-linked; K11170
dehydrogenase/reductase SDR family member X [EC:1.1.-.-]
Length=330
Score = 51.2 bits (121), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 35/98 (35%), Positives = 46/98 (46%), Gaps = 6/98 (6%)
Query 38 FCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLK 97
F RV IVTGG GIG+ TAK L R V+I + Q ++K+ E L
Sbjct 38 FPPRPDRVAIVTGGTDGIGYSTAKHLAR----LGMHVIIAGNNDSKAKQVVSKIKEETLN 93
Query 98 AEMELL--DITKKESRESFVAAIKSKYGHVDSLVNNAG 133
++E L D+ S FV K K + L+NNAG
Sbjct 94 DKVEFLYCDLASMTSIRQFVQKFKMKKIPLHVLINNAG 131
> mmu:80707 Wwox, 5330426P09Rik, 9030416C10Rik, WOX1; WW domain-containing
oxidoreductase
Length=414
Score = 49.7 bits (117), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 38/102 (37%), Positives = 55/102 (53%), Gaps = 7/102 (6%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEME-- 101
+V +VTG N GIGFETAK L G A V++ R+ +A++++ E KA++E
Sbjct 125 KVVLVTGANSGIGFETAKSFA--LHG--AHVILACRNLSRASEAVSRILEEWHKAKVEAM 180
Query 102 LLDITKKESRESFVAAIKSKYGHVDSLVNNAG-FAFKKAATE 142
LD+ S + F A K+K + LV NAG FA T+
Sbjct 181 TLDLAVLRSVQHFAEAFKAKNVSLHVLVCNAGTFALPWGLTK 222
> eco:b4266 idnO, ECK4259, JW4223, yjgU; 5-keto-D-gluconate-5-reductase
(EC:1.1.1.69); K00046 gluconate 5-dehydrogenase [EC:1.1.1.69]
Length=254
Score = 49.3 bits (116), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 38/151 (25%), Positives = 70/151 (46%), Gaps = 10/151 (6%)
Query 40 TMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAE 99
++A + ++TG +GIGF A L + A ++I E A+ KL EG++A
Sbjct 6 SLAGKNILITGSAQGIGFLLATGLGK----YGAQIIINDITAERAELAVEKLHQEGIQAV 61
Query 100 MELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGIN---- 155
++T K ++ V I+ G +D LVNNAG + TE + +N
Sbjct 62 AAPFNVTHKHEIDAAVEHIEKDIGPIDVLVNNAGIQRRHPFTEFPEQEWNDVIAVNQTAV 121
Query 156 YYATRDITLDMMGLFKPGSRIVNVASAAGEM 186
+ ++ +T M+ + +++N+ S E+
Sbjct 122 FLVSQAVTRHMVE--RKAGKVINICSMQSEL 150
> eco:b1093 fabG, ECK1079, JW1079; 3-oxoacyl-[acyl-carrier-protein]
reductase (EC:1.1.1.100); K00059 3-oxoacyl-[acyl-carrier
protein] reductase [EC:1.1.1.100]
Length=244
Score = 48.9 bits (115), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 34/92 (36%), Positives = 50/92 (54%), Gaps = 9/92 (9%)
Query 44 RVFIVTGGNKGIGFETAKKLC-RDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMEL 102
++ +VTG ++GIG A+ L R K VI + ENG QA++ K M
Sbjct 6 KIALVTGASRGIGRAIAETLAARGAK------VIGTATSENGAQAISDYLGANGKGLM-- 57
Query 103 LDITKKESRESFVAAIKSKYGHVDSLVNNAGF 134
L++T S ES + I++++G VD LVNNAG
Sbjct 58 LNVTDPASIESVLEKIRAEFGEVDILVNNAGI 89
> xla:398939 hsd17b6, MGC68605; hydroxysteroid (17-beta) dehydrogenase
6 homolog
Length=318
Score = 48.9 bits (115), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 52/178 (29%), Positives = 78/178 (43%), Gaps = 37/178 (20%)
Query 28 LPRFLRSKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQA 87
L R+ R + ++ + +TG + G G AK+L DK+ G+Q
Sbjct 14 LYRWNRQRKRIENLSDKYVFITGCDSGFGNVLAKQL----------------DKQ-GIQV 56
Query 88 LAKL----AAEGLKAEME------LLDITKKESRESFVAAIKSKYGH--VDSLVNNAGFA 135
LA AE LK+E ++++T +S +S A + G + LVNNAG+
Sbjct 57 LATCLTVKGAETLKSEASSRLRTVIMNVTDSQSVKSAAAWVTGIVGDAGLWGLVNNAGYG 116
Query 136 FKKAATEPVAVQAK----VTCGINYYATRDITLDMMGLF-KPGSRIVNVASAAGEMAL 188
F P Q K IN D+TL+++ L K RIVNV+S AG +A
Sbjct 117 F---VFSPTGWQKKEHFVKVLEINLLGMVDVTLNLLPLLRKAQGRIVNVSSNAGRLAF 171
> mmu:66065 Hsd17b14, 0610039E24Rik, Dhrs10, retSDR3; hydroxysteroid
(17-beta) dehydrogenase 14
Length=273
Score = 48.5 bits (114), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 39/146 (26%), Positives = 67/146 (45%), Gaps = 10/146 (6%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELL 103
+V +VTGG++GIG + R A VV +D+ G +L+
Sbjct 10 KVVVVTGGSRGIG----AAIVRAFVDSGAQVVFCDKDEAGGRALEQELSGTVFIPG---- 61
Query 104 DITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQA-KVTCGINYYATRD- 161
D+T++ ++ V+ S++GH+D +VNNAG+ E + Q + +N T
Sbjct 62 DVTQERDLQTLVSETLSRFGHLDCVVNNAGYHPPAQLPEETSAQGFRQLLEVNLLGTYTL 121
Query 162 ITLDMMGLFKPGSRIVNVASAAGEMA 187
I L + L K I+N++S G +
Sbjct 122 IKLALPHLRKSRGNIINISSLVGAIG 147
> hsa:51741 WWOX, D16S432E, FOR, FRA16D, HHCMA56, PRO0128, SDR41C1,
WOX1; WW domain containing oxidoreductase (EC:1.1.1.-)
Length=414
Score = 48.5 bits (114), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 37/102 (36%), Positives = 55/102 (53%), Gaps = 7/102 (6%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEME-- 101
+V +VTG N GIGFETAK L G A V++ R+ +A++++ E KA++E
Sbjct 125 KVVVVTGANSGIGFETAKSFA--LHG--AHVILACRNMARASEAVSRILEEWHKAKVEAM 180
Query 102 LLDITKKESRESFVAAIKSKYGHVDSLV-NNAGFAFKKAATE 142
LD+ S + F A K+K + LV N A FA + T+
Sbjct 181 TLDLALLRSVQHFAEAFKAKNVPLHVLVCNAATFALPWSLTK 222
> xla:399015 hypothetical protein MGC68519; K11154 retinol dehydrogenase
16 [EC:1.1.1.-]
Length=319
Score = 48.1 bits (113), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 50/166 (30%), Positives = 73/166 (43%), Gaps = 13/166 (7%)
Query 28 LPRFLRSKFTFCTMAQRVFIVTGGNKGIGFETAKKL-CRDLKGENAVVVITSRDKENGVQ 86
L R+ R K T + + ++TG + G G AK+L R LK V+ + E G +
Sbjct 15 LYRWNRQKQTLENLTNKYVLITGCDSGFGNLAAKQLDARGLK------VLAACLTEKGAE 68
Query 87 ALAKLAAEGLKAEMELLDITKKESRESFVAAIKSKYGHVD--SLVNNAGFAFKKAATEPV 144
L K + LK ++D++ ES G+ LVNNAG +
Sbjct 69 ELKKETSSRLKT--VIVDVSDSESVSKAAEWASQIVGNAGLWGLVNNAGIGLPVGPNAWM 126
Query 145 AVQAKV-TCGINYYATRDITLDMMGLF-KPGSRIVNVASAAGEMAL 188
+ V IN T D+TL ++ L K RIVNVAS G ++L
Sbjct 127 KKEHFVKVIDINLLGTVDVTLKLLPLIRKAKGRIVNVASIMGRISL 172
> xla:447183 dhrsx, MGC85576; dehydrogenase/reductase (SDR family)
X-linked; K11170 dehydrogenase/reductase SDR family member
X [EC:1.1.-.-]
Length=327
Score = 48.1 bits (113), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 47/167 (28%), Positives = 77/167 (46%), Gaps = 17/167 (10%)
Query 38 FCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLK 97
F + +V IVTGG KGIG TAK+L R V+I ++ G +A+ ++ +
Sbjct 36 FSSQNGKVAIVTGGAKGIGCSTAKQLSR----LGMHVIIAGNNEAEGNEAVTRIQQDTQN 91
Query 98 AEMELL--DITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGIN 155
++E L D+ +S FV +K + LVNNAG A + G+N
Sbjct 92 EKVEFLYCDLASMKSIRQFVQNFTAKNLCLHVLVNNAGVML--VPERKTADGFEEHFGLN 149
Query 156 Y---YATRDITLDMM---GLFKPGSRIVNVASA---AGEMALQEMSA 193
Y + ++ L M G +RI+ V+SA GE+ ++++
Sbjct 150 YLGHFLLTNLLLKTMKKSGTENLNARIITVSSATHYVGELNFDDLNS 196
> cel:R05D8.10 dhs-15; DeHydrogenases, Short chain family member
(dhs-15)
Length=278
Score = 48.1 bits (113), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 30/98 (30%), Positives = 53/98 (54%), Gaps = 7/98 (7%)
Query 42 AQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAE-- 99
+ +V I+TG + GIG TA L + E A V +T R E + + ++ G ++
Sbjct 5 SDKVAIITGSSSGIGRSTAVLLAQ----EGAKVTVTGRSSEKIQETVNEIHKNGGSSDNI 60
Query 100 -MELLDITKKESRESFVAAIKSKYGHVDSLVNNAGFAF 136
+ L D+ + E ++ + + S++G +D L+NNAG AF
Sbjct 61 NIVLGDLNESECQDELIKSTLSRFGKIDILINNAGAAF 98
> xla:398882 rdh16, MGC68820; retinol dehydrogenase 16 (all-trans)
Length=317
Score = 48.1 bits (113), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 44/163 (26%), Positives = 76/163 (46%), Gaps = 11/163 (6%)
Query 30 RFLRSKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALA 89
R+ R + + ++ +TG + G G AK+L +N + V+ + + G + L
Sbjct 16 RWNREREKLLNLTEKYVFITGCDSGFGNLLAKQL-----DKNGLHVLAACLTDKGAEELK 70
Query 90 KLAAEGLKAEMELLDITKKESRESFVAAIKSKYGH--VDSLVNNAGFAFKKAATEPVAVQ 147
K + L M L++ +S S + G+ + LVNNAG + A E +A +
Sbjct 71 KETSSRLNTVM--LNVADSQSVNSAAKWVSDIVGNKGLWGLVNNAGISNPIAPNEWLAKE 128
Query 148 AKV-TCGINYYATRDITLDMMGLF-KPGSRIVNVASAAGEMAL 188
+ +N D+TL ++ L K R+VNVAS AG ++L
Sbjct 129 DYLKVLNVNLLGVIDVTLKLLPLIRKARGRVVNVASVAGRVSL 171
> cel:C06E4.6 hypothetical protein
Length=274
Score = 48.1 bits (113), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 46/93 (49%), Gaps = 7/93 (7%)
Query 44 RVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDK---ENGVQALAKLAAEGLKAEM 100
+V I+TG + GIG TA L D A V IT RD E QA+ K
Sbjct 7 KVAIITGSSNGIGRATAVLLATD----GAKVTITGRDAARLEETRQAILKAGISATNVNS 62
Query 101 ELLDITKKESRESFVAAIKSKYGHVDSLVNNAG 133
+ D+T E ++ +++ K+G ++ L+NNAG
Sbjct 63 VVADVTTAEGQDLLISSTLDKFGKINILINNAG 95
> cel:F28H7.2 hypothetical protein
Length=284
Score = 47.8 bits (112), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 54/109 (49%), Gaps = 12/109 (11%)
Query 34 SKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAA 93
S+FT +V I+TG + GIG TA R L E A V +T R+ E + L
Sbjct 2 SRFT-----DKVAIITGSSNGIGQATA----RLLASEGAKVTVTGRNAERLEETKNILLG 52
Query 94 EGLKAEMELL---DITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKA 139
G+ L+ DIT++ +E+ + + K+G +D LVNNAG A
Sbjct 53 AGVPEGNVLVVVGDITQESVQENLIKSTLDKFGKIDILVNNAGAGIPDA 101
> mmu:15484 Hsd11b2, 11HSD2; hydroxysteroid 11-beta dehydrogenase
2 (EC:1.1.1.146); K00071 11beta-hydroxysteroid dehydrogenase
[EC:1.1.1.146]
Length=386
Score = 47.8 bits (112), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 48/158 (30%), Positives = 75/158 (47%), Gaps = 11/158 (6%)
Query 41 MAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEM 100
+A R ++TG + G G ETAKKL D G + + + ++ L L + LK +
Sbjct 80 VATRAVLITGCDTGFGKETAKKL--DAMGFTVLATVLDLNSPGALE-LRDLCSPRLK--L 134
Query 101 ELLDITKKE--SRESFVAAIKSKYGHVDSLVNNAGFAFKKAATE--PVAVQAKVTCGINY 156
+D+TK E SR + + + LVNNAG A E PVA K +N+
Sbjct 135 LQMDLTKAEDISRVLEITKAHTASTGLWGLVNNAGLNIVVADVELSPVATFRKCM-EVNF 193
Query 157 YATRDITLDMMGLFKPG-SRIVNVASAAGEMALQEMSA 193
+ ++T ++ L + RIV V S AG+M ++A
Sbjct 194 FGALELTKGLLPLLRHSRGRIVTVGSPAGDMPYPCLAA 231
> hsa:121214 SDR9C7, FLJ16333, MGC126600, MGC126602, RDHS, SDR-O,
SDRO; short chain dehydrogenase/reductase family 9C, member
7 (EC:1.1.1.-)
Length=313
Score = 47.8 bits (112), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 41/153 (26%), Positives = 74/153 (48%), Gaps = 11/153 (7%)
Query 40 TMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAE 99
++++ +TG + G G AK+L + + V+ + E G Q L + + L+
Sbjct 22 NLSEKYVFITGCDSGFGNLLAKQLV-----DRGMQVLAACFTEEGSQKLQRDTSYRLQTT 76
Query 100 MELLDITKKESRESFVAAIKSKYGH--VDSLVNNAGFAFKKAATEPVAVQAKV-TCGINY 156
LLD+TK ES ++ ++ K G + +LVNNAG E + V +N
Sbjct 77 --LLDVTKSESIKAAAQWVRDKVGEQGLWALVNNAGVGLPSGPNEWLTKDDFVKVINVNL 134
Query 157 YATRDITLDMMGLFKPG-SRIVNVASAAGEMAL 188
++TL M+ + K R+VN++S+ G +A+
Sbjct 135 VGLIEVTLHMLPMVKRARGRVVNMSSSGGRVAV 167
> cel:F36H9.3 dhs-13; DeHydrogenases, Short chain family member
(dhs-13); K11147 dehydrogenase/reductase SDR family member
4 [EC:1.1.-.-]
Length=257
Score = 47.8 bits (112), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 36/107 (33%), Positives = 49/107 (45%), Gaps = 13/107 (12%)
Query 26 SPLPRFLRSKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGV 85
SP +FL RV +VT KGIGF AK+L A VV+ SR KEN
Sbjct 3 SPATKFL---------TDRVALVTASTKGIGFAIAKQLG----AAGASVVVCSRKKENVD 49
Query 86 QALAKLAAEGLKAEMELLDITKKESRESFVAAIKSKYGHVDSLVNNA 132
+A+A L E + A + K R + ++ +D LV+NA
Sbjct 50 EAVAALRLENIDAHGTTAHVGNKSDRTKLIDFTLDRFTKLDILVSNA 96
> dre:553712 MGC112332; zgc:112332
Length=298
Score = 47.4 bits (111), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 46/147 (31%), Positives = 68/147 (46%), Gaps = 10/147 (6%)
Query 41 MAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEM 100
+ ++ I+TG N GIG ETA RDL A VV+ RD E A +L +
Sbjct 18 LDEKTVIITGANTGIGKETA----RDLARRGARVVMACRDLEKAEAARRELMDNSGNQNI 73
Query 101 EL--LDITKKESRESFVAAIKSKYGHVDSLVNNAGFAFKKAATEPVAVQAKVTCGINYYA 158
+ LD+ +S ++F I + V+ L+NNAG A ++ G+N+
Sbjct 74 VVKKLDLADTKSIKAFAELINKEEKQVNILINNAGIMM--CPYSKTADGFEMQFGVNHLG 131
Query 159 TRDITLDMMGLFKPG--SRIVNVASAA 183
+ ++ L K SRIVNVAS A
Sbjct 132 HFLLIYLLLDLLKKSTPSRIVNVASVA 158
> ath:AT2G29150 tropinone reductase, putative / tropine dehydrogenase,
putative; K08081 tropine dehydrogenase [EC:1.1.1.206]
Length=268
Score = 47.4 bits (111), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 31/100 (31%), Positives = 53/100 (53%), Gaps = 5/100 (5%)
Query 47 IVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQALAKLAAEGLKAEMELLDIT 106
+VTGG+KG+G + + +L A V +RD+ + L + A+G + + D++
Sbjct 22 LVTGGSKGLG----EAVVEELAMLGARVHTCARDETQLQERLREWQAKGFEVTTSVCDVS 77
Query 107 KKESRESFVAAIKSKY-GHVDSLVNNAGFAFKKAATEPVA 145
+E RE + + S + G ++ LVNNAG K +TE A
Sbjct 78 SREQREKLMETVSSVFQGKLNILVNNAGTGIIKPSTEYTA 117
> mmu:27400 Hsd17b6, 17betaHSD9, Hsd17b9, Rdh8; hydroxysteroid
(17-beta) dehydrogenase 6 (EC:1.1.1.62 1.1.1.63 1.1.1.105);
K13369 estradiol 17beta-dehydrogenase / testosterone 17beta-dehydrogenase
/ retinol dehydrogenase [EC:1.1.1.62 1.1.1.63
1.1.1.105]
Length=317
Score = 47.4 bits (111), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 47/165 (28%), Positives = 75/165 (45%), Gaps = 11/165 (6%)
Query 28 LPRFLRSKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKGENAVVVITSRDKENGVQA 87
L R+ R + + + +TG + G G A++L R + V+ + E G +
Sbjct 14 LLRWYRERQVVSHLQDKYVFITGCDSGFGNLLARQLDR-----RGMRVLAACLTEKGAEE 68
Query 88 LAKLAAEGLKAEMELLDITKKESRESFVAAIKSKYGH--VDSLVNNAGFAFKKAATEPVA 145
L ++ L E +LD+TK ES + +K + G + LVNNAG A E
Sbjct 69 LRNKTSDRL--ETVILDVTKTESIVAATQWVKERVGDRGLWGLVNNAGVLQPFAYIEWYR 126
Query 146 VQAKVTC-GINYYATRDITLDMMGLFKPG-SRIVNVASAAGEMAL 188
+ + +N +T+ M+ L K RIVNV+SA G +AL
Sbjct 127 PEDYMPIFQVNLIGLTQVTISMLFLVKKARGRIVNVSSALGRVAL 171
> dre:100318301 dhrsx; dehydrogenase/reductase (SDR family) X-linked
Length=324
Score = 47.0 bits (110), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 59/198 (29%), Positives = 91/198 (45%), Gaps = 32/198 (16%)
Query 11 KFSLYSIF-RSFICRFSPLPRFLRSKFTFCTMAQRVFIVTGGNKGIGFETAKKLCRDLKG 69
K LY +F +SF R LP +V IVTGG +G+G+E + R L
Sbjct 20 KVLLYQLFNKSF--RLPDLPE----------QNGKVAIVTGGTRGMGYE----ISRHLVS 63
Query 70 ENAVVVITSRDKENGVQALAKLAAEGLKAEMEL--LDITKKESRESFVAAIKSKYGHVDS 127
+ V+I ++E G+ A+ K+ E + ++E LD+ S FV +K +
Sbjct 64 LDMHVIIAGNEEEEGLAAVKKIQEELNQGKVEFMYLDLASLTSVRQFVQRYNAKGLPLHV 123
Query 128 LVNNAGFAF-KKAATEPVAVQAKVTCGINYYATRDITLDMMGLF----KPG--SRIVNVA 180
LVNNAG + TE ++ G+NY +T ++G KPG SRIV ++
Sbjct 124 LVNNAGVMLVPERRTED---GFELHFGLNYLGHFLLTNLLLGALRKTGKPGKCSRIVIMS 180
Query 181 SA---AGEMALQEMSAEL 195
SA G + L ++ L
Sbjct 181 SATHYGGRLTLDDLQGRL 198
Lambda K H
0.322 0.133 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 11167158100
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40