bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1393_orf2
Length=171
Score E
Sequences producing significant alignments: (Bits) Value
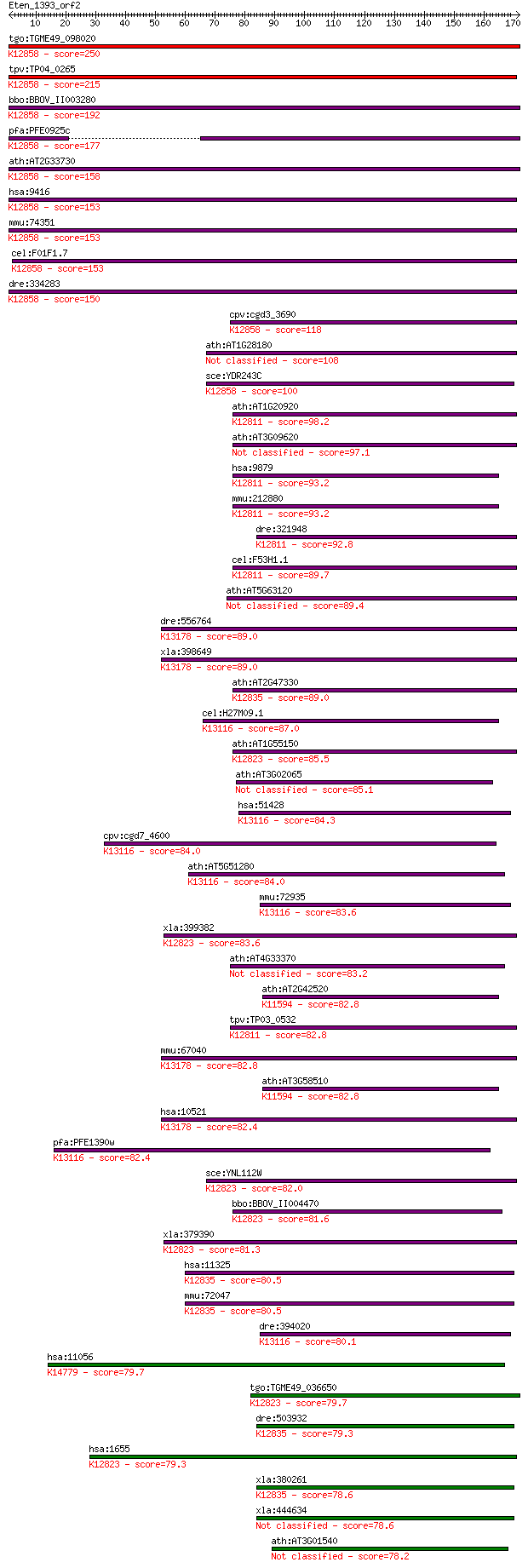
tgo:TGME49_098020 DEAD-box ATP-dependent RNA helicase, putativ... 250 2e-66
tpv:TP04_0265 small nuclear ribonucleoprotein; K12858 ATP-depe... 215 6e-56
bbo:BBOV_II003280 18.m06276; DEAD box RNA helicase; K12858 ATP... 192 3e-49
pfa:PFE0925c snrnp protein, putative; K12858 ATP-dependent RNA... 177 2e-44
ath:AT2G33730 DEAD box RNA helicase, putative; K12858 ATP-depe... 158 8e-39
hsa:9416 DDX23, MGC8416, PRPF28, U5-100K, U5-100KD, prp28; DEA... 153 3e-37
mmu:74351 Ddx23, 3110082M05Rik, 4921506D17Rik; DEAD (Asp-Glu-A... 153 3e-37
cel:F01F1.7 ddx-23; DEAD boX helicase homolog family member (d... 153 3e-37
dre:334283 ddx23, wu:fi39b12, zgc:63742; DEAD (Asp-Glu-Ala-Asp... 150 2e-36
cpv:cgd3_3690 U5 snRNP 100 kD protein ; K12858 ATP-dependent R... 118 8e-27
ath:AT1G28180 ATP binding / ATP-dependent helicase/ helicase/ ... 108 6e-24
sce:YDR243C PRP28; Prp28p (EC:3.6.1.-); K12858 ATP-dependent R... 100 2e-21
ath:AT1G20920 DEAD box RNA helicase, putative; K12811 ATP-depe... 98.2 1e-20
ath:AT3G09620 DEAD/DEAH box helicase, putative 97.1 3e-20
hsa:9879 DDX46, FLJ25329, KIAA0801, MGC9936, PRPF5, Prp5; DEAD... 93.2 4e-19
mmu:212880 Ddx46, 2200005K02Rik, 8430438J23Rik, AI325430, AI95... 93.2 4e-19
dre:321948 ddx46, fb39a03, wu:fb39a03; DEAD (Asp-Glu-Ala-Asp) ... 92.8 6e-19
cel:F53H1.1 hypothetical protein; K12811 ATP-dependent RNA hel... 89.7 4e-18
ath:AT5G63120 ethylene-responsive DEAD box RNA helicase, putat... 89.4 5e-18
dre:556764 similar to Probable RNA-dependent helicase p72 (DEA... 89.0 7e-18
xla:398649 ddx17, MGC80019; DEAD (Asp-Glu-Ala-Asp) box polypep... 89.0 7e-18
ath:AT2G47330 DEAD/DEAH box helicase, putative; K12835 ATP-dep... 89.0 8e-18
cel:H27M09.1 hypothetical protein; K13116 ATP-dependent RNA he... 87.0 3e-17
ath:AT1G55150 DEAD box RNA helicase, putative (RH20); K12823 A... 85.5 8e-17
ath:AT3G02065 DEAD/DEAH box helicase family protein 85.1 1e-16
hsa:51428 DDX41, ABS, MGC8828; DEAD (Asp-Glu-Ala-Asp) box poly... 84.3 2e-16
cpv:cgd7_4600 abstrakt protein SF II helicase + Znknuckle C2HC... 84.0 2e-16
ath:AT5G51280 DEAD-box protein abstrakt, putative; K13116 ATP-... 84.0 3e-16
mmu:72935 Ddx41, 2900024F02Rik, AA958953, ABS, AI324246; DEAD ... 83.6 3e-16
xla:399382 ddx5, MGC81559; DEAD (Asp-Glu-Ala-Asp) box polypept... 83.6 3e-16
ath:AT4G33370 DEAD-box protein abstrakt, putative 83.2 3e-16
ath:AT2G42520 DEAD box RNA helicase, putative; K11594 ATP-depe... 82.8 5e-16
tpv:TP03_0532 ATP-dependent RNA helicase; K12811 ATP-dependent... 82.8 5e-16
mmu:67040 Ddx17, 2610007K22Rik, A430025E01Rik, AI047725, C8092... 82.8 5e-16
ath:AT3G58510 DEAD box RNA helicase, putative (RH11); K11594 A... 82.8 6e-16
hsa:10521 DDX17, DKFZp761H2016, P72, RH70; DEAD (Asp-Glu-Ala-A... 82.4 7e-16
pfa:PFE1390w RNA helicase-1; K13116 ATP-dependent RNA helicase... 82.4 7e-16
sce:YNL112W DBP2; Dbp2p (EC:3.6.1.-); K12823 ATP-dependent RNA... 82.0 1e-15
bbo:BBOV_II004470 18.m06373; p68-like protein; K12823 ATP-depe... 81.6 1e-15
xla:379390 MGC53795; similar to DEAD/H (Asp-Glu-Ala-Asp/His) b... 81.3 2e-15
hsa:11325 DDX42, FLJ43179, RHELP, RNAHP, SF3b125; DEAD (Asp-Gl... 80.5 3e-15
mmu:72047 Ddx42, 1810047H21Rik, AW319508, AW556242, B430002H05... 80.5 3e-15
dre:394020 ddx41, MGC55896, wu:fb92e02, zgc:55896; DEAD (Asp-G... 80.1 3e-15
hsa:11056 DDX52, HUSSY19, ROK1; DEAD (Asp-Glu-Ala-Asp) box pol... 79.7 5e-15
tgo:TGME49_036650 DEAD/DEAH box helicase, putative (EC:5.99.1.... 79.7 5e-15
dre:503932 ddx42, im:7148194, zgc:111815; DEAD (Asp-Glu-Ala-As... 79.3 6e-15
hsa:1655 DDX5, DKFZp434E109, DKFZp686J01190, G17P1, HLR1, HUMP... 79.3 6e-15
xla:380261 ddx42, MGC54025; DEAD (Asp-Glu-Ala-Asp) box polypep... 78.6 9e-15
xla:444634 MGC84147 protein 78.6 1e-14
ath:AT3G01540 DRH1; DRH1 (DEAD BOX RNA HELICASE 1); ATP-depend... 78.2 1e-14
> tgo:TGME49_098020 DEAD-box ATP-dependent RNA helicase, putative
(EC:2.7.11.25); K12858 ATP-dependent RNA helicase DDX23/PRP28
[EC:3.6.4.13]
Length=1158
Score = 250 bits (638), Expect = 2e-66, Method: Compositional matrix adjust.
Identities = 130/171 (76%), Positives = 141/171 (82%), Gaps = 1/171 (0%)
Query 1 MDIREQRKANNFYDELVKRRQEYEQKQRQRGAEEAAAAAATAAAEAARASDKKKTKDLED 60
MDIREQRK NNFYDELVKRRQE+ QK AAAAAT A AAR + + ++ ED
Sbjct 637 MDIREQRKQNNFYDELVKRRQEH-QKAEASRGAAEAAAAATEAVRAARDAQASRLREKED 695
Query 61 EPELEGHWSTKKREEMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQA 120
+ GHW+TKKREEM +RDWRIFREDFEIY+KGGRVPPPIRTWAE+ LPWELIEAVK A
Sbjct 696 AEDNRGHWTTKKREEMNERDWRIFREDFEIYIKGGRVPPPIRTWAESALPWELIEAVKHA 755
Query 121 NYDRPTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPLT 171
NYDRPTPIQMQAIPIALE RDLIGIAETGSGKTAAFVLP+LTYVK LPPL
Sbjct 756 NYDRPTPIQMQAIPIALEQRDLIGIAETGSGKTAAFVLPMLTYVKGLPPLN 806
> tpv:TP04_0265 small nuclear ribonucleoprotein; K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=744
Score = 215 bits (547), Expect = 6e-56, Method: Compositional matrix adjust.
Identities = 107/171 (62%), Positives = 123/171 (71%), Gaps = 22/171 (12%)
Query 1 MDIREQRKANNFYDELVKRRQEYEQKQRQRGAEEAAAAAATAAAEAARASDKKKTKDLED 60
+D+REQRK NNFYDEL ++R E Q + E+
Sbjct 248 IDVREQRKKNNFYDELSRKRAELPQTTPKPPEPTPKH---------------------EE 286
Query 61 EPE-LEGHWSTKKREEMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQ 119
E E L HW+ KK EMT+RDWRIFREDFEIY+KGGRVPPPIRTWAE+ LPWEL+EA+K+
Sbjct 287 ESEVLSNHWTKKKLSEMTERDWRIFREDFEIYIKGGRVPPPIRTWAESPLPWELLEAIKK 346
Query 120 ANYDRPTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPL 170
A Y +PTPIQMQAIPIALEMRDLIGIA TGSGKTAAFVLP+LTYVK+LPPL
Sbjct 347 AGYIKPTPIQMQAIPIALEMRDLIGIAVTGSGKTAAFVLPMLTYVKKLPPL 397
> bbo:BBOV_II003280 18.m06276; DEAD box RNA helicase; K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=714
Score = 192 bits (489), Expect = 3e-49, Method: Compositional matrix adjust.
Identities = 94/171 (54%), Positives = 116/171 (67%), Gaps = 17/171 (9%)
Query 1 MDIREQRKANNFYDELVKRRQEYEQKQRQRGAEEAAAAAATAAAEAARASDKKKTKDLED 60
MD +EQRK +FYD+L K R + AA S++ +T+
Sbjct 213 MDPKEQRKHADFYDKLSKLR-----------------TGSDAAFSRDSRSNEDRTEPTSV 255
Query 61 EPELEGHWSTKKREEMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQA 120
+ +++ HWS K +E MT RDWRIFREDF+IY+KG RVPPP+RTWAE+ LP EL+ A+K A
Sbjct 256 DDDVDTHWSAKTKENMTQRDWRIFREDFDIYVKGTRVPPPMRTWAESNLPSELLRAIKDA 315
Query 121 NYDRPTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPLT 171
+ PTPIQMQAIPI L MRDLIG+AETGSGKT AFVLP+LTYVK LPPL
Sbjct 316 GFKSPTPIQMQAIPIGLGMRDLIGLAETGSGKTVAFVLPMLTYVKALPPLN 366
> pfa:PFE0925c snrnp protein, putative; K12858 ATP-dependent RNA
helicase DDX23/PRP28 [EC:3.6.4.13]
Length=1123
Score = 177 bits (448), Expect = 2e-44, Method: Composition-based stats.
Identities = 85/107 (79%), Positives = 96/107 (89%), Gaps = 0/107 (0%)
Query 65 EGHWSTKKREEMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDR 124
E HWS K REEMTDRDWRIFRED EIY+KGG VPPPIR W E+ L +L++A+K+A Y++
Sbjct 661 EKHWSQKSREEMTDRDWRIFREDNEIYIKGGVVPPPIRKWEESNLSNDLLKAIKKAKYEK 720
Query 125 PTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPLT 171
PTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLP+L+YVKQLPPLT
Sbjct 721 PTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPMLSYVKQLPPLT 767
Score = 34.7 bits (78), Expect = 0.14, Method: Composition-based stats.
Identities = 14/20 (70%), Positives = 18/20 (90%), Gaps = 0/20 (0%)
Query 1 MDIREQRKANNFYDELVKRR 20
+D+REQRK NNFYD+LV+ R
Sbjct 518 IDVREQRKKNNFYDKLVQNR 537
> ath:AT2G33730 DEAD box RNA helicase, putative; K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=733
Score = 158 bits (400), Expect = 8e-39, Method: Compositional matrix adjust.
Identities = 83/173 (47%), Positives = 114/173 (65%), Gaps = 14/173 (8%)
Query 1 MDIREQRKANNFYDELVKRRQEYEQKQRQRGAEEAAAAAATAAAEAAR--ASDKKKTKDL 58
MD REQ+K + E E + R + AAA+ R A+D + D+
Sbjct 222 MDRREQKKQAA--------KHEKEMRDEIRKKDGIVEKPEEAAAQRVREEAADTYDSFDM 273
Query 59 EDEPELEGHWSTKKREEMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVK 118
++ HWS K+ EEMT+RDWRIFREDF I KG R+P P+R+W E++L EL++AV+
Sbjct 274 R----VDRHWSDKRLEEMTERDWRIFREDFNISYKGSRIPRPMRSWEESKLTSELLKAVE 329
Query 119 QANYDRPTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPLT 171
+A Y +P+PIQM AIP+ L+ RD+IGIAETGSGKTAAFVLP+L Y+ +LPP++
Sbjct 330 RAGYKKPSPIQMAAIPLGLQQRDVIGIAETGSGKTAAFVLPMLAYISRLPPMS 382
> hsa:9416 DDX23, MGC8416, PRPF28, U5-100K, U5-100KD, prp28; DEAD
(Asp-Glu-Ala-Asp) box polypeptide 23 (EC:3.6.4.13); K12858
ATP-dependent RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=820
Score = 153 bits (386), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 76/171 (44%), Positives = 110/171 (64%), Gaps = 19/171 (11%)
Query 1 MDIREQ-RKANNFYDELVKRRQEYEQKQRQRGAEEAAAAAATAAAEAARASDKKKTKDLE 59
+D+++Q R+ + FY +L+++R+ E+K++ EEA R D+
Sbjct 307 IDLKQQKREQSRFYGDLMEKRRTLEEKEQ----EEARLRKLRKKEAKQRWDDR------- 355
Query 60 DEPELEGHWSTKKREEMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQ 119
HWS KK +EMTDRDWRIFRED+ I KGG++P PIR+W ++ LP ++E + +
Sbjct 356 -------HWSQKKLDEMTDRDWRIFREDYSITTKGGKIPNPIRSWKDSSLPPHILEVIDK 408
Query 120 ANYDRPTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPL 170
Y PTPIQ QAIPI L+ RD+IG+AETGSGKTAAF++PLL ++ LP +
Sbjct 409 CGYKEPTPIQRQAIPIGLQNRDIIGVAETGSGKTAAFLIPLLVWITTLPKI 459
> mmu:74351 Ddx23, 3110082M05Rik, 4921506D17Rik; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 23 (EC:3.6.1.-); K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=819
Score = 153 bits (386), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 76/171 (44%), Positives = 110/171 (64%), Gaps = 19/171 (11%)
Query 1 MDIREQ-RKANNFYDELVKRRQEYEQKQRQRGAEEAAAAAATAAAEAARASDKKKTKDLE 59
+D+++Q R+ + FY +L+++R+ E+K++ EEA R D+
Sbjct 306 IDLKQQKREQSRFYGDLMEKRRTLEEKEQ----EEARLRKLRKKEAKQRWDDR------- 354
Query 60 DEPELEGHWSTKKREEMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQ 119
HWS KK +EMTDRDWRIFRED+ I KGG++P PIR+W ++ LP ++E + +
Sbjct 355 -------HWSQKKLDEMTDRDWRIFREDYSITTKGGKIPNPIRSWKDSSLPPHILEVIDK 407
Query 120 ANYDRPTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPL 170
Y PTPIQ QAIPI L+ RD+IG+AETGSGKTAAF++PLL ++ LP +
Sbjct 408 CGYKEPTPIQRQAIPIGLQNRDIIGVAETGSGKTAAFLIPLLVWITTLPKI 458
> cel:F01F1.7 ddx-23; DEAD boX helicase homolog family member
(ddx-23); K12858 ATP-dependent RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=730
Score = 153 bits (386), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 78/170 (45%), Positives = 104/170 (61%), Gaps = 19/170 (11%)
Query 2 DIREQRK-ANNFYDELVKRRQEYEQKQRQRGAEEAAAAAATAAAEAARASDKKKTKDLED 60
D+ Q+K N+FY E+++ R+ ++K+++ E A R
Sbjct 218 DVNAQKKEKNSFYQEMMENRRTVDEKEQEMHRLEKELKKEKKVAHDDR------------ 265
Query 61 EPELEGHWSTKKREEMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQA 120
HW K+ EM+DRDWRIFREDF I +KGGRVP P+R W EA P E+ +AVK+
Sbjct 266 ------HWRMKELSEMSDRDWRIFREDFNISIKGGRVPRPLRNWEEAGFPDEVYQAVKEI 319
Query 121 NYDRPTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPL 170
Y PTPIQ QAIPI L+ RD+IG+AETGSGKTAAF+LPLL ++ LP +
Sbjct 320 GYLEPTPIQRQAIPIGLQNRDVIGVAETGSGKTAAFLLPLLVWITSLPKM 369
> dre:334283 ddx23, wu:fi39b12, zgc:63742; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 23 (EC:3.6.1.-); K12858 ATP-dependent RNA
helicase DDX23/PRP28 [EC:3.6.4.13]
Length=807
Score = 150 bits (379), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 76/171 (44%), Positives = 107/171 (62%), Gaps = 19/171 (11%)
Query 1 MDIREQ-RKANNFYDELVKRRQEYEQKQRQRGAEEAAAAAATAAAEAARASDKKKTKDLE 59
+D+++Q R + FY +L+++R+ E+K++ EE R D+
Sbjct 294 IDLKQQKRDQSRFYGDLMEKRRTNEEKEQ----EEQRLKKVRKKEAKQRWDDR------- 342
Query 60 DEPELEGHWSTKKREEMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQ 119
HWS KK +EMTDRDWRIFRED+ I KGG++P PIR W E LP ++E +++
Sbjct 343 -------HWSQKKLDEMTDRDWRIFREDYSITTKGGKIPNPIRNWKEYSLPPHILEVIEK 395
Query 120 ANYDRPTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPL 170
Y PTPIQ QAIPI L+ RD+IG+AETGSGKTAAF++PLL ++ LP +
Sbjct 396 CGYKDPTPIQRQAIPIGLQNRDIIGVAETGSGKTAAFLIPLLVWITTLPKI 446
> cpv:cgd3_3690 U5 snRNP 100 kD protein ; K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=529
Score = 118 bits (296), Expect = 8e-27, Method: Composition-based stats.
Identities = 52/96 (54%), Positives = 71/96 (73%), Gaps = 0/96 (0%)
Query 75 EMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIP 134
+MT+RDW+IFRED+ I ++G VP PIR W + + E ++ Y++PTPIQMQ IP
Sbjct 115 DMTERDWKIFREDYSINVRGKDVPNPIRNWKDCHVLEIQTELIRNIGYEKPTPIQMQCIP 174
Query 135 IALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPL 170
I L++RD+IGIAETGSGKT AF++PL++YV P L
Sbjct 175 IGLKLRDMIGIAETGSGKTIAFLIPLISYVGNKPIL 210
> ath:AT1G28180 ATP binding / ATP-dependent helicase/ helicase/
nucleic acid binding
Length=614
Score = 108 bits (271), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 50/104 (48%), Positives = 63/104 (60%), Gaps = 25/104 (24%)
Query 67 HWSTKKREEMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPT 126
HWS KK EEM +RDWRIF+EDF I +G ++P P+R W E
Sbjct 192 HWSEKKLEEMNERDWRIFKEDFNISYRGSKIPHPMRNWEE-------------------- 231
Query 127 PIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPL 170
IP+ LE RD+IGI+ TGSGKTAAFVLP+L Y+ +LPP+
Sbjct 232 -----TIPLGLEQRDVIGISATGSGKTAAFVLPMLAYISRLPPM 270
> sce:YDR243C PRP28; Prp28p (EC:3.6.1.-); K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=588
Score = 100 bits (250), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 49/108 (45%), Positives = 65/108 (60%), Gaps = 5/108 (4%)
Query 67 HWSTKKREEMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAE-LPWELIEAVKQ-ANYDR 124
HW+ K EM +RDWRI +ED+ I KGG V P+R W E +P +L+ + Q +
Sbjct 137 HWTEKSLHEMNERDWRILKEDYAIVTKGGTVENPLRNWEELNIIPRDLLRVIIQELRFPS 196
Query 125 PTPIQMQAIPIALEM---RDLIGIAETGSGKTAAFVLPLLTYVKQLPP 169
PTPIQ IP M RD +G+A TGSGKT AFV+P+L + + PP
Sbjct 197 PTPIQRITIPNVCNMKQYRDFLGVASTGSGKTLAFVIPILIKMSRSPP 244
> ath:AT1G20920 DEAD box RNA helicase, putative; K12811 ATP-dependent
RNA helicase DDX46/PRP5 [EC:3.6.4.13]
Length=828
Score = 98.2 bits (243), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 41/95 (43%), Positives = 65/95 (68%), Gaps = 0/95 (0%)
Query 76 MTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIPI 135
MT + +R++ E+ + G VP PI+ W + L ++++ +K+ NY++P PIQ QA+PI
Sbjct 165 MTQEEVNTYRKELELKVHGKDVPRPIKFWHQTGLTSKILDTMKKLNYEKPMPIQTQALPI 224
Query 136 ALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPL 170
+ RD IG+A+TGSGKT FVLP+L ++K PP+
Sbjct 225 IMSGRDCIGVAKTGSGKTLGFVLPMLRHIKDQPPV 259
> ath:AT3G09620 DEAD/DEAH box helicase, putative
Length=989
Score = 97.1 bits (240), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 41/95 (43%), Positives = 64/95 (67%), Gaps = 0/95 (0%)
Query 76 MTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIPI 135
MT +R++ E+ + G VP PI+ W + L ++++ +K+ NY++P PIQ QA+PI
Sbjct 370 MTQDAVNAYRKELELKVHGKDVPRPIQFWHQTGLTSKILDTLKKLNYEKPMPIQAQALPI 429
Query 136 ALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPL 170
+ RD IG+A+TGSGKT FVLP+L ++K PP+
Sbjct 430 IMSGRDCIGVAKTGSGKTLGFVLPMLRHIKDQPPV 464
> hsa:9879 DDX46, FLJ25329, KIAA0801, MGC9936, PRPF5, Prp5; DEAD
(Asp-Glu-Ala-Asp) box polypeptide 46 (EC:3.6.4.13); K12811
ATP-dependent RNA helicase DDX46/PRP5 [EC:3.6.4.13]
Length=1031
Score = 93.2 bits (230), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 39/90 (43%), Positives = 62/90 (68%), Gaps = 1/90 (1%)
Query 76 MTDRDWRIFREDFE-IYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIP 134
M+ + +FR + E I +KG P PI++W + + +++ ++K+ Y++PTPIQ QAIP
Sbjct 345 MSQEEVNVFRLEMEGITVKGKGCPKPIKSWVQCGISMKILNSLKKHGYEKPTPIQTQAIP 404
Query 135 IALEMRDLIGIAETGSGKTAAFVLPLLTYV 164
+ RDLIGIA+TGSGKT AF+LP+ ++
Sbjct 405 AIMSGRDLIGIAKTGSGKTIAFLLPMFRHI 434
> mmu:212880 Ddx46, 2200005K02Rik, 8430438J23Rik, AI325430, AI957095,
MGC116676, MGC31579, mKIAA0801; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 46 (EC:3.6.4.13); K12811 ATP-dependent RNA
helicase DDX46/PRP5 [EC:3.6.4.13]
Length=1031
Score = 93.2 bits (230), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 39/90 (43%), Positives = 62/90 (68%), Gaps = 1/90 (1%)
Query 76 MTDRDWRIFREDFE-IYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIP 134
M+ + +FR + E I +KG P PI++W + + +++ ++K+ Y++PTPIQ QAIP
Sbjct 345 MSQEEVNVFRLEMEGITVKGKGCPKPIKSWVQCGISMKILNSLKKHGYEKPTPIQTQAIP 404
Query 135 IALEMRDLIGIAETGSGKTAAFVLPLLTYV 164
+ RDLIGIA+TGSGKT AF+LP+ ++
Sbjct 405 AIMSGRDLIGIAKTGSGKTIAFLLPMFRHI 434
> dre:321948 ddx46, fb39a03, wu:fb39a03; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 46 (EC:3.6.4.13); K12811 ATP-dependent RNA
helicase DDX46/PRP5 [EC:3.6.4.13]
Length=1018
Score = 92.8 bits (229), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 41/88 (46%), Positives = 61/88 (69%), Gaps = 1/88 (1%)
Query 84 FREDFE-IYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIPIALEMRDL 142
+R + E I +KG P PI+TW + + +++ A+K+ NY++PTPIQ QAIP + RDL
Sbjct 321 YRLELEGISVKGKGCPKPIKTWVQCGISMKVLNALKKHNYEKPTPIQAQAIPAIMSGRDL 380
Query 143 IGIAETGSGKTAAFVLPLLTYVKQLPPL 170
IGIA+TGSGKT AF+LP+ ++ P+
Sbjct 381 IGIAKTGSGKTIAFLLPMFRHILDQRPV 408
> cel:F53H1.1 hypothetical protein; K12811 ATP-dependent RNA helicase
DDX46/PRP5 [EC:3.6.4.13]
Length=970
Score = 89.7 bits (221), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 41/96 (42%), Positives = 63/96 (65%), Gaps = 1/96 (1%)
Query 76 MTDRDWRIFREDFE-IYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIP 134
MT + + +RE+ + I +KG P PI+TWA+ + +++ +K+ Y +PT IQ QAIP
Sbjct 277 MTKAEVKAYREELDSITVKGIDCPKPIKTWAQCGVNLKMMNVLKKFEYSKPTSIQAQAIP 336
Query 135 IALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPL 170
+ RD+IGIA+TGSGKT AF+LP+ ++ P L
Sbjct 337 SIMSGRDVIGIAKTGSGKTLAFLLPMFRHILDQPEL 372
> ath:AT5G63120 ethylene-responsive DEAD box RNA helicase, putative
(RH30)
Length=484
Score = 89.4 bits (220), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 43/97 (44%), Positives = 66/97 (68%), Gaps = 0/97 (0%)
Query 74 EEMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAI 133
+ MT++D ++R + +I ++G VP P++ + +A P ++EA+ + + PTPIQ Q
Sbjct 137 QAMTEQDVAMYRTERDISVEGRDVPKPMKMFQDANFPDNILEAIAKLGFTEPTPIQAQGW 196
Query 134 PIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPL 170
P+AL+ RDLIGIAETGSGKT A++LP L +V P L
Sbjct 197 PMALKGRDLIGIAETGSGKTLAYLLPALVHVSAQPRL 233
> dre:556764 similar to Probable RNA-dependent helicase p72 (DEAD-box
protein p72) (DEAD-box protein 17); K13178 ATP-dependent
RNA helicase DDX17 [EC:3.6.4.13]
Length=671
Score = 89.0 bits (219), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 43/121 (35%), Positives = 71/121 (58%), Gaps = 2/121 (1%)
Query 52 KKKTKDLEDEPELEGHWSTKKRE--EMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAEL 109
+KK DL+ P+ E ++ + E M+ D +R EI ++G P P+ + +A+
Sbjct 43 RKKKWDLDQLPKFEKNFYNENPEVHHMSQYDVEEYRRKREITVRGSGCPKPVTNFHQAQF 102
Query 110 PWELIEAVKQANYDRPTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPP 169
P +++ + Q N+ PT IQ Q P+AL RD++GIA+TGSGKT A++LP + ++ P
Sbjct 103 PQYVMDVLLQQNFKEPTAIQAQGFPLALSGRDMVGIAQTGSGKTLAYLLPAIVHINHQPY 162
Query 170 L 170
L
Sbjct 163 L 163
> xla:398649 ddx17, MGC80019; DEAD (Asp-Glu-Ala-Asp) box polypeptide
17; K13178 ATP-dependent RNA helicase DDX17 [EC:3.6.4.13]
Length=610
Score = 89.0 bits (219), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 44/121 (36%), Positives = 69/121 (57%), Gaps = 2/121 (1%)
Query 52 KKKTKDLEDEPELEGHWSTKKRE--EMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAEL 109
+KK DL + P+ E ++ T+ E MT D R EI ++G P P+ + +A
Sbjct 30 RKKRWDLNELPKFEKNFYTEHPEVARMTQHDVEELRRKKEITIRGVNCPKPLYAFHQANF 89
Query 110 PWELIEAVKQANYDRPTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPP 169
P +++ + + PTPIQ Q P+AL RD++GIA+TGSGKT A++LP + ++ P
Sbjct 90 PQYVLDVLLDQRFKEPTPIQCQGFPLALSGRDMVGIAQTGSGKTLAYLLPAMVHINHQPY 149
Query 170 L 170
L
Sbjct 150 L 150
> ath:AT2G47330 DEAD/DEAH box helicase, putative; K12835 ATP-dependent
RNA helicase DDX42 [EC:3.6.4.13]
Length=760
Score = 89.0 bits (219), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 40/95 (42%), Positives = 64/95 (67%), Gaps = 0/95 (0%)
Query 76 MTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIPI 135
MT+++ +R+ I + G V P++T+ + +++ A+K+ Y++PT IQ QA+PI
Sbjct 202 MTEQETTDYRQRLGIRVSGFDVHRPVKTFEDCGFSSQIMSAIKKQAYEKPTAIQCQALPI 261
Query 136 ALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPL 170
L RD+IGIA+TGSGKTAAFVLP++ ++ P L
Sbjct 262 VLSGRDVIGIAKTGSGKTAAFVLPMIVHIMDQPEL 296
> cel:H27M09.1 hypothetical protein; K13116 ATP-dependent RNA
helicase DDX41 [EC:3.6.4.13]
Length=630
Score = 87.0 bits (214), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 44/100 (44%), Positives = 60/100 (60%), Gaps = 6/100 (6%)
Query 66 GHWSTKKREEMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAV-KQANYDR 124
GH + +E D+ I R+ I +G +PPPI ++ E + P L+E + KQ
Sbjct 158 GHIRRQSQE-----DYEIQRKRLGISCEGDHIPPPIGSFLEMKFPKSLLEFMQKQKGIVT 212
Query 125 PTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYV 164
PT IQ+Q IP+AL RD+IGIA TGSGKT FVLPL+ +
Sbjct 213 PTAIQIQGIPVALSGRDMIGIASTGSGKTMTFVLPLVMFC 252
> ath:AT1G55150 DEAD box RNA helicase, putative (RH20); K12823
ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=501
Score = 85.5 bits (210), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 41/95 (43%), Positives = 64/95 (67%), Gaps = 0/95 (0%)
Query 76 MTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIPI 135
MTD + +R+ EI ++G +P P++++ + P ++E VK+A + PTPIQ Q P+
Sbjct 73 MTDTEVEEYRKLREITVEGKDIPKPVKSFRDVGFPDYVLEEVKKAGFTEPTPIQSQGWPM 132
Query 136 ALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPL 170
A++ RDLIGIAETGSGKT +++LP + +V P L
Sbjct 133 AMKGRDLIGIAETGSGKTLSYLLPAIVHVNAQPML 167
> ath:AT3G02065 DEAD/DEAH box helicase family protein
Length=505
Score = 85.1 bits (209), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 38/88 (43%), Positives = 60/88 (68%), Gaps = 2/88 (2%)
Query 77 TDRDWRIFREDFEIYLKG--GRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIP 134
+ D ++ R +I+++G VPPP+ T+ LP +L+ ++ A YD PTPIQMQAIP
Sbjct 83 SSHDAQLLRRKLDIHVQGQGSAVPPPVLTFTSCGLPPKLLLNLETAGYDFPTPIQMQAIP 142
Query 135 IALEMRDLIGIAETGSGKTAAFVLPLLT 162
AL + L+ A+TGSGKTA+F++P+++
Sbjct 143 AALTGKSLLASADTGSGKTASFLVPIIS 170
> hsa:51428 DDX41, ABS, MGC8828; DEAD (Asp-Glu-Ala-Asp) box polypeptide
41 (EC:3.6.4.13); K13116 ATP-dependent RNA helicase
DDX41 [EC:3.6.4.13]
Length=622
Score = 84.3 bits (207), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 39/95 (41%), Positives = 60/95 (63%), Gaps = 5/95 (5%)
Query 78 DRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIPIAL 137
+R R+ R+ + I ++G +PPPI+++ E + P ++ +K+ PTPIQ+Q IP L
Sbjct 158 ERHERV-RKKYHILVEGDGIPPPIKSFKEMKFPAAILRGLKKKGIHHPTPIQIQGIPTIL 216
Query 138 EMRDLIGIAETGSGKTAAFVLPLLTYV----KQLP 168
RD+IGIA TGSGKT F LP++ + K+LP
Sbjct 217 SGRDMIGIAFTGSGKTLVFTLPVIMFCLEQEKRLP 251
> cpv:cgd7_4600 abstrakt protein SF II helicase + Znknuckle C2HC
(PA) ; K13116 ATP-dependent RNA helicase DDX41 [EC:3.6.4.13]
Length=570
Score = 84.0 bits (206), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 45/132 (34%), Positives = 68/132 (51%), Gaps = 1/132 (0%)
Query 33 EEAAAAAATAAAEAARASDKKKTKDLEDEPELEGHWST-KKREEMTDRDWRIFREDFEIY 91
EE AA + A + + K + E W KK ++ + + R I
Sbjct 38 EEKLLAAVNQSYNAPLKAVHEIAKGITFSKREETSWRVPKKYSSLSASECQDLRSRLLIV 97
Query 92 LKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIPIALEMRDLIGIAETGSG 151
+ G VPPPI ++ + P E+++A+ +P+ IQMQ +PI L RDLIG+A TGSG
Sbjct 98 VNGSDVPPPILSFKDMGFPQEILDALASKGISKPSQIQMQGLPIILMGRDLIGLAFTGSG 157
Query 152 KTAAFVLPLLTY 163
KT FVLP++ +
Sbjct 158 KTIVFVLPMIMF 169
> ath:AT5G51280 DEAD-box protein abstrakt, putative; K13116 ATP-dependent
RNA helicase DDX41 [EC:3.6.4.13]
Length=591
Score = 84.0 bits (206), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 39/106 (36%), Positives = 63/106 (59%), Gaps = 0/106 (0%)
Query 61 EPELEGHWSTKKREEMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQA 120
EP L G +M+ + + R+ + I + G +PPPI+ + + + P +++ +K+
Sbjct 105 EPLLTGWKPPLHIRKMSSKQRDLIRKQWHIIVNGDDIPPPIKNFKDMKFPRPVLDTLKEK 164
Query 121 NYDRPTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQ 166
+PTPIQ+Q +P+ L RD+IGIA TGSGKT FVLP++ Q
Sbjct 165 GIVQPTPIQVQGLPVILAGRDMIGIAFTGSGKTLVFVLPMIMIALQ 210
> mmu:72935 Ddx41, 2900024F02Rik, AA958953, ABS, AI324246; DEAD
(Asp-Glu-Ala-Asp) box polypeptide 41 (EC:3.6.4.13); K13116
ATP-dependent RNA helicase DDX41 [EC:3.6.4.13]
Length=622
Score = 83.6 bits (205), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 37/88 (42%), Positives = 56/88 (63%), Gaps = 4/88 (4%)
Query 85 REDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIPIALEMRDLIG 144
R+ + I ++G +PPPI+++ E + P ++ +K+ PTPIQ+Q IP L RD+IG
Sbjct 164 RKKYHILVEGDGIPPPIKSFKEMKFPAAILRGLKKKGILHPTPIQIQGIPTILSGRDMIG 223
Query 145 IAETGSGKTAAFVLPLLTYV----KQLP 168
IA TGSGKT F LP++ + K+LP
Sbjct 224 IAFTGSGKTLVFTLPVIMFCLEQEKRLP 251
> xla:399382 ddx5, MGC81559; DEAD (Asp-Glu-Ala-Asp) box polypeptide
5; K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=608
Score = 83.6 bits (205), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 42/120 (35%), Positives = 72/120 (60%), Gaps = 2/120 (1%)
Query 53 KKTKDLEDEPELEGHWSTKKREEM--TDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELP 110
KK +L++ P+ E ++ + + + T ++ +R EI ++G P PI + EA P
Sbjct 41 KKKWNLDELPKFEKNFYQEHPDVVRRTPQECDQYRRSKEITVRGINCPKPILNFNEASFP 100
Query 111 WELIEAVKQANYDRPTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPL 170
++EA+K+ N+ PTPIQ Q P+AL D++G+A TGSGKT +++LP + ++ P L
Sbjct 101 ANVMEAIKRQNFTEPTPIQGQGWPVALSGLDMVGVAMTGSGKTLSYLLPGIVHINHQPFL 160
> ath:AT4G33370 DEAD-box protein abstrakt, putative
Length=542
Score = 83.2 bits (204), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 56/92 (60%), Gaps = 0/92 (0%)
Query 75 EMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIP 134
+M+ + + R+ + I + G +PPPI+ + + + P L+ +K PTPIQ+Q +P
Sbjct 70 KMSTKQMDLIRKQWHITVNGEDIPPPIKNFMDMKFPSPLLRMLKDKGIMHPTPIQVQGLP 129
Query 135 IALEMRDLIGIAETGSGKTAAFVLPLLTYVKQ 166
+ L RD+IGIA TGSGKT FVLP++ Q
Sbjct 130 VVLSGRDMIGIAFTGSGKTLVFVLPMIILALQ 161
> ath:AT2G42520 DEAD box RNA helicase, putative; K11594 ATP-dependent
RNA helicase [EC:3.6.4.13]
Length=633
Score = 82.8 bits (203), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 38/79 (48%), Positives = 52/79 (65%), Gaps = 0/79 (0%)
Query 86 EDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIPIALEMRDLIGI 145
ED I G VPPP+ T+AE +L L +++ Y +PTP+Q AIPI LE RDL+
Sbjct 143 EDIPIETSGDNVPPPVNTFAEIDLGEALNLNIRRCKYVKPTPVQRHAIPILLEGRDLMAC 202
Query 146 AETGSGKTAAFVLPLLTYV 164
A+TGSGKTAAF P+++ +
Sbjct 203 AQTGSGKTAAFCFPIISGI 221
> tpv:TP03_0532 ATP-dependent RNA helicase; K12811 ATP-dependent
RNA helicase DDX46/PRP5 [EC:3.6.4.13]
Length=894
Score = 82.8 bits (203), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 40/97 (41%), Positives = 63/97 (64%), Gaps = 1/97 (1%)
Query 75 EMTDRDWRIFRE-DFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAI 133
M + + FR+ + I + G + P PI ++++ LP +++ +++ Y++P PIQMQ I
Sbjct 389 NMGEHEVDAFRKANGNIRVYGKKCPRPISSFSQCGLPDPILKILEKREYEKPFPIQMQCI 448
Query 134 PIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPL 170
P + RD+IGIAETGSGKT AF+LP + +V PPL
Sbjct 449 PALMCGRDVIGIAETGSGKTLAFLLPGIRHVLDQPPL 485
> mmu:67040 Ddx17, 2610007K22Rik, A430025E01Rik, AI047725, C80929,
Gm926, MGC79147, p72; DEAD (Asp-Glu-Ala-Asp) box polypeptide
17 (EC:3.6.4.13); K13178 ATP-dependent RNA helicase DDX17
[EC:3.6.4.13]
Length=650
Score = 82.8 bits (203), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 43/122 (35%), Positives = 70/122 (57%), Gaps = 3/122 (2%)
Query 52 KKKTKDLEDEPELEGHWSTKKRE--EMTDRDWRIFREDFEIYLKGGRVPP-PIRTWAEAE 108
+KK DL + P+ E ++ + E +T + R EI ++GG V P P+ + A
Sbjct 39 RKKKWDLSELPKFEKNFYVEHPEVARLTPYEVDELRRKKEITVRGGDVCPKPVFAFHHAN 98
Query 109 LPWELIEAVKQANYDRPTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLP 168
P +++ + ++ PTPIQ Q P+AL RD++GIA+TGSGKT A++LP + ++ P
Sbjct 99 FPQYVMDVLMDQHFTEPTPIQCQGFPLALSGRDMVGIAQTGSGKTLAYLLPAIVHINHQP 158
Query 169 PL 170
L
Sbjct 159 YL 160
> ath:AT3G58510 DEAD box RNA helicase, putative (RH11); K11594
ATP-dependent RNA helicase [EC:3.6.4.13]
Length=612
Score = 82.8 bits (203), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 37/79 (46%), Positives = 52/79 (65%), Gaps = 0/79 (0%)
Query 86 EDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIPIALEMRDLIGI 145
ED + GG VPPP+ T+A+ +L L +++ Y RPTP+Q AIPI L RDL+
Sbjct 135 EDIPVETSGGDVPPPVNTFADIDLGDALNLNIRRCKYVRPTPVQRHAIPILLAERDLMAC 194
Query 146 AETGSGKTAAFVLPLLTYV 164
A+TGSGKTAAF P+++ +
Sbjct 195 AQTGSGKTAAFCFPIISGI 213
> hsa:10521 DDX17, DKFZp761H2016, P72, RH70; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 17 (EC:3.6.4.13); K13178 ATP-dependent
RNA helicase DDX17 [EC:3.6.4.13]
Length=731
Score = 82.4 bits (202), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 43/122 (35%), Positives = 70/122 (57%), Gaps = 3/122 (2%)
Query 52 KKKTKDLEDEPELEGHWSTKKRE--EMTDRDWRIFREDFEIYLKGGRVPP-PIRTWAEAE 108
+KK DL + P+ E ++ + E +T + R EI ++GG V P P+ + A
Sbjct 118 RKKKWDLSELPKFEKNFYVEHPEVARLTPYEVDELRRKKEITVRGGDVCPKPVFAFHHAN 177
Query 109 LPWELIEAVKQANYDRPTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLP 168
P +++ + ++ PTPIQ Q P+AL RD++GIA+TGSGKT A++LP + ++ P
Sbjct 178 FPQYVMDVLMDQHFTEPTPIQCQGFPLALSGRDMVGIAQTGSGKTLAYLLPAIVHINHQP 237
Query 169 PL 170
L
Sbjct 238 YL 239
> pfa:PFE1390w RNA helicase-1; K13116 ATP-dependent RNA helicase
DDX41 [EC:3.6.4.13]
Length=665
Score = 82.4 bits (202), Expect = 7e-16, Method: Composition-based stats.
Identities = 49/147 (33%), Positives = 80/147 (54%), Gaps = 1/147 (0%)
Query 16 LVKRRQEYEQKQRQRGAEEAAAAAATAAAEAARASDKKKTKDLEDEPELEGHWSTKKREE 75
L K+ E ++ + R EE A + A A S K++ K + + +E W K+ +
Sbjct 130 LEKKNDEIDETEEIRKREEKLLAQVSKALNAPLQSVKERAKGIVYKENVESIWKLPKKYK 189
Query 76 MTDRDW-RIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIP 134
+ + + R F I + G +P PI+ + + + P +++ +++ N +PT IQMQ +P
Sbjct 190 LLKKSYVEKIRRIFYIDVNGDDIPAPIKNFKDMKFPKAILKGLRKKNIKKPTQIQMQGLP 249
Query 135 IALEMRDLIGIAETGSGKTAAFVLPLL 161
L RD+IGIA TGSGKT FVLPL+
Sbjct 250 SILLGRDIIGIAFTGSGKTIVFVLPLI 276
> sce:YNL112W DBP2; Dbp2p (EC:3.6.1.-); K12823 ATP-dependent RNA
helicase DDX5/DBP2 [EC:3.6.4.13]
Length=546
Score = 82.0 bits (201), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 41/104 (39%), Positives = 62/104 (59%), Gaps = 3/104 (2%)
Query 67 HWSTKKREEMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPT 126
H S + R +D + FR++ E+ + G +P PI T+ EA P ++ VK +D+PT
Sbjct 81 HESVRDR---SDSEIAQFRKENEMTISGHDIPKPITTFDEAGFPDYVLNEVKAEGFDKPT 137
Query 127 PIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPL 170
IQ Q P+AL RD++GIA TGSGKT ++ LP + ++ P L
Sbjct 138 GIQCQGWPMALSGRDMVGIAATGSGKTLSYCLPGIVHINAQPLL 181
> bbo:BBOV_II004470 18.m06373; p68-like protein; K12823 ATP-dependent
RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=529
Score = 81.6 bits (200), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 40/91 (43%), Positives = 59/91 (64%), Gaps = 1/91 (1%)
Query 76 MTDRDWRIFREDFEIYLKGGR-VPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIP 134
M+ D R++ EI + GR VP P+ ++ P +++A++ A + PTPIQ+Q P
Sbjct 81 MSSADVDRVRKEREITIIAGRDVPKPVVSFEHTSFPDYILKAIRAAGFTAPTPIQVQGWP 140
Query 135 IALEMRDLIGIAETGSGKTAAFVLPLLTYVK 165
IAL RD+IGIAETGSGKT AF+LP + ++
Sbjct 141 IALSGRDVIGIAETGSGKTLAFLLPAVVHIN 171
> xla:379390 MGC53795; similar to DEAD/H (Asp-Glu-Ala-Asp/His)
box polypeptide 5 (RNA helicase, 68kDa); K12823 ATP-dependent
RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=607
Score = 81.3 bits (199), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 42/124 (33%), Positives = 70/124 (56%), Gaps = 10/124 (8%)
Query 53 KKTKDLEDEPELEGHWSTKKREEMTDRDWRI------FREDFEIYLKGGRVPPPIRTWAE 106
KK +L++ P+ E ++ +E+ D R +R EI ++G P P+ + E
Sbjct 39 KKKWNLDELPKFEKNFY----QELPDVSRRTPQECDQYRRSKEITVRGLNCPKPVLNFHE 94
Query 107 AELPWELIEAVKQANYDRPTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQ 166
A P ++E +K+ N+ PTPIQ Q P+AL D++G+A TGSGKT +++LP + ++
Sbjct 95 ASFPANVMEVIKRLNFTEPTPIQGQGWPVALSGLDMVGVAMTGSGKTLSYLLPGIVHINH 154
Query 167 LPPL 170
P L
Sbjct 155 QPFL 158
> hsa:11325 DDX42, FLJ43179, RHELP, RNAHP, SF3b125; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 42 (EC:3.6.4.13); K12835 ATP-dependent
RNA helicase DDX42 [EC:3.6.4.13]
Length=938
Score = 80.5 bits (197), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 41/115 (35%), Positives = 66/115 (57%), Gaps = 5/115 (4%)
Query 60 DEPELEGHWSTKKRE--EMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAV 117
D P E ++ + E +T + R + + G P P ++A +L+ +
Sbjct 209 DYPPFEKNFYNEHEEITNLTPQQLIDLRHKLNLRVSGAAPPRPGSSFAHFGFDEQLMHQI 268
Query 118 KQANYDRPTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYV---KQLPP 169
+++ Y +PTPIQ Q +P+AL RD+IGIA+TGSGKTAAF+ P+L ++ K+L P
Sbjct 269 RKSEYTQPTPIQCQGVPVALSGRDMIGIAKTGSGKTAAFIWPMLIHIMDQKELEP 323
> mmu:72047 Ddx42, 1810047H21Rik, AW319508, AW556242, B430002H05Rik,
RHELP, RNAHP, SF3b125; DEAD (Asp-Glu-Ala-Asp) box polypeptide
42 (EC:3.6.4.13); K12835 ATP-dependent RNA helicase
DDX42 [EC:3.6.4.13]
Length=929
Score = 80.5 bits (197), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 41/115 (35%), Positives = 66/115 (57%), Gaps = 5/115 (4%)
Query 60 DEPELEGHWSTKKRE--EMTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAV 117
D P E ++ + E +T + R + + G P P ++A +L+ +
Sbjct 209 DYPPFEKNFYNEHEEITNLTPQQLIDLRHKLNLRVSGAAPPRPGSSFAHFGFDEQLMHQI 268
Query 118 KQANYDRPTPIQMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYV---KQLPP 169
+++ Y +PTPIQ Q +P+AL RD+IGIA+TGSGKTAAF+ P+L ++ K+L P
Sbjct 269 RKSEYTQPTPIQCQGVPVALSGRDMIGIAKTGSGKTAAFIWPMLIHIMDQKELEP 323
> dre:394020 ddx41, MGC55896, wu:fb92e02, zgc:55896; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 41 (EC:3.6.4.13); K13116 ATP-dependent
RNA helicase DDX41 [EC:3.6.4.13]
Length=306
Score = 80.1 bits (196), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 36/88 (40%), Positives = 56/88 (63%), Gaps = 4/88 (4%)
Query 85 REDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIPIALEMRDLIG 144
R+ + I ++G +P PI+++ E + P +++ +K+ PTPIQ+Q IP L RD+IG
Sbjct 155 RKKYHILVEGEGIPAPIKSFREMKFPQAILKGLKKKGIVHPTPIQIQGIPTILSGRDMIG 214
Query 145 IAETGSGKTAAFVLPLLTYV----KQLP 168
IA TGSGKT F LP++ + K+LP
Sbjct 215 IAFTGSGKTLVFTLPIIMFCLEQEKRLP 242
> hsa:11056 DDX52, HUSSY19, ROK1; DEAD (Asp-Glu-Ala-Asp) box polypeptide
52 (EC:3.6.4.13); K14779 ATP-dependent RNA helicase
DDX52/ROK1 [EC:3.6.4.13]
Length=599
Score = 79.7 bits (195), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 50/158 (31%), Positives = 85/158 (53%), Gaps = 9/158 (5%)
Query 14 DELVKRRQEYEQKQRQRGAEEAAAAAATAAAEAARASDKKKTKDLEDEP-ELEGHWSTKK 72
+ L +R++E +K+R+ E A+ A + + + K +ED+ + E ++ K
Sbjct 76 ESLTERKREQSKKKRKTMTSEIASQEEGATIQWMSSVEAK----IEDKKVQRESKLTSGK 131
Query 73 REEMTDRDWRIFREDFEIYLKGGRVPPPIRTWA----EAELPWELIEAVKQANYDRPTPI 128
E + R +I+++G +P PI T+ E ++ L++ + A + PTPI
Sbjct 132 LENLRKEKINFLRNKHKIHVQGTDLPDPIATFQQLDQEYKINSRLLQNILDAGFQMPTPI 191
Query 129 QMQAIPIALEMRDLIGIAETGSGKTAAFVLPLLTYVKQ 166
QMQAIP+ L R+L+ A TGSGKT AF +P+L +KQ
Sbjct 192 QMQAIPVMLHGRELLASAPTGSGKTLAFSIPILMQLKQ 229
> tgo:TGME49_036650 DEAD/DEAH box helicase, putative (EC:5.99.1.3);
K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=550
Score = 79.7 bits (195), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 38/90 (42%), Positives = 54/90 (60%), Gaps = 0/90 (0%)
Query 82 RIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIPIALEMRD 141
RI R + + G VP P+ T+ P +++ + Q + +PT IQ+Q PIAL RD
Sbjct 109 RIRRANEITIVHGHNVPKPVPTFEYTSFPSYILDVINQTGFQKPTAIQVQGWPIALSGRD 168
Query 142 LIGIAETGSGKTAAFVLPLLTYVKQLPPLT 171
+IGIAETGSGKT AF+LP + ++ P L
Sbjct 169 MIGIAETGSGKTLAFLLPAIVHINAQPYLN 198
> dre:503932 ddx42, im:7148194, zgc:111815; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 42 (EC:3.6.1.-); K12835 ATP-dependent RNA
helicase DDX42 [EC:3.6.4.13]
Length=908
Score = 79.3 bits (194), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 37/89 (41%), Positives = 55/89 (61%), Gaps = 3/89 (3%)
Query 84 FREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIPIALEMRDLI 143
R + + G P P ++A +L+ ++++ Y +PTPIQ Q +PIAL RD I
Sbjct 237 LRRKLNLKVSGAAPPKPATSFAHFGFDEQLMHQIRKSEYTQPTPIQCQGVPIALSGRDAI 296
Query 144 GIAETGSGKTAAFVLPLLTYV---KQLPP 169
GIA+TGSGKTAAF+ P+L ++ K+L P
Sbjct 297 GIAKTGSGKTAAFIWPILVHIMDQKELEP 325
> hsa:1655 DDX5, DKFZp434E109, DKFZp686J01190, G17P1, HLR1, HUMP68,
p68; DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (EC:3.6.4.13);
K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=614
Score = 79.3 bits (194), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 44/155 (28%), Positives = 78/155 (50%), Gaps = 12/155 (7%)
Query 28 RQRGAEEAAAAAATAAAEAARASDKK----------KTKDLEDEPELEGHWSTKKRE--E 75
R RG + A + A S KK K +L++ P+ E ++ + +
Sbjct 8 RDRGRDRGFGAPRFGGSRAGPLSGKKFGNPGEKLVKKKWNLDELPKFEKNFYQEHPDLAR 67
Query 76 MTDRDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIPI 135
T ++ +R EI ++G P P+ + EA P +++ + + N+ PT IQ Q P+
Sbjct 68 RTAQEVETYRRSKEITVRGHNCPKPVLNFYEANFPANVMDVIARQNFTEPTAIQAQGWPV 127
Query 136 ALEMRDLIGIAETGSGKTAAFVLPLLTYVKQLPPL 170
AL D++G+A+TGSGKT +++LP + ++ P L
Sbjct 128 ALSGLDMVGVAQTGSGKTLSYLLPAIVHINHQPFL 162
> xla:380261 ddx42, MGC54025; DEAD (Asp-Glu-Ala-Asp) box polypeptide
42 (EC:3.6.4.13); K12835 ATP-dependent RNA helicase DDX42
[EC:3.6.4.13]
Length=947
Score = 78.6 bits (192), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 36/89 (40%), Positives = 55/89 (61%), Gaps = 3/89 (3%)
Query 84 FREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIPIALEMRDLI 143
R + + G P ++A +L+ ++++ Y +PTPIQ Q IP+AL RD+I
Sbjct 232 LRHKLNLRVSGAAPPRLCSSFAHFGFDEQLLHQIRKSEYTQPTPIQCQGIPVALSGRDMI 291
Query 144 GIAETGSGKTAAFVLPLLTYV---KQLPP 169
GIA+TGSGKTAAF+ P+L ++ K+L P
Sbjct 292 GIAKTGSGKTAAFIWPILVHIMDQKELQP 320
> xla:444634 MGC84147 protein
Length=450
Score = 78.6 bits (192), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 36/89 (40%), Positives = 55/89 (61%), Gaps = 3/89 (3%)
Query 84 FREDFEIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIPIALEMRDLI 143
R + + G P ++A +L+ ++++ Y +PTPIQ Q IP+AL RD+I
Sbjct 233 LRHKLNLRVSGAAAPRLCSSFAHFGFDEQLMHQIRKSEYTKPTPIQCQGIPVALSGRDMI 292
Query 144 GIAETGSGKTAAFVLPLLTYV---KQLPP 169
GIA+TGSGKTAAF+ P+L ++ K+L P
Sbjct 293 GIAKTGSGKTAAFIWPILVHIMDQKELQP 321
> ath:AT3G01540 DRH1; DRH1 (DEAD BOX RNA HELICASE 1); ATP-dependent
RNA helicase/ ATPase
Length=619
Score = 78.2 bits (191), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 33/79 (41%), Positives = 54/79 (68%), Gaps = 0/79 (0%)
Query 89 EIYLKGGRVPPPIRTWAEAELPWELIEAVKQANYDRPTPIQMQAIPIALEMRDLIGIAET 148
EI + GG+VPPP+ ++ P EL+ V A + PTPIQ Q+ PIA++ RD++ IA+T
Sbjct 145 EITVSGGQVPPPLMSFEATGFPPELLREVLSAGFSAPTPIQAQSWPIAMQGRDIVAIAKT 204
Query 149 GSGKTAAFVLPLLTYVKQL 167
GSGKT +++P +++++
Sbjct 205 GSGKTLGYLIPGFLHLQRI 223
Lambda K H
0.314 0.130 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4276754328
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40