bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1416_orf3
Length=269
Score E
Sequences producing significant alignments: (Bits) Value
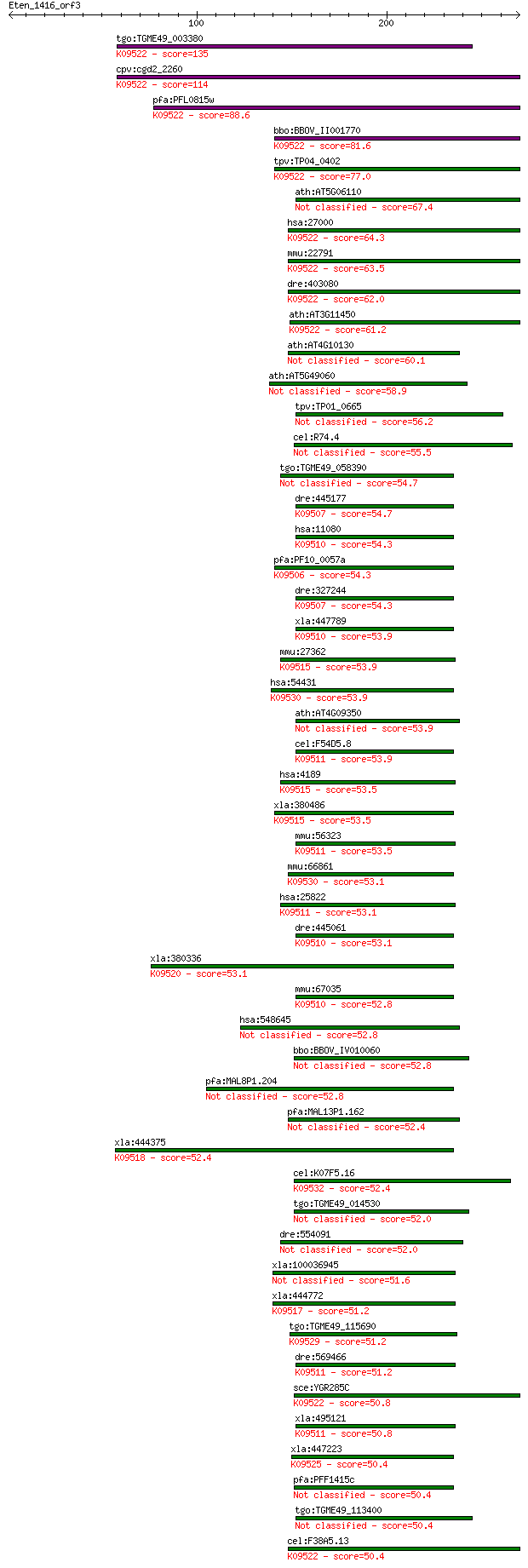
tgo:TGME49_003380 DnaJ domain-containing protein ; K09522 DnaJ... 135 2e-31
cpv:cgd2_2260 zuotin related factor-1 like protein with a DNAJ... 114 4e-25
pfa:PFL0815w DNA-binding chaperone, putative; K09522 DnaJ homo... 88.6 2e-17
bbo:BBOV_II001770 18.m06137; myb-like DNA-binding/DnaJ domain ... 81.6 3e-15
tpv:TP04_0402 DNA-binding chaperone; K09522 DnaJ homolog subfa... 77.0 7e-14
ath:AT5G06110 DNAJ heat shock N-terminal domain-containing pro... 67.4 6e-11
hsa:27000 DNAJC2, MPHOSPH11, MPP11, ZRF1, ZUO1; DnaJ (Hsp40) h... 64.3 4e-10
mmu:22791 Dnajc2, AU020218, MIDA1, Zrf1, Zrf2; DnaJ (Hsp40) ho... 63.5 8e-10
dre:403080 dnajc2, zgc:85671, zrf1; DnaJ (Hsp40) homolog, subf... 62.0 2e-09
ath:AT3G11450 DNAJ heat shock N-terminal domain-containing pro... 61.2 4e-09
ath:AT4G10130 DNAJ heat shock N-terminal domain-containing pro... 60.1 9e-09
ath:AT5G49060 DNAJ heat shock N-terminal domain-containing pro... 58.9 2e-08
tpv:TP01_0665 chaperone protein DnaJ 56.2 1e-07
cel:R74.4 dnj-16; DNaJ domain (prokaryotic heat shock protein)... 55.5 2e-07
tgo:TGME49_058390 DnaJ protein, putative 54.7 3e-07
dre:445177 dnajb1a, dnajb1, hspf1, zf-Hsp40, zgc:101068; DnaJ ... 54.7 4e-07
hsa:11080 DNAJB4, DNAJW, DjB4, HLJ1; DnaJ (Hsp40) homolog, sub... 54.3 4e-07
pfa:PF10_0057a DNAJ protein, putative; K09506 DnaJ homolog sub... 54.3 4e-07
dre:327244 dnajb1b, dnj-13C, fd19c10, wu:fd19c10, zgc:55492; D... 54.3 5e-07
xla:447789 dnajb4, MGC83507; DnaJ (Hsp40) homolog, subfamily B... 53.9 6e-07
mmu:27362 Dnajb9, AA408011, AA673251, AA673481, AW556981, ERdj... 53.9 6e-07
hsa:54431 DNAJC10, DKFZp434J1813, ERdj5, JPDI, MGC104194; DnaJ... 53.9 6e-07
ath:AT4G09350 DNAJ heat shock N-terminal domain-containing pro... 53.9 6e-07
cel:F54D5.8 dnj-13; DNaJ domain (prokaryotic heat shock protei... 53.9 7e-07
hsa:4189 DNAJB9, DKFZp564F1862, ERdj4, MDG-1, MDG1, MST049, MS... 53.5 7e-07
xla:380486 dnajb9, MGC53411; DnaJ (Hsp40) homolog, subfamily B... 53.5 8e-07
mmu:56323 Dnajb5, 1110058L06Rik, AI462558, Hsc40, Hsp40-3; Dna... 53.5 9e-07
mmu:66861 Dnajc10, 1200006L06Rik, D2Ertd706e, ERdj5, JPDI; Dna... 53.1 9e-07
hsa:25822 DNAJB5, Hsc40, KIAA1045; DnaJ (Hsp40) homolog, subfa... 53.1 1e-06
dre:445061 dnajb4, zgc:91922; DnaJ (Hsp40) homolog, subfamily ... 53.1 1e-06
xla:380336 dnajb14, MGC53932, flj14281; DnaJ (Hsp40) homolog, ... 53.1 1e-06
mmu:67035 Dnajb4, 1700029A20Rik, 2010306G19Rik, 5730460G06Rik;... 52.8 1e-06
hsa:548645 DNAJC25, bA16L21.2.1; DnaJ (Hsp40) homolog, subfami... 52.8 1e-06
bbo:BBOV_IV010060 23.m06223; heat shock protein DNAJ 52.8 1e-06
pfa:MAL8P1.204 DNAJ protein, putative 52.8 1e-06
pfa:MAL13P1.162 DNAJ like protein, putative 52.4 2e-06
xla:444375 dnajb12, MGC82876; DnaJ (Hsp40) homolog, subfamily ... 52.4 2e-06
cel:K07F5.16 hypothetical protein; K09532 DnaJ homolog subfami... 52.4 2e-06
tgo:TGME49_014530 DnaJ domain-containing protein 52.0 2e-06
dre:554091 zgc:110447 52.0 2e-06
xla:100036945 hypothetical protein LOC100036945 51.6 3e-06
xla:444772 dnajb11, MGC81924; DnaJ (Hsp40) homolog, subfamily ... 51.2 3e-06
tgo:TGME49_115690 DnaJ domain-containing protein ; K09529 DnaJ... 51.2 3e-06
dre:569466 novel protein similar to vertebrate DnaJ (Hsp40) ho... 51.2 4e-06
sce:YGR285C ZUO1; Zuo1p; K09522 DnaJ homolog subfamily C member 2 50.8
xla:495121 dnajb5; DnaJ (Hsp40) homolog, subfamily B, member 5... 50.8 5e-06
xla:447223 dnajc5g, MGC82663, csp, dnajc5, dnajc5a; DnaJ (Hsp4... 50.4 6e-06
pfa:PFF1415c DNAJ domain protein, putative 50.4 6e-06
tgo:TGME49_113400 DnaJ domain-containing protein 50.4 7e-06
cel:F38A5.13 dnj-11; DNaJ domain (prokaryotic heat shock prote... 50.4 7e-06
> tgo:TGME49_003380 DnaJ domain-containing protein ; K09522 DnaJ
homolog subfamily C member 2
Length=684
Score = 135 bits (339), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 87/209 (41%), Positives = 125/209 (59%), Gaps = 25/209 (11%)
Query 58 PLIALSICTITKKVEPAGRSFFDKYEKRTPSKHR-------RKQDASPTPPPEEDEEYNE 110
P + S+CT ++VE AGR+F +++E+ PS+ R +D E DE+++
Sbjct 33 PQVLCSLCTAQRRVEAAGRAFLERFER--PSRKAGAGVSPGRAKDKDAKRGDESDEDFSA 90
Query 111 RTEKWRAKSKNKSPGGGGGGSSSSWASGTHLKRILAYNQTLYQILGVEDGASLDEIKKQY 170
E+ R KSK K G GG + +AS T LKR+L QTLY++LGV +G + +EIKKQY
Sbjct 91 VDEE-RWKSKRKGKTAGSGGGGACFASSTRLKRLLGEAQTLYEVLGVHEGTTTEEIKKQY 149
Query 171 RKKVLEYHPDKA---KSTATSPSGETTTATPTSAA------AEGLPSSE------HGAFL 215
R+ VLE+HPDKA +S +S G + ++P SA +G +SE H FL
Sbjct 150 RRLVLEHHPDKAVTRRSPGSSEEGASGRSSPHSAKEKREELTDGQATSEASADAGHARFL 209
Query 216 KVQEAYEALSDSNFRKQYDSSLPFDESIP 244
K+QEAYEAL+D+ FR+QYDS+LP+ P
Sbjct 210 KIQEAYEALTDTEFRRQYDSALPWSSRRP 238
> cpv:cgd2_2260 zuotin related factor-1 like protein with a DNAJ
domain at the N-terminus and 2 SANT domains ; K09522 DnaJ
homolog subfamily C member 2
Length=677
Score = 114 bits (284), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 75/219 (34%), Positives = 110/219 (50%), Gaps = 31/219 (14%)
Query 58 PLIALSIC--TITKKVEPAGRSFFDKYEKRTPSK---HRRKQDASPTPPPEEDEEYNERT 112
P+I S+ KK+EP G +FF +YE + +K + K + P + Y++
Sbjct 24 PVIQSSVAEFAFKKKIEPVGEAFFYRYEVKYSNKINFLKEKIKIESSIPI--NNIYSQ-- 79
Query 113 EKWRAKSKNKSPGGGGGGSSSSWASGTHLKRILAYNQTLYQILGVEDGASLDEIKKQYRK 172
E+ + KSK S G + S LK ++ +TLY+ LG+++ + EIK+ YR+
Sbjct 80 EECKKKSKKISGNKSGNKLAKGVLSLARLKELVEEKETLYEKLGLDENVCVKEIKQAYRR 139
Query 173 KVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSEHGAFLKVQEAYEALSDSNFRKQ 232
VL +HPDK K ++ E FLK+QEAYE LSD N R
Sbjct 140 LVLSHHPDKNKENSSDARSE--------------------EFLKIQEAYEILSDKNLRHA 179
Query 233 YDSSLPFDESIPD--AAECEDGAKFYETFGAAFHRNARW 269
YDS+LPFD+SIP +E D +F + F F RN+RW
Sbjct 180 YDSALPFDDSIPSVYVSENNDFYEFKDFFSPIFRRNSRW 218
> pfa:PFL0815w DNA-binding chaperone, putative; K09522 DnaJ homolog
subfamily C member 2
Length=939
Score = 88.6 bits (218), Expect = 2e-17, Method: Composition-based stats.
Identities = 67/227 (29%), Positives = 96/227 (42%), Gaps = 46/227 (20%)
Query 77 SFFDKYEKRTPSKHRRKQDASPTPPPEEDEEYNERTEKWRAKSKNKSPGGGGGGSSSSWA 136
+ +DK R+K + E+DEE N+ + + K N + G +
Sbjct 116 NIYDKNISNDKENKRKKAN-------EKDEEMNK--DDYDNKENNNN---GADNIKGQFK 163
Query 137 SGTHL-KRILAYNQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTT 195
G++L K+ + N +Y +LGVE+ L+ IK Y+K +L +HPDK K TA E
Sbjct 164 KGSNLIKKCIDQNIDVYDVLGVEETDDLETIKSCYKKLILLFHPDKNKGTAYLNEKEKEK 223
Query 196 ATPTSAAA--------------------------------EGLPSSEH-GAFLKVQEAYE 222
E L + E FLK+Q++Y
Sbjct 224 EKKKGKNKKKKNYMNDANNNNNNNNNNEKDFLYFIEKYNIEKLTNDEKKNIFLKIQDSYT 283
Query 223 ALSDSNFRKQYDSSLPFDESIPDAAECEDGAKFYETFGAAFHRNARW 269
LSD RKQYDSS+PFDE IP + E+ FY F RNA+W
Sbjct 284 ILSDKILRKQYDSSIPFDERIPTLTQLEEAKNFYNFLRPVFKRNAKW 330
> bbo:BBOV_II001770 18.m06137; myb-like DNA-binding/DnaJ domain
containing protein; K09522 DnaJ homolog subfamily C member
2
Length=647
Score = 81.6 bits (200), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 46/131 (35%), Positives = 64/131 (48%), Gaps = 6/131 (4%)
Query 141 LKRILAYNQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGE--TTTATP 198
++ ++ NQ Y++L V D L +IK Y++ VL HPDKA +T E
Sbjct 133 VRDFVSKNQNAYELLDVCDSDDLSKIKANYKRIVLLLHPDKAGNTRVPEDMEHYVNKYRI 192
Query 199 TSAAAEGLPSSEHGAFLKVQEAYEALSDSNFRKQYDSSLPFDESIPDAAECEDGAKFYET 258
E L F+ +Q+A+ +SD R +YD SLPFDE+IP E + FY
Sbjct 193 RHLGDEQLKQQ----FILLQDAFTIMSDPQLRHEYDCSLPFDETIPTKEEAKLADDFYGL 248
Query 259 FGAAFHRNARW 269
F F NARW
Sbjct 249 FAPVFELNARW 259
> tpv:TP04_0402 DNA-binding chaperone; K09522 DnaJ homolog subfamily
C member 2
Length=674
Score = 77.0 bits (188), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 42/129 (32%), Positives = 66/129 (51%), Gaps = 3/129 (2%)
Query 141 LKRILAYNQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTS 200
++ +A + Y++L D +++IK YR+ VL HPDK ++ +
Sbjct 133 VRNFIAKEKNAYELLNCTDSDGINKIKANYRRLVLLLHPDKGQNQKVPEDLKDYEKKYKV 192
Query 201 AAAEGLPSSEHGAFLKVQEAYEALSDSNFRKQYDSSLPFDESIPDAAECEDGAKFYETFG 260
+ SE FL +Q+A+ L+D + R +YDS+LPFDESIP E + F+ FG
Sbjct 193 STISRQEKSE--LFLLLQDAFTILNDVDMRLEYDSNLPFDESIPTHEEAK-RKDFFLLFG 249
Query 261 AAFHRNARW 269
F N+RW
Sbjct 250 PIFQMNSRW 258
> ath:AT5G06110 DNAJ heat shock N-terminal domain-containing protein
/ cell division protein-related
Length=663
Score = 67.4 bits (163), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 43/121 (35%), Positives = 63/121 (52%), Gaps = 14/121 (11%)
Query 152 YQILGVEDG---ASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPS 208
Y +LG+ + A+ D+I+K YR L++HPDK + T A + + S
Sbjct 102 YALLGLGNLRYLATDDQIRKSYRDAALKHHPDKLATLLLL----EETEEAKQAKKDEIES 157
Query 209 SEHGAFLKVQEAYEALSDSNFRKQYDSSLPFDESIPDAAECEDGAKFYETFGAAFHRNAR 268
F +QEAYE L DS R+ +DS+ FD+ +P +D F++ FG AF RNAR
Sbjct 158 H----FKLIQEAYEVLMDSTKRRIFDSTDEFDDKVPTDCAPQD---FFKVFGPAFKRNAR 210
Query 269 W 269
W
Sbjct 211 W 211
> hsa:27000 DNAJC2, MPHOSPH11, MPP11, ZRF1, ZUO1; DnaJ (Hsp40)
homolog, subfamily C, member 2; K09522 DnaJ homolog subfamily
C member 2
Length=568
Score = 64.3 bits (155), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 43/126 (34%), Positives = 63/126 (50%), Gaps = 23/126 (18%)
Query 148 NQTLYQILG---VEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAE 204
NQ Y +LG V A+ +IK ++ VL++HPDK K AA E
Sbjct 86 NQDHYAVLGLGHVRYKATQRQIKAAHKAMVLKHHPDKRK-----------------AAGE 128
Query 205 GLPSSEHGAFLKVQEAYEALSDSNFRKQYDSSLP-FDESIPDAAECEDGAKFYETFGAAF 263
+ ++ F + +AYE LSD R+ ++S P FD S+P +E +D F+E F F
Sbjct 129 PIKEGDNDYFTCITKAYEMLSDPVKRRAFNSVDPTFDNSVPSKSEAKDN--FFEVFTPVF 186
Query 264 HRNARW 269
RN+RW
Sbjct 187 ERNSRW 192
> mmu:22791 Dnajc2, AU020218, MIDA1, Zrf1, Zrf2; DnaJ (Hsp40)
homolog, subfamily C, member 2; K09522 DnaJ homolog subfamily
C member 2
Length=621
Score = 63.5 bits (153), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 42/126 (33%), Positives = 63/126 (50%), Gaps = 23/126 (18%)
Query 148 NQTLYQILG---VEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAE 204
NQ Y +LG V A+ +IK ++ VL++HPDK K AA E
Sbjct 86 NQDHYAVLGLGHVRYTATQRQIKAAHKAMVLKHHPDKRK-----------------AAGE 128
Query 205 GLPSSEHGAFLKVQEAYEALSDSNFRKQYDSSLP-FDESIPDAAECEDGAKFYETFGAAF 263
+ ++ F + +AYE LSD R+ ++S P FD S+P +E +D F++ F F
Sbjct 129 PIKEGDNDYFTCITKAYEMLSDPVKRRAFNSVDPTFDNSVPSKSEAKDN--FFQVFSPVF 186
Query 264 HRNARW 269
RN+RW
Sbjct 187 ERNSRW 192
> dre:403080 dnajc2, zgc:85671, zrf1; DnaJ (Hsp40) homolog, subfamily
C, member 2; K09522 DnaJ homolog subfamily C member
2
Length=618
Score = 62.0 bits (149), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 43/126 (34%), Positives = 62/126 (49%), Gaps = 23/126 (18%)
Query 148 NQTLYQILG---VEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAE 204
NQ Y +LG V A+ +IK ++ VL++HPDK K AA E
Sbjct 83 NQDHYAVLGLAHVRYKATQKQIKAAHKAMVLKHHPDKRK-----------------AAGE 125
Query 205 GLPSSEHGAFLKVQEAYEALSDSNFRKQYDSSLP-FDESIPDAAECEDGAKFYETFGAAF 263
+ ++ F + +A E LSD R+ +DS P FD ++P AE ++ F+E F F
Sbjct 126 QIVEGDNDYFTCITKAIEILSDPVKRRAFDSVDPTFDNAVPTKAEGKEN--FFEVFAPVF 183
Query 264 HRNARW 269
RNARW
Sbjct 184 ERNARW 189
> ath:AT3G11450 DNAJ heat shock N-terminal domain-containing protein
/ cell division protein-related; K09522 DnaJ homolog
subfamily C member 2
Length=663
Score = 61.2 bits (147), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 41/124 (33%), Positives = 60/124 (48%), Gaps = 23/124 (18%)
Query 149 QTLYQILGVEDG---ASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEG 205
Q Y +LG+ + A+ D+I+K YR+ L++HPDK + E
Sbjct 123 QDHYALLGLSNLRYLATEDQIRKSYREAALKHHPDKLAKAKEAKKDEI------------ 170
Query 206 LPSSEHGAFLKVQEAYEALSDSNFRKQYDSSLPFDESIPDAAECEDGAKFYETFGAAFHR 265
F +QEAYE L D R+ +DS+ FD+ +P +D F++ FG AF R
Sbjct 171 -----ESRFKAIQEAYEVLMDPTRRRIFDSTDEFDDEVPSDCLPQD---FFKVFGPAFKR 222
Query 266 NARW 269
NARW
Sbjct 223 NARW 226
> ath:AT4G10130 DNAJ heat shock N-terminal domain-containing protein
Length=174
Score = 60.1 bits (144), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 33/90 (36%), Positives = 49/90 (54%), Gaps = 19/90 (21%)
Query 148 NQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLP 207
++T Y+IL V++ AS +EI+ YR +L HPDK +T+ S
Sbjct 9 HETYYEILSVKEDASYEEIRNSYRSAILHSHPDKLNNTSRS------------------- 49
Query 208 SSEHGAFLKVQEAYEALSDSNFRKQYDSSL 237
SS+ FLK+Q+A+E LSD+ R YD+ L
Sbjct 50 SSDDEKFLKIQKAWEVLSDAELRVVYDNDL 79
> ath:AT5G49060 DNAJ heat shock N-terminal domain-containing protein
Length=354
Score = 58.9 bits (141), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 39/104 (37%), Positives = 49/104 (47%), Gaps = 23/104 (22%)
Query 138 GTHLKRILAYNQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTAT 197
L R + N Y ILG+E S+DEI+K YRK L+ HPDK K+
Sbjct 87 NVDLVRNIIRNNDYYAILGLEKNCSVDEIRKAYRKLSLKVHPDKNKA------------- 133
Query 198 PTSAAAEGLPSSEHGAFLKVQEAYEALSDSNFRKQYDSSLPFDE 241
P SE AF KV +A+ LSD N R+Q+D DE
Sbjct 134 ---------PGSEE-AFKKVSKAFTCLSDGNSRRQFDQVGIVDE 167
> tpv:TP01_0665 chaperone protein DnaJ
Length=229
Score = 56.2 bits (134), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 36/112 (32%), Positives = 56/112 (50%), Gaps = 24/112 (21%)
Query 152 YQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSEH 211
Y++LGV A D IKKQYRK ++YHPDK +P + +A
Sbjct 10 YKLLGVSPDADEDAIKKQYRKLAMKYHPDK------NPHNKEKSAE-------------- 49
Query 212 GAFLKVQEAYEALSDSNFRKQYDSSLP--FDESIPDAAECEDGAKFYE-TFG 260
F K+ +AYE LSD R+ YD+++ FD++ + +D + ++ FG
Sbjct 50 -MFKKISQAYEVLSDKRKRRNYDNNVNFGFDQNYANNFNFDDAQRIFQMVFG 100
> cel:R74.4 dnj-16; DNaJ domain (prokaryotic heat shock protein)
family member (dnj-16)
Length=378
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 41/116 (35%), Positives = 52/116 (44%), Gaps = 23/116 (19%)
Query 151 LYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSE 210
YQ+LGVE AS EIK YRK L+YHPD+ P A A+
Sbjct 18 FYQLLGVEKMASEAEIKSAYRKLALKYHPDR---------------NPNDAHAQ------ 56
Query 211 HGAFLKVQEAYEALSDSNFRKQYDSSLPFDESIP-DAAECEDGAKFYETFGAAFHR 265
F KV AY LSD N R+QYD S P + + + + + FGA F +
Sbjct 57 -EEFKKVSIAYSVLSDPNKRRQYDVSGPSENQLDFEGFDVSEMGGVGRVFGALFSK 111
> tgo:TGME49_058390 DnaJ protein, putative
Length=397
Score = 54.7 bits (130), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 34/91 (37%), Positives = 47/91 (51%), Gaps = 23/91 (25%)
Query 144 ILAYNQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAA 203
+LA Q LY +LGV+ AS DEIKK YRK ++YHPDK K
Sbjct 44 VLAAKQNLYSVLGVKRNASADEIKKAYRKLSMKYHPDKNKE------------------- 84
Query 204 EGLPSSEHGAFLKVQEAYEALSDSNFRKQYD 234
P++E F ++ AYE L+++ R+ YD
Sbjct 85 ---PNAE-AKFKEISFAYEILNNAEKRQVYD 111
> dre:445177 dnajb1a, dnajb1, hspf1, zf-Hsp40, zgc:101068; DnaJ
(Hsp40) homolog, subfamily B, member 1a; K09507 DnaJ homolog
subfamily B member 1
Length=335
Score = 54.7 bits (130), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 34/83 (40%), Positives = 44/83 (53%), Gaps = 23/83 (27%)
Query 152 YQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSEH 211
Y+ILG+E GAS +EIKK YRK+ L +HPDK K SA AE
Sbjct 6 YRILGIEKGASDEEIKKAYRKQALRFHPDKNK----------------SAGAE------- 42
Query 212 GAFLKVQEAYEALSDSNFRKQYD 234
F ++ EAY+ LSD+ + YD
Sbjct 43 DKFKEIAEAYDVLSDAKKKDIYD 65
> hsa:11080 DNAJB4, DNAJW, DjB4, HLJ1; DnaJ (Hsp40) homolog, subfamily
B, member 4; K09510 DnaJ homolog subfamily B member
4
Length=337
Score = 54.3 bits (129), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 44/83 (53%), Gaps = 23/83 (27%)
Query 152 YQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSEH 211
Y ILG+E GAS ++IKK YRK+ L++HPDK KS P +E
Sbjct 6 YCILGIEKGASDEDIKKAYRKQALKFHPDKNKS----------------------PQAEE 43
Query 212 GAFLKVQEAYEALSDSNFRKQYD 234
F +V EAYE LSD R+ YD
Sbjct 44 -KFKEVAEAYEVLSDPKKREIYD 65
> pfa:PF10_0057a DNAJ protein, putative; K09506 DnaJ homolog subfamily
A member 5
Length=796
Score = 54.3 bits (129), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 34/94 (36%), Positives = 45/94 (47%), Gaps = 21/94 (22%)
Query 141 LKRILAYNQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTS 200
+++ L Y Y+IL V+ AS+D+IKK Y+K VL YHPDK T
Sbjct 4 IEKCLCY----YKILNVDTDASVDDIKKSYKKIVLLYHPDKNTHLCDEDKKRYT------ 53
Query 201 AAAEGLPSSEHGAFLKVQEAYEALSDSNFRKQYD 234
F K+QEAYE L++ RK YD
Sbjct 54 -----------NIFRKIQEAYECLTNEVQRKWYD 76
> dre:327244 dnajb1b, dnj-13C, fd19c10, wu:fd19c10, zgc:55492;
DnaJ (Hsp40) homolog, subfamily B, member 1b; K09507 DnaJ homolog
subfamily B member 1
Length=337
Score = 54.3 bits (129), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 34/83 (40%), Positives = 43/83 (51%), Gaps = 23/83 (27%)
Query 152 YQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSEH 211
Y +LG++ GAS DEIKK YRK+ L+YHPDK K SA AE
Sbjct 6 YSVLGIQKGASDDEIKKAYRKQALKYHPDKNK----------------SAGAE------- 42
Query 212 GAFLKVQEAYEALSDSNFRKQYD 234
F ++ EAY+ LSD + YD
Sbjct 43 EKFKEIAEAYDVLSDPKKKDIYD 65
> xla:447789 dnajb4, MGC83507; DnaJ (Hsp40) homolog, subfamily
B, member 4; K09510 DnaJ homolog subfamily B member 4
Length=339
Score = 53.9 bits (128), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 44/83 (53%), Gaps = 23/83 (27%)
Query 152 YQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSEH 211
Y ILG+E GAS D+IKK YRK+ L++HPDK K SA AE
Sbjct 6 YSILGIEKGASEDDIKKAYRKQALKWHPDKNK----------------SAHAE------- 42
Query 212 GAFLKVQEAYEALSDSNFRKQYD 234
F ++ EAYE LSD ++ YD
Sbjct 43 EKFKEIAEAYEVLSDPKKKEVYD 65
> mmu:27362 Dnajb9, AA408011, AA673251, AA673481, AW556981, ERdj4,
Mdg1, mDj7; DnaJ (Hsp40) homolog, subfamily B, member 9;
K09515 DnaJ homolog subfamily B member 9
Length=222
Score = 53.9 bits (128), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 49/92 (53%), Gaps = 24/92 (26%)
Query 144 ILAYNQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAA 203
ILA +++ Y ILGV AS +IKK + K ++YHPDK KS
Sbjct 21 ILA-SKSYYDILGVPKSASERQIKKAFHKLAMKYHPDKNKS------------------- 60
Query 204 EGLPSSEHGAFLKVQEAYEALSDSNFRKQYDS 235
P +E F ++ EAYE LSD+N RK+YD+
Sbjct 61 ---PDAE-AKFREIAEAYETLSDANSRKEYDT 88
> hsa:54431 DNAJC10, DKFZp434J1813, ERdj5, JPDI, MGC104194; DnaJ
(Hsp40) homolog, subfamily C, member 10; K09530 DnaJ homolog
subfamily C member 10
Length=793
Score = 53.9 bits (128), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 31/96 (32%), Positives = 46/96 (47%), Gaps = 22/96 (22%)
Query 139 THLKRILAYNQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATP 198
++ ++ +Q Y +LGV AS EI++ ++K L+ HPDK P
Sbjct 24 VYMAILVGTDQDFYSLLGVSKTASSREIRQAFKKLALKLHPDK---------------NP 68
Query 199 TSAAAEGLPSSEHGAFLKVQEAYEALSDSNFRKQYD 234
+ A HG FLK+ AYE L D + RK+YD
Sbjct 69 NNPNA-------HGDFLKINRAYEVLKDEDLRKKYD 97
> ath:AT4G09350 DNAJ heat shock N-terminal domain-containing protein
Length=249
Score = 53.9 bits (128), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 42/86 (48%), Gaps = 21/86 (24%)
Query 152 YQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSEH 211
YQ LGV A L+EIK YR+ EYHPD TT+ P A+E
Sbjct 108 YQFLGVSTDADLEEIKSAYRRLSKEYHPD-------------TTSLPLKTASE------- 147
Query 212 GAFLKVQEAYEALSDSNFRKQYDSSL 237
F+K++E Y LSD R+ YD +L
Sbjct 148 -KFMKLREVYNVLSDEETRRFYDWTL 172
> cel:F54D5.8 dnj-13; DNaJ domain (prokaryotic heat shock protein)
family member (dnj-13); K09511 DnaJ homolog subfamily B
member 5
Length=331
Score = 53.9 bits (128), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 33/83 (39%), Positives = 42/83 (50%), Gaps = 23/83 (27%)
Query 152 YQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSEH 211
Y++LG+ GA+ DEIKK YRK L+YHPDK K A AE
Sbjct 6 YKVLGISKGATDDEIKKAYRKMALKYHPDKNK----------------EAGAE------- 42
Query 212 GAFLKVQEAYEALSDSNFRKQYD 234
F ++ EAY+ LSD +K YD
Sbjct 43 NKFKEIAEAYDVLSDDKKKKIYD 65
> hsa:4189 DNAJB9, DKFZp564F1862, ERdj4, MDG-1, MDG1, MST049,
MSTP049; DnaJ (Hsp40) homolog, subfamily B, member 9; K09515
DnaJ homolog subfamily B member 9
Length=223
Score = 53.5 bits (127), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 49/92 (53%), Gaps = 24/92 (26%)
Query 144 ILAYNQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAA 203
ILA +++ Y ILGV AS +IKK + K ++YHPDK KS
Sbjct 21 ILA-SKSYYDILGVPKSASERQIKKAFHKLAMKYHPDKNKS------------------- 60
Query 204 EGLPSSEHGAFLKVQEAYEALSDSNFRKQYDS 235
P +E F ++ EAYE LSD+N RK+YD+
Sbjct 61 ---PDAE-AKFREIAEAYETLSDANRRKEYDT 88
> xla:380486 dnajb9, MGC53411; DnaJ (Hsp40) homolog, subfamily
B, member 9; K09515 DnaJ homolog subfamily B member 9
Length=221
Score = 53.5 bits (127), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 35/94 (37%), Positives = 46/94 (48%), Gaps = 23/94 (24%)
Query 141 LKRILAYNQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTS 200
+ I+ +T Y ILGV AS +IKK + K ++YHPDK K SP ET
Sbjct 17 ISEIILAKKTYYDILGVPKNASERQIKKAFHKLAMKYHPDKNK----SPDAET------- 65
Query 201 AAAEGLPSSEHGAFLKVQEAYEALSDSNFRKQYD 234
F ++ EAYE LSD + RK+YD
Sbjct 66 ------------KFREIAEAYETLSDESKRKEYD 87
> mmu:56323 Dnajb5, 1110058L06Rik, AI462558, Hsc40, Hsp40-3; DnaJ
(Hsp40) homolog, subfamily B, member 5; K09511 DnaJ homolog
subfamily B member 5
Length=348
Score = 53.5 bits (127), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 33/84 (39%), Positives = 42/84 (50%), Gaps = 23/84 (27%)
Query 152 YQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSEH 211
Y+ILG+ GA+ DEIKK YRK L+YHPDK K P++E
Sbjct 6 YKILGIPSGANEDEIKKAYRKMALKYHPDKNKE----------------------PNAEE 43
Query 212 GAFLKVQEAYEALSDSNFRKQYDS 235
F ++ EAY+ LSD R YD
Sbjct 44 -KFKEIAEAYDVLSDPKKRSLYDQ 66
> mmu:66861 Dnajc10, 1200006L06Rik, D2Ertd706e, ERdj5, JPDI; DnaJ
(Hsp40) homolog, subfamily C, member 10; K09530 DnaJ homolog
subfamily C member 10
Length=793
Score = 53.1 bits (126), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 31/87 (35%), Positives = 42/87 (48%), Gaps = 22/87 (25%)
Query 148 NQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLP 207
+Q Y +LGV AS EI++ ++K L+ HPDK P + A
Sbjct 33 DQNFYSLLGVSKTASSREIRQAFKKLALKLHPDK---------------NPNNPNA---- 73
Query 208 SSEHGAFLKVQEAYEALSDSNFRKQYD 234
HG FLK+ AYE L D + RK+YD
Sbjct 74 ---HGDFLKINRAYEVLKDEDLRKKYD 97
> hsa:25822 DNAJB5, Hsc40, KIAA1045; DnaJ (Hsp40) homolog, subfamily
B, member 5; K09511 DnaJ homolog subfamily B member 5
Length=382
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/92 (35%), Positives = 44/92 (47%), Gaps = 23/92 (25%)
Query 144 ILAYNQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAA 203
+ + Y+ILG+ GA+ DEIKK YRK L+YHPDK K
Sbjct 32 VAVMGKDYYKILGIPSGANEDEIKKAYRKMALKYHPDKNKE------------------- 72
Query 204 EGLPSSEHGAFLKVQEAYEALSDSNFRKQYDS 235
P++E F ++ EAY+ LSD R YD
Sbjct 73 ---PNAEE-KFKEIAEAYDVLSDPKKRGLYDQ 100
> dre:445061 dnajb4, zgc:91922; DnaJ (Hsp40) homolog, subfamily
B, member 4; K09510 DnaJ homolog subfamily B member 4
Length=340
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 44/83 (53%), Gaps = 23/83 (27%)
Query 152 YQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSEH 211
Y+ILG+ GAS D+IKK YRK+ L++HPDK K +A AE
Sbjct 6 YKILGITKGASDDDIKKAYRKQALKWHPDKNK----------------AANAE------- 42
Query 212 GAFLKVQEAYEALSDSNFRKQYD 234
F +V EAYE LSD R+ YD
Sbjct 43 EKFKEVAEAYEVLSDPKKREIYD 65
> xla:380336 dnajb14, MGC53932, flj14281; DnaJ (Hsp40) homolog,
subfamily B, member 14; K09520 DnaJ homolog subfamily B member
14
Length=371
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 50/166 (30%), Positives = 74/166 (44%), Gaps = 37/166 (22%)
Query 76 RSFFDKYEKRTPSKHRR-------KQDASPTPPPEEDEEYNERTEKWRAKSKNKSPGGGG 128
R FF K E+ PS R K D + P + E+ ++ TE+ K++ S G G
Sbjct 28 RRFFSKAERLYPSSEARVLLDALEKNDTAGNGP--QSEKMSKSTEQ--PKAEKDSSGDTG 83
Query 129 GGSSSSWASGTHLKRILAYNQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATS 188
G + G ++RI +T Y++LGV A +++KK YRK L++HPDK + +
Sbjct 84 KGHTQDQVDG--VQRIKKC-KTYYEVLGVSPDAGEEDLKKAYRKLALKFHPDKNHAPGAT 140
Query 189 PSGETTTATPTSAAAEGLPSSEHGAFLKVQEAYEALSDSNFRKQYD 234
AF K+ AY LS+ RKQYD
Sbjct 141 E-----------------------AFKKIGNAYAVLSNPEKRKQYD 163
> mmu:67035 Dnajb4, 1700029A20Rik, 2010306G19Rik, 5730460G06Rik;
DnaJ (Hsp40) homolog, subfamily B, member 4; K09510 DnaJ
homolog subfamily B member 4
Length=337
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 32/83 (38%), Positives = 44/83 (53%), Gaps = 23/83 (27%)
Query 152 YQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSEH 211
Y ILG++ GA+ +++KK YRK+ L++HPDK KS P +E
Sbjct 6 YHILGIDKGATDEDVKKAYRKQALKFHPDKNKS----------------------PQAEE 43
Query 212 GAFLKVQEAYEALSDSNFRKQYD 234
F +V EAYE LSD R+ YD
Sbjct 44 -KFKEVAEAYEVLSDPKKREIYD 65
> hsa:548645 DNAJC25, bA16L21.2.1; DnaJ (Hsp40) homolog, subfamily
C, member 25
Length=360
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 40/130 (30%), Positives = 52/130 (40%), Gaps = 27/130 (20%)
Query 123 SPGGGGGGSSSSWA---------------SGTHLKRILAYNQTLYQILGVEDGASLDEIK 167
SPG G G + W +G ++ + + Y++LGV A EI
Sbjct 7 SPGWGAGAAGRRWWMLLAPLLPALLLVRPAGALVEGLYCGTRDCYEVLGVSRSAGKAEIA 66
Query 168 KQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSEHGAFLKVQEAYEALSDS 227
+ YR+ YHPD+ + P E TP SA AFL V AYE L D
Sbjct 67 RAYRQLARRYHPDRYRP---QPGDEGPGRTPQSAEE---------AFLLVATAYETLKDE 114
Query 228 NFRKQYDSSL 237
RK YD L
Sbjct 115 ETRKDYDYML 124
> bbo:BBOV_IV010060 23.m06223; heat shock protein DNAJ
Length=192
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/92 (36%), Positives = 47/92 (51%), Gaps = 21/92 (22%)
Query 151 LYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSE 210
Y++LGV+ AS DEIKK+Y+ +++HPDK P + A ++E
Sbjct 16 FYKVLGVKPDASDDEIKKKYKALAIKWHPDK---------------NPNNLAE----ATE 56
Query 211 HGAFLKVQEAYEALSDSNFRKQYDSSLPFDES 242
F K+ AYE LSD R+ YD PFD S
Sbjct 57 --MFQKISTAYETLSDPQKRRDYDCFSPFDTS 86
> pfa:MAL8P1.204 DNAJ protein, putative
Length=961
Score = 52.8 bits (125), Expect = 1e-06, Method: Composition-based stats.
Identities = 39/130 (30%), Positives = 55/130 (42%), Gaps = 30/130 (23%)
Query 105 DEEYNERTEKWRAKSKNKSPGGGGGGSSSSWASGTHLKRILAYNQTLYQILGVEDGASLD 164
DEE +E +K K+ + GG S S +S K Y IL V+ AS
Sbjct 500 DEEAHELFSNKNSKQKDNNNDNAYGGCSRSRSSCVDTK--------YYDILNVKPYASFK 551
Query 165 EIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSEHGAFLKVQEAYEAL 224
EIK + K L+YHPDK ++ + F K+ EAY+ L
Sbjct 552 EIKDSFYKLALKYHPDKNENNIEAKI----------------------MFQKINEAYQIL 589
Query 225 SDSNFRKQYD 234
SD + R++YD
Sbjct 590 SDEDQRRKYD 599
> pfa:MAL13P1.162 DNAJ like protein, putative
Length=247
Score = 52.4 bits (124), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 32/90 (35%), Positives = 46/90 (51%), Gaps = 21/90 (23%)
Query 148 NQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLP 207
N+ Y IL ++ ++ +EIK+ YRK L+YHPD+ +P+
Sbjct 41 NKNFYDILNIKKDSNKNEIKQAYRKLALKYHPDR------NPNNR--------------K 80
Query 208 SSEHGAFLKVQEAYEALSDSNFRKQYDSSL 237
SE F ++ EAYE LSD N +K YDS L
Sbjct 81 ESEQ-KFREITEAYETLSDDNKKKMYDSQL 109
> xla:444375 dnajb12, MGC82876; DnaJ (Hsp40) homolog, subfamily
B, member 12; K09518 DnaJ homolog subfamily B member 12
Length=373
Score = 52.4 bits (124), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 52/187 (27%), Positives = 79/187 (42%), Gaps = 33/187 (17%)
Query 57 EPLIALSICTITKKVEPAGRSFFDKYEKRTPSKHRRK--QDASPTPPPEED-----EEYN 109
E IA+++ I + F +K ++ P++ R + S PE + E N
Sbjct 9 ERCIAIAVGAIKSQNTEKALRFLEKAQRLFPTERARALYESLSQKSQPENNQSDSTETTN 68
Query 110 ERTEKWRAKSKNKSPGGGGGGSSSSWASGT--HLKRILAYNQTLYQILGVEDGASLDEIK 167
R K A S + G GG + S+ +KRI + Y+ILGV A+ D++K
Sbjct 69 PRLRKNAADSTPSANGEAGGETGKSYTPDQLEAVKRI-KQCKDYYEILGVTREATEDDLK 127
Query 168 KQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSEHGAFLKVQEAYEALSDS 227
K YRK L++HPDK + + AF + AY LS++
Sbjct 128 KSYRKLALKFHPDKNYAPGATE-----------------------AFKAIGNAYAVLSNA 164
Query 228 NFRKQYD 234
RKQYD
Sbjct 165 EKRKQYD 171
> cel:K07F5.16 hypothetical protein; K09532 DnaJ homolog subfamily
C member 12
Length=155
Score = 52.4 bits (124), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 35/116 (30%), Positives = 56/116 (48%), Gaps = 29/116 (25%)
Query 151 LYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSE 210
LY++LG ++ +S+D+I +YR +V + HPDK K T+ + +
Sbjct 16 LYRLLGCDESSSIDQITAEYRARVRDCHPDKVKEHNTNSTEK------------------ 57
Query 211 HGAFLKVQEAYEALSDSNFRKQYDSSL--PFDESIPDAAECEDGAKFYETFGAAFH 264
F+++Q AY L+ + RK YDS L PF S D + +D TF + H
Sbjct 58 ---FMELQNAYSILTSDSRRKTYDSWLHSPFPVSFEDFTKSQD------TFQMSTH 104
> tgo:TGME49_014530 DnaJ domain-containing protein
Length=714
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 32/92 (34%), Positives = 43/92 (46%), Gaps = 21/92 (22%)
Query 151 LYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSE 210
Y++LGV+ A DEIKK YRK L++HPD+ +P
Sbjct 403 FYEVLGVKKDAGTDEIKKAYRKLALKWHPDR------NPDNRQQA--------------- 441
Query 211 HGAFLKVQEAYEALSDSNFRKQYDSSLPFDES 242
F V EAY+ LS+S R+QYD+ F S
Sbjct 442 EAQFRLVSEAYQTLSNSEKRQQYDAMRQFGAS 473
> dre:554091 zgc:110447
Length=199
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 38/98 (38%), Positives = 48/98 (48%), Gaps = 26/98 (26%)
Query 144 ILAYNQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAA 203
ILA + Y+ILGV AS +IKK + K + +HPDK KS
Sbjct 21 ILA-EKDYYEILGVPKDASDRQIKKAFHKLAMRFHPDKNKS------------------- 60
Query 204 EGLPSSEHGAFLKVQEAYEALSDSNFRKQYDS--SLPF 239
P +E F ++ EAYE LSD N RK+YD S PF
Sbjct 61 ---PDAE-AKFREIAEAYETLSDDNRRKEYDQTRSRPF 94
> xla:100036945 hypothetical protein LOC100036945
Length=358
Score = 51.6 bits (122), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 33/96 (34%), Positives = 46/96 (47%), Gaps = 22/96 (22%)
Query 140 HLKRILAYNQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPT 199
+L +++ + Y+ILGV GA++ EIKK YRK L+ HPD+ P
Sbjct 15 YLMVVVSGGRDFYKILGVSRGATVKEIKKAYRKLALQLHPDR---------------NPD 59
Query 200 SAAAEGLPSSEHGAFLKVQEAYEALSDSNFRKQYDS 235
A+ F + AYE LSD RKQYD+
Sbjct 60 DPNAQ-------DKFQDLGAAYEVLSDEEKRKQYDT 88
> xla:444772 dnajb11, MGC81924; DnaJ (Hsp40) homolog, subfamily
B, member 11; K09517 DnaJ homolog subfamily B member 11
Length=360
Score = 51.2 bits (121), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 33/96 (34%), Positives = 46/96 (47%), Gaps = 22/96 (22%)
Query 140 HLKRILAYNQTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPT 199
+L +++ + Y+ILGV GA++ EIKK YRK L+ HPD+ P
Sbjct 17 YLMVVVSGGRDFYKILGVSRGATVKEIKKAYRKLALQLHPDR---------------NPD 61
Query 200 SAAAEGLPSSEHGAFLKVQEAYEALSDSNFRKQYDS 235
A+ F + AYE LSD RKQYD+
Sbjct 62 DPNAQ-------DKFQDLGAAYEVLSDEEKRKQYDT 90
> tgo:TGME49_115690 DnaJ domain-containing protein ; K09529 DnaJ
homolog subfamily C member 9
Length=293
Score = 51.2 bits (121), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 33/88 (37%), Positives = 43/88 (48%), Gaps = 0/88 (0%)
Query 149 QTLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPS 208
+TLY++L V AS EI K YR L HPDK + T+ +A E L S
Sbjct 20 KTLYELLRVSPSASQSEITKAYRNLALLLHPDKVVHRLEQQAEATSKGEGKAADLEELKS 79
Query 209 SEHGAFLKVQEAYEALSDSNFRKQYDSS 236
F ++Q AYE L D RK+YD +
Sbjct 80 EATRHFQELQAAYEVLKDPKRRKRYDET 107
> dre:569466 novel protein similar to vertebrate DnaJ (Hsp40)
homolog, subfamily B, member 5 (DNAJB5); K09511 DnaJ homolog
subfamily B member 5
Length=360
Score = 51.2 bits (121), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 32/84 (38%), Positives = 42/84 (50%), Gaps = 23/84 (27%)
Query 152 YQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSEH 211
Y+ILG+ G++ DEIKK YRK L++HPDK K P++E
Sbjct 6 YKILGIPSGSNEDEIKKAYRKMALKFHPDKNKD----------------------PNAEE 43
Query 212 GAFLKVQEAYEALSDSNFRKQYDS 235
F ++ EAYE LSD R YD
Sbjct 44 -KFKEIAEAYEVLSDPKKRVIYDQ 66
> sce:YGR285C ZUO1; Zuo1p; K09522 DnaJ homolog subfamily C member
2
Length=433
Score = 50.8 bits (120), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 42/122 (34%), Positives = 59/122 (48%), Gaps = 24/122 (19%)
Query 151 LYQILGVED---GASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLP 207
LY +G+ A+ +I K +RK+V++YHPDK SAA L
Sbjct 98 LYAAMGLSKLRFRATESQIIKAHRKQVVKYHPDK-----------------QSAAGGSL- 139
Query 208 SSEHGAFLKVQEAYEALSDSNFRKQYDSSLPFDESIPDAAECEDGAKFYETFGAAFHRNA 267
+ G F +Q+A+E L+DSN R QYDS F +P + D FYE +G F A
Sbjct 140 -DQDGFFKIIQKAFETLTDSNKRAQYDSC-DFVADVPPPKKGTD-YDFYEAWGPVFEAEA 196
Query 268 RW 269
R+
Sbjct 197 RF 198
> xla:495121 dnajb5; DnaJ (Hsp40) homolog, subfamily B, member
5; K09511 DnaJ homolog subfamily B member 5
Length=348
Score = 50.8 bits (120), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 34/84 (40%), Positives = 41/84 (48%), Gaps = 23/84 (27%)
Query 152 YQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSEH 211
Y+ILG+ GA+ DEIKK YRK L+YHPDK K A AE
Sbjct 6 YKILGLASGANEDEIKKAYRKMALKYHPDKNK----------------DANAE------- 42
Query 212 GAFLKVQEAYEALSDSNFRKQYDS 235
F ++ EAY+ LSD R YD
Sbjct 43 DKFKEIAEAYDVLSDPKKRAVYDQ 66
> xla:447223 dnajc5g, MGC82663, csp, dnajc5, dnajc5a; DnaJ (Hsp40)
homolog, subfamily C, member 5 gamma; K09525 DnaJ homolog
subfamily C member 5
Length=184
Score = 50.4 bits (119), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 32/85 (37%), Positives = 42/85 (49%), Gaps = 22/85 (25%)
Query 150 TLYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSS 209
+LY +LG++ GAS D+IKK YRK L+YHPDK P +A
Sbjct 17 SLYVVLGLQKGASPDDIKKAYRKLALKYHPDK------------NPDNPEAAE------- 57
Query 210 EHGAFLKVQEAYEALSDSNFRKQYD 234
F ++ A+ LSD N RK YD
Sbjct 58 ---KFKEINSAHSTLSDENRRKIYD 79
> pfa:PFF1415c DNAJ domain protein, putative
Length=380
Score = 50.4 bits (119), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 32/84 (38%), Positives = 40/84 (47%), Gaps = 23/84 (27%)
Query 151 LYQILGVEDGASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAEGLPSSE 210
LY +LGV+ AS D+IKK YRK +YHPDKAK ++
Sbjct 48 LYDVLGVDKNASSDDIKKSYRKLSKKYHPDKAKDKNSN---------------------- 85
Query 211 HGAFLKVQEAYEALSDSNFRKQYD 234
F ++ EAYE L D RK YD
Sbjct 86 -NKFSEIAEAYEILGDEEKRKIYD 108
> tgo:TGME49_113400 DnaJ domain-containing protein
Length=401
Score = 50.4 bits (119), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 34/96 (35%), Positives = 45/96 (46%), Gaps = 27/96 (28%)
Query 152 YQILGVEDGASLDEIKKQYRKKVLEYHPDK---AKSTATSPSGETTTATPTSAAAEGLPS 208
Y++LGV A+ DEIKK YRK + +HPDK K AT+
Sbjct 120 YEVLGVAKTATADEIKKSYRKLAIRWHPDKNIDKKDEATA-------------------- 159
Query 209 SEHGAFLKVQEAYEALSDSNFRKQYDSSLPFDESIP 244
F ++ EAYE LSD R++YD S F + P
Sbjct 160 ----RFKEISEAYEVLSDPEKRRRYDLSDSFGVNGP 191
> cel:F38A5.13 dnj-11; DNaJ domain (prokaryotic heat shock protein)
family member (dnj-11); K09522 DnaJ homolog subfamily
C member 2
Length=589
Score = 50.4 bits (119), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 37/127 (29%), Positives = 59/127 (46%), Gaps = 25/127 (19%)
Query 148 NQTLYQILGVED---GASLDEIKKQYRKKVLEYHPDKAKSTATSPSGETTTATPTSAAAE 204
NQ Y++LG+ A+ DEI+ YR+KVL++HPDK K
Sbjct 96 NQDHYKVLGLSKLRWQATSDEIRFCYRQKVLKHHPDKKKH-------------------R 136
Query 205 GLPSSEHGAFLKVQEAYEALSDSNFRKQ-YDS-SLPFDESIPDAAECEDGAKFYETFGAA 262
G+ + F + +AYE + S+ ++Q +DS F++ IP+ + FY
Sbjct 137 GIVMEKEEYFTCITKAYEQVGMSDVKRQAFDSVDHKFNDIIPNEKSI-NHNNFYNELAPV 195
Query 263 FHRNARW 269
F N+RW
Sbjct 196 FQLNSRW 202
Lambda K H
0.311 0.128 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10082348456
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40