bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
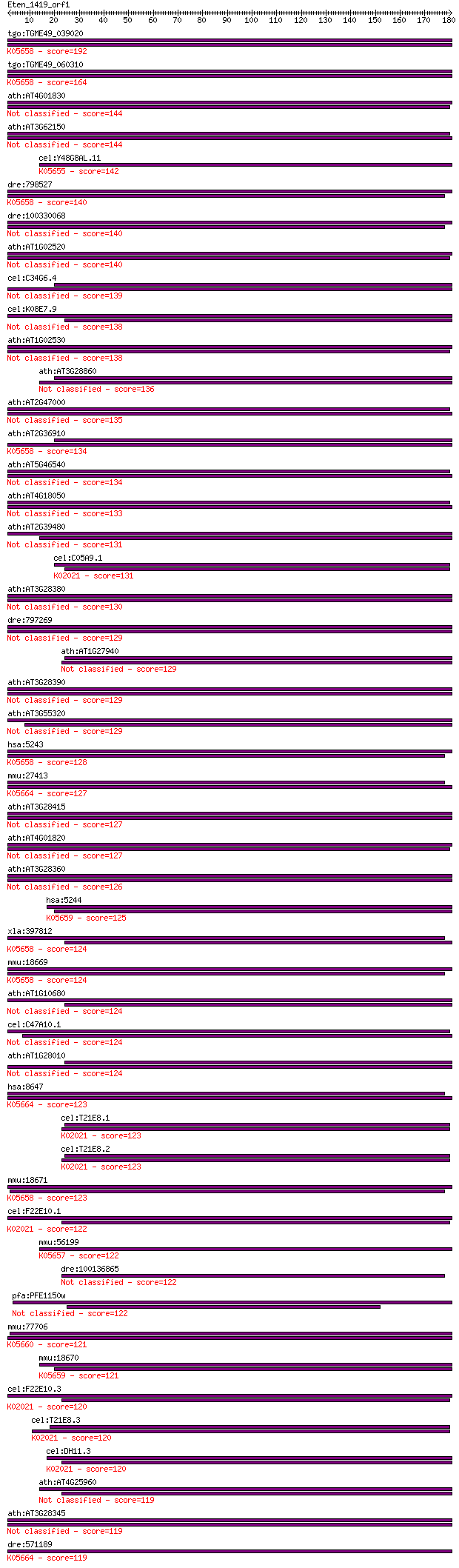
Query= Eten_1419_orf1
Length=180
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_039020 ATP-binding cassette protein subfamily B mem... 192 4e-49
tgo:TGME49_060310 ATP-binding cassette protein subfamily B mem... 164 1e-40
ath:AT4G01830 PGP5; PGP5 (P-GLYCOPROTEIN 5); ATPase, coupled t... 144 1e-34
ath:AT3G62150 PGP21; PGP21 (P-GLYCOPROTEIN 21); ATPase, couple... 144 2e-34
cel:Y48G8AL.11 haf-6; HAlF transporter (PGP related) family me... 142 9e-34
dre:798527 abcb5, im:7158730, mdr1, wu:fc18f02, wu:fi81f06; AT... 140 2e-33
dre:100330068 ATP-binding cassette, subfamily B, member 1B-like 140 2e-33
ath:AT1G02520 PGP11; PGP11 (P-GLYCOPROTEIN 11); ATPase, couple... 140 2e-33
cel:C34G6.4 pgp-2; P-GlycoProtein related family member (pgp-2) 139 7e-33
cel:K08E7.9 pgp-1; P-GlycoProtein related family member (pgp-1) 138 1e-32
ath:AT1G02530 PGP12; PGP12 (P-GLYCOPROTEIN 12); ATPase, couple... 138 1e-32
ath:AT3G28860 ABCB19; ABCB19; ATPase, coupled to transmembrane... 136 3e-32
ath:AT2G47000 ABCB4; ABCB4 (ATP BINDING CASSETTE SUBFAMILY B4)... 135 6e-32
ath:AT2G36910 ABCB1; ABCB1 (ATP BINDING CASSETTE SUBFAMILY B1)... 134 1e-31
ath:AT5G46540 PGP7; PGP7 (P-GLYCOPROTEIN 7); ATPase, coupled t... 134 1e-31
ath:AT4G18050 PGP9; PGP9 (P-GLYCOPROTEIN 9); ATPase, coupled t... 133 3e-31
ath:AT2G39480 PGP6; PGP6 (P-GLYCOPROTEIN 6); ATPase, coupled t... 131 1e-30
cel:C05A9.1 pgp-5; P-GlycoProtein related family member (pgp-5... 131 2e-30
ath:AT3G28380 PGP17; PGP17 (P-GLYCOPROTEIN 17); ATP binding / ... 130 2e-30
dre:797269 abcb11a; ATP-binding cassette, sub-family B (MDR/TA... 129 6e-30
ath:AT1G27940 PGP13; PGP13 (P-GLYCOPROTEIN 13); ATPase, couple... 129 7e-30
ath:AT3G28390 PGP18; PGP18 (P-GLYCOPROTEIN 18); ATPase, couple... 129 7e-30
ath:AT3G55320 PGP20; PGP20 (P-GLYCOPROTEIN 20); ATPase, couple... 129 8e-30
hsa:5243 ABCB1, ABC20, CD243, CLCS, GP170, MDR1, MGC163296, P-... 128 1e-29
mmu:27413 Abcb11, ABC16, Bsep, Lith1, PFIC2, PGY4, SPGP; ATP-b... 127 1e-29
ath:AT3G28415 P-glycoprotein, putative 127 2e-29
ath:AT4G01820 PGP3; PGP3 (P-GLYCOPROTEIN 3); ATPase, coupled t... 127 2e-29
ath:AT3G28360 PGP16; PGP16 (P-GLYCOPROTEIN 16); ATPase, couple... 126 4e-29
hsa:5244 ABCB4, ABC21, GBD1, MDR2, MDR2/3, MDR3, PFIC-3, PGY3;... 125 9e-29
xla:397812 abcb1, xemdr; ATP-binding cassette, sub-family B (M... 124 1e-28
mmu:18669 Abcb1b, Abcb1, Mdr1, Mdr1b, Pgy-1, Pgy1, mdr; ATP-bi... 124 1e-28
ath:AT1G10680 PGP10; PGP10 (P-GLYCOPROTEIN 10); ATPase, couple... 124 2e-28
cel:C47A10.1 pgp-9; P-GlycoProtein related family member (pgp-9) 124 2e-28
ath:AT1G28010 PGP14; PGP14 (P-GLYCOPROTEIN 14); ATPase, couple... 124 2e-28
hsa:8647 ABCB11, ABC16, BRIC2, BSEP, PFIC-2, PFIC2, PGY4, SPGP... 123 3e-28
cel:T21E8.1 pgp-6; P-GlycoProtein related family member (pgp-6... 123 3e-28
cel:T21E8.2 pgp-7; P-GlycoProtein related family member (pgp-7... 123 3e-28
mmu:18671 Abcb1a, Abcb4, Evi32, Mdr1a, Mdr3, P-gp, Pgp, Pgy-3,... 123 4e-28
cel:F22E10.1 pgp-12; P-GlycoProtein related family member (pgp... 122 6e-28
mmu:56199 Abcb10, AU016331, Abc-me, Abcb12, C76534; ATP-bindin... 122 7e-28
dre:100136865 abcb4, zgc:172149; ATP-binding cassette, sub-fam... 122 8e-28
pfa:PFE1150w PfMDR1, Pgh-1; multidrug resistance protein 122 9e-28
mmu:77706 Abcb5, 9230106F14Rik; ATP-binding cassette, sub-fami... 121 1e-27
mmu:18670 Abcb4, Mdr2, Pgy-2, Pgy2, mdr-2; ATP-binding cassett... 121 1e-27
cel:F22E10.3 pgp-14; P-GlycoProtein related family member (pgp... 120 2e-27
cel:T21E8.3 pgp-8; P-GlycoProtein related family member (pgp-8... 120 3e-27
cel:DH11.3 pgp-11; P-GlycoProtein related family member (pgp-1... 120 4e-27
ath:AT4G25960 PGP2; PGP2 (P-GLYCOPROTEIN 2); ATPase, coupled t... 119 4e-27
ath:AT3G28345 ABC transporter family protein 119 6e-27
dre:571189 abcb11b; ATP-binding cassette, sub-family B (MDR/TA... 119 7e-27
> tgo:TGME49_039020 ATP-binding cassette protein subfamily B member
2 (EC:3.6.3.30 3.6.3.44); K05658 ATP-binding cassette,
subfamily B (MDR/TAP), member 1
Length=1407
Score = 192 bits (489), Expect = 4e-49, Method: Compositional matrix adjust.
Identities = 99/183 (54%), Positives = 132/183 (72%), Gaps = 3/183 (1%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+DRPS+ID RD+GGKV+ + F G + V+++ F YP RP+ VY+ L+F +K GE VALV
Sbjct 1104 LDRPSKIDSRDTGGKVIDAKDFSGLVRVEKVRFRYPERPNIPVYQELSFEMKPGETVALV 1163
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEP-NKEAER-VHIS-EGRILLDGDD 117
GASGCGKST+VQL+ERFYDL+++ V+ + S+ P +A R V + GRI LDG D
Sbjct 1164 GASGCGKSTVVQLLERFYDLENSSGVETGASSDPSGPRTADATREVELPRNGRITLDGID 1223
Query 118 LRELNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFP 177
+RE+N+ S+R GLV QEP+LF M++ +NI +KP AT EE+ AAKLANA FISTFP
Sbjct 1224 IREINIQSLRSLIGLVGQEPVLFSMSVADNIRYAKPEATLEEVVHAAKLANADSFISTFP 1283
Query 178 AGY 180
GY
Sbjct 1284 DGY 1286
Score = 106 bits (264), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 66/181 (36%), Positives = 99/181 (54%), Gaps = 29/181 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I+R S ID G+ + +I + + F+YP R + +++ L ++ G+ +ALV
Sbjct 422 IERQSPIDPLSPEGR--KDVLVQADIRFENVVFSYPARKEKKIFNGLNLTLPAGKTIALV 479
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G+SG GKSTIVQ+++R YD P+ EG I + +++
Sbjct 480 GSSGSGKSTIVQMLQRLYD-----------------PD---------EGCIFIGDIPIKD 513
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAI-SKPGATQEEIREAAKLANAAGFISTFPAG 179
+N+ +R Q+G+VSQE LF ++I ENIA+ + TQEEI EAAK ANA FIS FP
Sbjct 514 INIQYLRAQQGIVSQEAKLFSISIGENIALGADHPVTQEEIEEAAKKANAHDFISQFPDK 573
Query 180 Y 180
Y
Sbjct 574 Y 574
> tgo:TGME49_060310 ATP-binding cassette protein subfamily B member
1 (EC:3.6.3.44); K05658 ATP-binding cassette, subfamily
B (MDR/TAP), member 1
Length=1345
Score = 164 bits (416), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 88/181 (48%), Positives = 113/181 (62%), Gaps = 8/181 (4%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKG-EIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVAL 59
IDR S ID RD+GG+ S G +I + F YPTR VYR L+F+IK GE+VAL
Sbjct 1050 IDRTSAIDTRDTGGRKFSDNANTGVDIAFTDVQFRYPTRTTLPVYRGLSFTIKAGESVAL 1109
Query 60 VGASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLR 119
VG SGCGKST +QL+ERFYDL+S+ K++ R G I G +LR
Sbjct 1110 VGPSGCGKSTAIQLLERFYDLQSSVAANACDA-------KDSRRKTKGGGSITFAGTELR 1162
Query 120 ELNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAG 179
+ NV S+R+Q G V QEP+LF+M++E+NI S P A+Q +I EAAK A A FI +FP G
Sbjct 1163 DANVSSLREQMGFVGQEPVLFNMSVEDNIRFSCPDASQTDIIEAAKQAQADSFIRSFPQG 1222
Query 180 Y 180
Y
Sbjct 1223 Y 1223
Score = 108 bits (270), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 73/181 (40%), Positives = 101/181 (55%), Gaps = 29/181 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I+R S++D D G L G+I + + F YP+RP+ V ++L I G++VALV
Sbjct 368 INRKSQLDPLDESG--LKDVNLTGDIVYENVTFAYPSRPEKTVLKDLNLRIPAGKSVALV 425
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SGCGKSTIVQL++R Y EP S G + + L+E
Sbjct 426 GGSGCGKSTIVQLLQRQY-----------------EP---------SAGTVRVGNTPLKE 459
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAI-SKPGATQEEIREAAKLANAAGFISTFPAG 179
+N+ ++R+ G+VSQEP LF T+EENIA+ S+ T E+ AA+ ANAAGFIS F
Sbjct 460 INLATLREGLGVVSQEPRLFSATVEENIALGSRSRVTHAEVEAAAQKANAAGFISDFAEK 519
Query 180 Y 180
Y
Sbjct 520 Y 520
> ath:AT4G01830 PGP5; PGP5 (P-GLYCOPROTEIN 5); ATPase, coupled
to transmembrane movement of substances
Length=1230
Score = 144 bits (364), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 85/182 (46%), Positives = 111/182 (60%), Gaps = 30/182 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
IDR S+ID RD G VL E KG+I++ I+FTY TRPD +V+R+L SI+ G+ VALV
Sbjct 962 IDRISKIDSRDESGMVL--ENVKGDIELCHISFTYQTRPDVQVFRDLCLSIRAGQTVALV 1019
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L++RFYD P+ G I LDG +L++
Sbjct 1020 GESGSGKSTVISLLQRFYD-----------------PD---------SGHITLDGVELKK 1053
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPG--ATQEEIREAAKLANAAGFISTFPA 178
L + +RQQ GLV QEP+LF+ TI NIA K G AT+ EI A++LANA FIS+
Sbjct 1054 LRLKWLRQQMGLVGQEPVLFNDTIRANIAYGKGGEEATEAEIIAASELANAHRFISSIQK 1113
Query 179 GY 180
GY
Sbjct 1114 GY 1115
Score = 135 bits (340), Expect = 7e-32, Method: Compositional matrix adjust.
Identities = 78/179 (43%), Positives = 100/179 (55%), Gaps = 28/179 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I+R ID D GKVL E +GEI++ + F+YP RP V+ + I G ALV
Sbjct 330 IEREPLIDTFDLNGKVL--EDIRGEIELRDVCFSYPARPKEEVFGGFSLLIPSGTTTALV 387
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L+ERFYD PN G++L+DG DL+E
Sbjct 388 GESGSGKSTVISLIERFYD-----------------PN---------SGQVLIDGVDLKE 421
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAG 179
+ +R + GLVSQEP+LF +I ENI K GAT EEI+ A+KLANAA FI P G
Sbjct 422 FQLKWIRGKIGLVSQEPVLFSSSIMENIGYGKEGATVEEIQAASKLANAAKFIDKLPLG 480
> ath:AT3G62150 PGP21; PGP21 (P-GLYCOPROTEIN 21); ATPase, coupled
to transmembrane movement of substances
Length=1292
Score = 144 bits (362), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 77/179 (43%), Positives = 107/179 (59%), Gaps = 28/179 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I R EID D+ GKVL + +G+I+++ +NF+YP RP+ +++R + SI G VALV
Sbjct 380 IKRKPEIDASDTTGKVL--DDIRGDIELNNVNFSYPARPEEQIFRGFSLSISSGSTVALV 437
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST+V L+ERFYD +S G + +DG +L+E
Sbjct 438 GQSGSGKSTVVSLIERFYDPQS--------------------------GEVRIDGINLKE 471
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAG 179
+ +R + GLVSQEP+LF +I+ENIA K AT EEIR+A +LANA+ FI P G
Sbjct 472 FQLKWIRSKIGLVSQEPVLFTSSIKENIAYGKENATVEEIRKATELANASKFIDKLPQG 530
Score = 142 bits (358), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 80/181 (44%), Positives = 112/181 (61%), Gaps = 29/181 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
IDR S+ID D G+VL + KG+I++ I+F YP+RPD +++++L SI+ G+ +ALV
Sbjct 1025 IDRESKIDPSDESGRVL--DNVKGDIELRHISFKYPSRPDVQIFQDLCLSIRAGKTIALV 1082
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L++RFYD P+ G+I LDG +++
Sbjct 1083 GESGSGKSTVIALLQRFYD-----------------PD---------SGQITLDGVEIKT 1116
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPG-ATQEEIREAAKLANAAGFISTFPAG 179
L + +RQQ GLVSQEP+LF+ TI NIA K G AT+ EI AA+L+NA GFIS G
Sbjct 1117 LQLKWLRQQTGLVSQEPVLFNETIRANIAYGKGGDATETEIVSAAELSNAHGFISGLQQG 1176
Query 180 Y 180
Y
Sbjct 1177 Y 1177
> cel:Y48G8AL.11 haf-6; HAlF transporter (PGP related) family
member (haf-6); K05655 ATP-binding cassette, subfamily B (MDR/TAP),
member 8
Length=668
Score = 142 bits (357), Expect = 9e-34, Method: Compositional matrix adjust.
Identities = 76/168 (45%), Positives = 101/168 (60%), Gaps = 27/168 (16%)
Query 14 GKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQL 73
G + + + G+I + ++F+YPTRP + V+ NLT SI G+ VAL G SG GKSTI L
Sbjct 413 GVCIPYHSLWGDIKFEDVSFSYPTRPGHTVFENLTLSIPAGQVVALCGPSGEGKSTITHL 472
Query 74 VERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQE-GL 132
+ERFY+ KS GR+ LDG DLRELNV +R Q GL
Sbjct 473 LERFYEPKS--------------------------GRVTLDGRDLRELNVEWLRGQVIGL 506
Query 133 VSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
+SQEP+LF ++EENI +P AT EE+REAA+ A+ F+S FP+GY
Sbjct 507 ISQEPVLFATSVEENIRYGRPDATDEEVREAARAAHVDEFVSRFPSGY 554
> dre:798527 abcb5, im:7158730, mdr1, wu:fc18f02, wu:fi81f06;
ATP-binding cassette, sub-family B (MDR/TAP), member 5; K05658
ATP-binding cassette, subfamily B (MDR/TAP), member 1
Length=1340
Score = 140 bits (354), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 80/182 (43%), Positives = 106/182 (58%), Gaps = 30/182 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+++ EID+ D G+ S TF G ID + F+YPTRP+ +V + L S+++G+ +ALV
Sbjct 1074 LEKKPEIDIYDESGERPS--TFSGNIDFKDVQFSYPTRPNVKVLQGLNVSVRQGQTLALV 1131
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G+SGCGKST +QL+ERFYD P G++L+DG D +
Sbjct 1132 GSSGCGKSTTIQLLERFYD--------PAG------------------GQVLVDGRDSKS 1165
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAI--SKPGATQEEIREAAKLANAAGFISTFPA 178
+N+ +R Q GLVSQEPILFD TI ENI + TQEEI EAAK AN FI T P
Sbjct 1166 VNLAWLRTQMGLVSQEPILFDCTISENIQYGDNSQTVTQEEIEEAAKKANIHNFILTLPD 1225
Query 179 GY 180
Y
Sbjct 1226 KY 1227
Score = 124 bits (312), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 70/177 (39%), Positives = 94/177 (53%), Gaps = 28/177 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
ID P ID G + +G+I+ INF YP+R D + + ++ + G+ +ALV
Sbjct 415 IDMPRPIDSSSKEGH--KPDRVRGDIEFKNINFNYPSRKDVTILQGMSLKVPHGKTIALV 472
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
GASGCGKST +QL++RFYD S G + LDG D+R
Sbjct 473 GASGCGKSTTIQLLQRFYDPDS--------------------------GEVTLDGHDIRS 506
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFP 177
LNV +R+ G+VSQEP+LF TI ENI + AT +I +A K ANA FIS P
Sbjct 507 LNVRWLRENMGIVSQEPVLFGTTIAENIRYGREDATDADIEQAIKEANAYDFISKLP 563
> dre:100330068 ATP-binding cassette, subfamily B, member 1B-like
Length=1063
Score = 140 bits (354), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 80/182 (43%), Positives = 106/182 (58%), Gaps = 30/182 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+++ EID+ D G+ S TF G ID + F+YPTRP+ +V + L S+++G+ +ALV
Sbjct 797 LEKKPEIDIYDESGERPS--TFSGNIDFKDVQFSYPTRPNVKVLQGLNVSVRQGQTLALV 854
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G+SGCGKST +QL+ERFYD P G++L+DG D +
Sbjct 855 GSSGCGKSTTIQLLERFYD--------PAG------------------GQVLVDGRDSKS 888
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAI--SKPGATQEEIREAAKLANAAGFISTFPA 178
+N+ +R Q GLVSQEPILFD TI ENI + TQEEI EAAK AN FI T P
Sbjct 889 VNLAWLRTQMGLVSQEPILFDCTISENIQYGDNSRTVTQEEIEEAAKKANIHNFILTLPD 948
Query 179 GY 180
Y
Sbjct 949 KY 950
Score = 124 bits (312), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 70/177 (39%), Positives = 94/177 (53%), Gaps = 28/177 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
ID P ID G + +G+I+ INF YP+R D + + ++ + G+ +ALV
Sbjct 140 IDMPRPIDSSSKEGH--KPDRVRGDIEFKNINFNYPSRKDVTILQGMSLKVPHGKTIALV 197
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
GASGCGKST +QL++RFYD S G + LDG D+R
Sbjct 198 GASGCGKSTTIQLLQRFYDPDS--------------------------GEVTLDGHDIRS 231
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFP 177
LNV +R+ G+VSQEP+LF TI ENI + AT +I +A K ANA FIS P
Sbjct 232 LNVRWLRENMGIVSQEPVLFGTTIAENIRYGREDATDADIEQAIKEANAYDFISKLP 288
> ath:AT1G02520 PGP11; PGP11 (P-GLYCOPROTEIN 11); ATPase, coupled
to transmembrane movement of substances
Length=1278
Score = 140 bits (354), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 81/183 (44%), Positives = 109/183 (59%), Gaps = 31/183 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
IDR S+ID D G VL E KG+I++ ++FTYP RPD +++R+L +I+ G+ VALV
Sbjct 1009 IDRKSKIDSSDETGTVL--ENVKGDIELRHLSFTYPARPDIQIFRDLCLTIRAGKTVALV 1066
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L++RFYD P+ G I LDG +L++
Sbjct 1067 GESGSGKSTVISLLQRFYD-----------------PD---------SGHITLDGVELKK 1100
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISK---PGATQEEIREAAKLANAAGFISTFP 177
L + +RQQ GLV QEP+LF+ TI NIA K AT+ EI AA+LANA FIS+
Sbjct 1101 LQLKWLRQQMGLVGQEPVLFNDTIRANIAYGKGSEEAATESEIIAAAELANAHKFISSIQ 1160
Query 178 AGY 180
GY
Sbjct 1161 QGY 1163
Score = 134 bits (336), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 75/179 (41%), Positives = 99/179 (55%), Gaps = 28/179 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I R ID D GKVL E +G+I++ ++F+YP RPD ++ + I G ALV
Sbjct 358 IKRKPLIDAYDVNGKVL--EDIRGDIELKDVHFSYPARPDEEIFDGFSLFIPSGATAALV 415
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L+ERFYD KS G +L+DG +L+E
Sbjct 416 GESGSGKSTVISLIERFYDPKS--------------------------GAVLIDGVNLKE 449
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAG 179
+ +R + GLVSQEP+LF +I ENIA K AT EEI+ A +LANAA FI P G
Sbjct 450 FQLKWIRSKIGLVSQEPVLFSSSIMENIAYGKENATVEEIKAATELANAAKFIDKLPQG 508
> cel:C34G6.4 pgp-2; P-GlycoProtein related family member (pgp-2)
Length=1272
Score = 139 bits (349), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 72/161 (44%), Positives = 98/161 (60%), Gaps = 26/161 (16%)
Query 20 ETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYD 79
+ KG+I ++F YP+R D V + ++ +K G+ +ALVG+SGCGKSTIV L++RFYD
Sbjct 393 DNMKGDISFKDVHFRYPSRKDIHVLKGISLELKAGDKIALVGSSGCGKSTIVNLLQRFYD 452
Query 80 LKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPIL 139
P K GR+L+DG DLRE+NV S+R+Q G+VSQEP+L
Sbjct 453 -----------------PTK---------GRVLIDGVDLREVNVHSLREQIGIVSQEPVL 486
Query 140 FDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
FD TI ENI + AT +++ EA K+ANA FI P GY
Sbjct 487 FDGTIYENIKMGNEHATHDQVVEACKMANANDFIKRLPDGY 527
Score = 130 bits (326), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 76/181 (41%), Positives = 100/181 (55%), Gaps = 30/181 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I+ P+ ID G V + G I + + F YPTR D +V + T IK G+ VALV
Sbjct 1006 IEHPTPIDSLSDSGIV---KPITGNISIRNVFFNYPTRKDTKVLQGFTLDIKAGKTVALV 1062
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SGCGKSTI+ L+ERFY N++ +G I++DGD++R
Sbjct 1063 GHSGCGKSTIMGLLERFY-------------------NQD-------KGMIMIDGDNIRN 1096
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAI-SKPGATQEEIREAAKLANAAGFISTFPAG 179
LN+ S+R+Q +VSQEP LFD TI ENI + T +EI EAAK+AN FI P G
Sbjct 1097 LNISSLREQVCIVSQEPTLFDCTIGENICYGTNRNVTYQEIVEAAKMANIHNFILGLPDG 1156
Query 180 Y 180
Y
Sbjct 1157 Y 1157
> cel:K08E7.9 pgp-1; P-GlycoProtein related family member (pgp-1)
Length=1321
Score = 138 bits (347), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 72/180 (40%), Positives = 104/180 (57%), Gaps = 28/180 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+DR ID G+ KG+I V+ ++FTYP+RPD + R + + G+ VALV
Sbjct 393 LDRKPVIDSSSKAGR--KDMKIKGDITVENVHFTYPSRPDVPILRGMNLRVNAGQTVALV 450
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G+SGCGKSTI+ L+ R+YD + +G+I +DG D+R+
Sbjct 451 GSSGCGKSTIISLLLRYYD--------------------------VLKGKITIDGVDVRD 484
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
+N+ +R+ +VSQEP LF+ TIEENI++ K G T+EE+ A K+ANA FI T P GY
Sbjct 485 INLEFLRKNVAVVSQEPALFNCTIEENISLGKEGITREEMVAACKMANAEKFIKTLPNGY 544
Score = 110 bits (276), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 59/159 (37%), Positives = 84/159 (52%), Gaps = 28/159 (17%)
Query 24 GEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYDLKST 83
G++ + F YP RP+ + + L+FS++ G+ +ALVG SGCGKST+V L+ERFYD
Sbjct 1075 GKVIFKNVRFAYPERPEIEILKGLSFSVEPGQTLALVGPSGCGKSTVVALLERFYDTLG- 1133
Query 84 YKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPILFDMT 143
G I +DG +++ LN R Q +VSQEP LFD +
Sbjct 1134 -------------------------GEIFIDGSEIKTLNPEHTRSQIAIVSQEPTLFDCS 1168
Query 144 IEENI--AISKPGATQEEIREAAKLANAAGFISTFPAGY 180
I ENI + T ++ EAA+LAN FI+ P G+
Sbjct 1169 IAENIIYGLDPSSVTMAQVEEAARLANIHNFIAELPEGF 1207
> ath:AT1G02530 PGP12; PGP12 (P-GLYCOPROTEIN 12); ATPase, coupled
to transmembrane movement of substances
Length=1273
Score = 138 bits (347), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 80/183 (43%), Positives = 109/183 (59%), Gaps = 31/183 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
IDR S+ID D G VL E KG+I++ ++FTYP RP +++R+L +I+ G+ VALV
Sbjct 1004 IDRKSKIDSSDETGTVL--ENVKGDIELRHLSFTYPARPGIQIFRDLCLTIRAGKTVALV 1061
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L++RFYD P+ G+I LDG +L++
Sbjct 1062 GESGSGKSTVISLLQRFYD-----------------PD---------SGQITLDGVELKK 1095
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISK---PGATQEEIREAAKLANAAGFISTFP 177
L + +RQQ GLV QEP+LF+ TI NIA K AT+ EI AA+LANA FIS+
Sbjct 1096 LQLKWLRQQMGLVGQEPVLFNDTIRANIAYGKGSEEAATESEIIAAAELANAHKFISSIQ 1155
Query 178 AGY 180
GY
Sbjct 1156 QGY 1158
Score = 129 bits (323), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 71/179 (39%), Positives = 98/179 (54%), Gaps = 28/179 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I R ID D GKVL +G+I++ ++F+YP RPD ++ + I G ALV
Sbjct 345 IKRKPLIDAYDVNGKVLGD--IRGDIELKDVHFSYPARPDEEIFDGFSLFIPSGATAALV 402
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L+ERFYD K+ G +L+DG +L+E
Sbjct 403 GESGSGKSTVINLIERFYDPKA--------------------------GEVLIDGINLKE 436
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAG 179
+ +R + GLV QEP+LF +I ENIA K AT +EI+ A +LANAA FI+ P G
Sbjct 437 FQLKWIRSKIGLVCQEPVLFSSSIMENIAYGKENATLQEIKVATELANAAKFINNLPQG 495
> ath:AT3G28860 ABCB19; ABCB19; ATPase, coupled to transmembrane
movement of substances / auxin efflux transmembrane transporter
Length=1252
Score = 136 bits (343), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 69/161 (42%), Positives = 99/161 (61%), Gaps = 26/161 (16%)
Query 20 ETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYD 79
ET +G+I+ ++F YP+RPD V+R+ I+ G + ALVGASG GKS+++ ++ERFYD
Sbjct 1004 ETIRGDIEFRHVDFAYPSRPDVMVFRDFNLRIRAGHSQALVGASGSGKSSVIAMIERFYD 1063
Query 80 LKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPIL 139
P++ G++++DG D+R LN+ S+R + GLV QEP L
Sbjct 1064 --------PLA------------------GKVMIDGKDIRRLNLKSLRLKIGLVQQEPAL 1097
Query 140 FDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
F TI +NIA K GAT+ E+ +AA+ ANA GFIS P GY
Sbjct 1098 FAATIFDNIAYGKDGATESEVIDAARAANAHGFISGLPEGY 1138
Score = 114 bits (285), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 70/167 (41%), Positives = 91/167 (54%), Gaps = 28/167 (16%)
Query 14 GKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQL 73
GK L + G I+ + F+YP+RPD ++RN G+ VA+VG SG GKST+V L
Sbjct 355 GKCL--DQVHGNIEFKDVTFSYPSRPDVMIFRNFNIFFPSGKTVAVVGGSGSGKSTVVSL 412
Query 74 VERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLV 133
+ERFYD PN G+ILLDG +++ L + +R+Q GLV
Sbjct 413 IERFYD-----------------PN---------SGQILLDGVEIKTLQLKFLREQIGLV 446
Query 134 SQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
+QEP LF TI ENI KP AT E+ AA ANA FI+ P GY
Sbjct 447 NQEPALFATTILENILYGKPDATMVEVEAAASAANAHSFITLLPKGY 493
> ath:AT2G47000 ABCB4; ABCB4 (ATP BINDING CASSETTE SUBFAMILY B4);
ATPase, coupled to transmembrane movement of substances
/ xenobiotic-transporting ATPase
Length=1286
Score = 135 bits (341), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 74/179 (41%), Positives = 104/179 (58%), Gaps = 28/179 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I+R ID + GKVL + KG+I++ + FTYP RPD +++R + I G VALV
Sbjct 361 IERRPNIDSYSTNGKVL--DDIKGDIELKDVYFTYPARPDEQIFRGFSLFISSGTTVALV 418
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST+V L+ERFYD ++ G +L+DG +L+E
Sbjct 419 GQSGSGKSTVVSLIERFYDPQA--------------------------GDVLIDGINLKE 452
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAG 179
+ +R + GLVSQEP+LF +I++NIA K AT EEI+ AA+LANA+ F+ P G
Sbjct 453 FQLKWIRSKIGLVSQEPVLFTASIKDNIAYGKEDATTEEIKAAAELANASKFVDKLPQG 511
Score = 134 bits (337), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 77/181 (42%), Positives = 109/181 (60%), Gaps = 29/181 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+DR S+ID G+VL + KG+I++ ++F YP RPD +++++L SI+ G+ VALV
Sbjct 1019 MDRESKIDPSVESGRVL--DNVKGDIELRHVSFKYPARPDVQIFQDLCLSIRAGKTVALV 1076
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L++RFYD P+ G I LDG +++
Sbjct 1077 GESGSGKSTVIALLQRFYD-----------------PD---------SGEITLDGVEIKS 1110
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPG-ATQEEIREAAKLANAAGFISTFPAG 179
L + +RQQ GLVSQEPILF+ TI NIA K G A++ EI +A+L+NA GFIS G
Sbjct 1111 LRLKWLRQQTGLVSQEPILFNETIRANIAYGKGGDASESEIVSSAELSNAHGFISGLQQG 1170
Query 180 Y 180
Y
Sbjct 1171 Y 1171
> ath:AT2G36910 ABCB1; ABCB1 (ATP BINDING CASSETTE SUBFAMILY B1);
ATPase, coupled to transmembrane movement of substances
/ auxin efflux transmembrane transporter/ calmodulin binding;
K05658 ATP-binding cassette, subfamily B (MDR/TAP), member
1
Length=1286
Score = 134 bits (338), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 68/161 (42%), Positives = 97/161 (60%), Gaps = 26/161 (16%)
Query 20 ETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYD 79
++ G +++ ++F+YP+RPD ++ N S+ G+ +ALVG+SG GKST+V L+ERFYD
Sbjct 362 DSVTGLVELKNVDFSYPSRPDVKILNNFCLSVPAGKTIALVGSSGSGKSTVVSLIERFYD 421
Query 80 LKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPIL 139
PN G++LLDG DL+ L + +RQQ GLVSQEP L
Sbjct 422 -----------------PN---------SGQVLLDGQDLKTLKLRWLRQQIGLVSQEPAL 455
Query 140 FDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
F +I+ENI + +P A Q EI EAA++ANA FI P G+
Sbjct 456 FATSIKENILLGRPDADQVEIEEAARVANAHSFIIKLPDGF 496
Score = 132 bits (333), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 69/180 (38%), Positives = 107/180 (59%), Gaps = 27/180 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+DR +EI+ D + + +GE+++ I+F+YP+RPD +++R+L+ + G+ +ALV
Sbjct 1000 LDRKTEIEPDDPDTTPVP-DRLRGEVELKHIDFSYPSRPDIQIFRDLSLRARAGKTLALV 1058
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SGCGKS+++ L++RFY EP S GR+++DG D+R+
Sbjct 1059 GPSGCGKSSVISLIQRFY-----------------EP---------SSGRVMIDGKDIRK 1092
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
N+ ++R+ +V QEP LF TI ENIA AT+ EI +AA LA+A FIS P GY
Sbjct 1093 YNLKAIRKHIAIVPQEPCLFGTTIYENIAYGHECATEAEIIQAATLASAHKFISALPEGY 1152
> ath:AT5G46540 PGP7; PGP7 (P-GLYCOPROTEIN 7); ATPase, coupled
to transmembrane movement of substances
Length=1248
Score = 134 bits (337), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 75/179 (41%), Positives = 100/179 (55%), Gaps = 28/179 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I R +ID D G+VL E KG+I++ + F YP RPD +++ + ++ G VALV
Sbjct 334 IKRKPKIDAYDMSGEVL--EEIKGDIELRDVYFRYPARPDVQIFVGFSLTVPNGMTVALV 391
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L+ERFYD +S G +L+DG DL++
Sbjct 392 GQSGSGKSTVISLIERFYDPES--------------------------GEVLIDGIDLKK 425
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAG 179
V +R + GLVSQEPILF TI ENI K A+ +EIR A KLANA+ FI P G
Sbjct 426 FQVKWIRSKIGLVSQEPILFATTIRENIVYGKKDASDQEIRTALKLANASNFIDKLPQG 484
Score = 129 bits (323), Expect = 8e-30, Method: Compositional matrix adjust.
Identities = 74/181 (40%), Positives = 103/181 (56%), Gaps = 29/181 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+D +ID G +L G+I++ ++F YP RPD +++ +L +I G+ VALV
Sbjct 982 LDSKPKIDSSSEKGTIL--PIVHGDIELQHVSFRYPMRPDIQIFSDLCLTISSGQTVALV 1039
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L+ERFYD P+ G+ILLD +++
Sbjct 1040 GESGSGKSTVISLLERFYD-----------------PD---------SGKILLDQVEIQS 1073
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISK-PGATQEEIREAAKLANAAGFISTFPAG 179
L + +R+Q GLVSQEP+LF+ TI NIA K GAT+EEI AAK AN FIS+ P G
Sbjct 1074 LKLSWLREQMGLVSQEPVLFNETIGSNIAYGKIGGATEEEIITAAKAANVHNFISSLPQG 1133
Query 180 Y 180
Y
Sbjct 1134 Y 1134
> ath:AT4G18050 PGP9; PGP9 (P-GLYCOPROTEIN 9); ATPase, coupled
to transmembrane movement of substances
Length=1236
Score = 133 bits (334), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 74/179 (41%), Positives = 102/179 (56%), Gaps = 28/179 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I R +ID D G VL E +G+I++ + F YP RPD +++ + + G+ VALV
Sbjct 332 IKRSPKIDAYDMSGSVL--EDIRGDIELKDVYFRYPARPDVQIFAGFSLFVPNGKTVALV 389
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L+ERFYD +S G++L+D DL++
Sbjct 390 GQSGSGKSTVISLIERFYDPES--------------------------GQVLIDNIDLKK 423
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAG 179
L + +R + GLVSQEP+LF TI+ENIA K AT +EIR A +LANAA FI P G
Sbjct 424 LQLKWIRSKIGLVSQEPVLFATTIKENIAYGKEDATDQEIRTAIELANAAKFIDKLPQG 482
Score = 117 bits (292), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 74/181 (40%), Positives = 102/181 (56%), Gaps = 29/181 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+D +ID G L + G+I+ ++F YP RPD +++R+L +I G+ VALV
Sbjct 970 LDSTPKIDSSSDEGTTL--QNVNGDIEFRHVSFRYPMRPDVQIFRDLCLTIPSGKTVALV 1027
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ ++ERFY+ S G+IL+D +++
Sbjct 1028 GESGSGKSTVISMIERFYNPDS--------------------------GKILIDQVEIQT 1061
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKP-GATQEEIREAAKLANAAGFISTFPAG 179
+ +RQQ GLVSQEPILF+ TI NIA K GAT+EEI AAK ANA FIS+ P G
Sbjct 1062 FKLSWLRQQMGLVSQEPILFNETIRSNIAYGKTGGATEEEIIAAAKAANAHNFISSLPQG 1121
Query 180 Y 180
Y
Sbjct 1122 Y 1122
> ath:AT2G39480 PGP6; PGP6 (P-GLYCOPROTEIN 6); ATPase, coupled
to transmembrane movement of substances
Length=1407
Score = 131 bits (329), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 72/180 (40%), Positives = 102/180 (56%), Gaps = 28/180 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
IDR I+ D+ LS G I++ I+F YPTRP+ V N + + G+ VA+V
Sbjct 1135 IDRVPTIEPDDTSA--LSPPNVYGSIELKNIDFCYPTRPEVLVLSNFSLKVNGGQTVAVV 1192
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKSTI+ L+ER+YD PV+ G++LLDG DL+
Sbjct 1193 GVSGSGKSTIISLIERYYD--------PVA------------------GQVLLDGRDLKS 1226
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
N+ +R GL+ QEPI+F TI ENI ++ A++ E++EAA++ANA FIS+ P GY
Sbjct 1227 YNLRWLRSHMGLIQQEPIIFSTTIRENIIYARHNASEAEMKEAARIANAHHFISSLPHGY 1286
Score = 97.8 bits (242), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 61/167 (36%), Positives = 92/167 (55%), Gaps = 29/167 (17%)
Query 14 GKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQL 73
G +LS +G I+ + F+Y +RP+ + ++ +AVALVG +G GKS+I+ L
Sbjct 402 GIILS--AVQGNIEFRNVYFSYLSRPEIPILSGFYLTVPAKKAVALVGRNGSGKSSIIPL 459
Query 74 VERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLV 133
+ERFYD P + G +LLDG++++ L + +R Q GLV
Sbjct 460 MERFYD--------P------------------TLGEVLLDGENIKNLKLEWLRSQIGLV 493
Query 134 SQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
+QEP L ++I ENIA + AT ++I EAAK A+A FIS+ GY
Sbjct 494 TQEPALLSLSIRENIAYGR-DATLDQIEEAAKKAHAHTFISSLEKGY 539
> cel:C05A9.1 pgp-5; P-GlycoProtein related family member (pgp-5);
K02021 putative ABC transport system ATP-binding protein
Length=1252
Score = 131 bits (329), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 66/160 (41%), Positives = 97/160 (60%), Gaps = 26/160 (16%)
Query 20 ETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYD 79
+TF+G I + F+YPTRPD V + ++F+++ GE +ALVGASG GKST++QL+ +Y+
Sbjct 368 DTFQGIISFKNVLFSYPTRPDVPVLKEISFNVQGGECIALVGASGSGKSTVIQLLLHYYN 427
Query 80 LKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPIL 139
+ S GRI +DG+D+ +N+ +RQ G+V QEP+L
Sbjct 428 IDS--------------------------GRISIDGNDIYNINIKQLRQAMGVVFQEPVL 461
Query 140 FDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAG 179
F+ +IEENI KP AT++EI +A K ANA F+ FP G
Sbjct 462 FNTSIEENIRFGKPDATEQEIIDALKNANAFDFVCNFPDG 501
Score = 97.1 bits (240), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 54/157 (34%), Positives = 84/157 (53%), Gaps = 27/157 (17%)
Query 24 GEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYDLKST 83
GEI++ ++F Y R D + ++ + G +ALVG SG GKSTI+ L+ERFY
Sbjct 1003 GEIELKNVSFEYAQRSDKMILDGVSLKLPAGRTLALVGPSGSGKSTIISLLERFY----- 1057
Query 84 YKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPILFDMT 143
H +G + +D +++ ++N+ +R+ LVSQEP+LF+ +
Sbjct 1058 ---------------------HAVDGEVKIDEENVVDVNLHHLRESVSLVSQEPVLFNCS 1096
Query 144 IEENIAIS-KPGATQEEIREAAKLANAAGFISTFPAG 179
I+EN A+Q EI +A K+ANA F+S FP G
Sbjct 1097 IKENFLFGISHNASQLEIDQALKVANAFSFVSQFPQG 1133
> ath:AT3G28380 PGP17; PGP17 (P-GLYCOPROTEIN 17); ATP binding
/ ATPase/ ATPase, coupled to transmembrane movement of substances
/ nucleoside-triphosphatase/ nucleotide binding
Length=1240
Score = 130 bits (327), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 73/180 (40%), Positives = 102/180 (56%), Gaps = 28/180 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I R +ID G++L E KGE++ + + FTY +RP+ ++ +L I G+ VALV
Sbjct 336 IKRVPDIDSNKKEGQIL--ERMKGEVEFNHVKFTYLSRPETTIFDDLCLKIPAGKTVALV 393
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L++RFYD P++ G IL+DG + +
Sbjct 394 GGSGSGKSTVISLLQRFYD--------PIA------------------GEILIDGVSIDK 427
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
L V +R Q GLVSQEP+LF +I ENI K A+ +E+ EAAK +NA FIS FP GY
Sbjct 428 LQVNWLRSQMGLVSQEPVLFATSITENILFGKEDASLDEVVEAAKASNAHTFISQFPLGY 487
Score = 115 bits (288), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 72/183 (39%), Positives = 99/183 (54%), Gaps = 32/183 (17%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+DR + I+ ++ G V E KG+I ++F YPTRPD ++ N + I EG++ A+V
Sbjct 972 LDRCTTIEPKNPDGYV--AEKIKGQITFLNVDFAYPTRPDVVIFENFSIEIDEGKSTAIV 1029
Query 61 GASGCGKSTIVQLVERFYD-LKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLR 119
G SG GKSTI+ L+ERFYD LK T K+ DG D+R
Sbjct 1030 GTSGSGKSTIIGLIERFYDPLKGTVKI---------------------------DGRDIR 1062
Query 120 ELNVVSVRQQEGLVSQEPILFDMTIEENIAI--SKPGATQEEIREAAKLANAAGFISTFP 177
++ S+R+ LVSQEP+LF TI ENI + + EI EAAK ANA FI++
Sbjct 1063 SYHLRSLRKYISLVSQEPMLFAGTIRENIMYGGTSDKIDESEIIEAAKAANAHDFITSLS 1122
Query 178 AGY 180
GY
Sbjct 1123 NGY 1125
> dre:797269 abcb11a; ATP-binding cassette, sub-family B (MDR/TAP),
member 11a
Length=1325
Score = 129 bits (323), Expect = 6e-30, Method: Compositional matrix adjust.
Identities = 73/180 (40%), Positives = 97/180 (53%), Gaps = 28/180 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
IDR EID G L + KG+I+ +NF YP+RP+ ++ +L +K GE A V
Sbjct 395 IDREPEIDCFSDEGHTL--DKVKGDIEFHSVNFNYPSRPEVKILDDLNIVVKAGETTAFV 452
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GK+T +QL++RFYD P SEG + LDG D+R
Sbjct 453 GPSGSGKTTTIQLIQRFYD--------P------------------SEGMVSLDGHDIRS 486
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
LN+ +R G+V QEP+LF TI ENI + G T +EI EAAK ANA FI + P +
Sbjct 487 LNIQWLRSLIGVVEQEPVLFATTIAENIRYGRAGVTMQEIIEAAKQANAYNFIMSLPQTF 546
Score = 119 bits (298), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 73/182 (40%), Positives = 100/182 (54%), Gaps = 30/182 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+DR +I+V + G+ S FKG+++ FTYP+RPD +V R L S+ G+ +A V
Sbjct 1059 LDRVPKINVSKTEGQ--SWNDFKGKVEFKGCRFTYPSRPDVQVLRGLVVSVHPGQTLAFV 1116
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G+SGCGKST VQL+ERFYD P+ EG++L+DG
Sbjct 1117 GSSGCGKSTSVQLLERFYD-----------------PD---------EGQVLIDGRPSDS 1150
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGAT--QEEIREAAKLANAAGFISTFPA 178
++V +R Q G+VSQEP+LFD +I ENI T EEI +AAK A F+ T P
Sbjct 1151 ISVPFLRSQIGIVSQEPVLFDCSIAENIQYGDNSRTVSMEEIIDAAKKAYLHDFVMTLPD 1210
Query 179 GY 180
Y
Sbjct 1211 KY 1212
> ath:AT1G27940 PGP13; PGP13 (P-GLYCOPROTEIN 13); ATPase, coupled
to transmembrane movement of substances
Length=1245
Score = 129 bits (323), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 69/157 (43%), Positives = 91/157 (57%), Gaps = 27/157 (17%)
Query 24 GEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYDLKST 83
G I+ +++F YP+RP N V+ NL+F+I+ G+ A VG SG GKSTI+ +V+RFY
Sbjct 370 GRIEFQKVSFAYPSRP-NMVFENLSFTIRSGKTFAFVGPSGSGKSTIISMVQRFY----- 423
Query 84 YKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPILFDMT 143
EPN G ILLDG+D++ L + R+Q GLVSQEP LF T
Sbjct 424 ------------EPN---------SGEILLDGNDIKSLKLKWFREQLGLVSQEPALFATT 462
Query 144 IEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
I NI + K A ++I EAAK ANA FI + P GY
Sbjct 463 IASNILLGKENANMDQIIEAAKAANADSFIKSLPNGY 499
Score = 114 bits (286), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 62/158 (39%), Positives = 88/158 (55%), Gaps = 26/158 (16%)
Query 23 KGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYDLKS 82
KG+I+ ++F YPTRP+ +++NL + G+++A+VG SG GKST++ L+ RFYD
Sbjct 1001 KGDIEFRNVSFVYPTRPEIDIFKNLNLRVSAGKSLAVVGPSGSGKSTVIGLIMRFYD--- 1057
Query 83 TYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPILFDM 142
P S G + +DG D++ LN+ S+R++ LV QEP LF
Sbjct 1058 --------------P---------SNGNLCIDGQDIKTLNLRSLRKKLALVQQEPALFST 1094
Query 143 TIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
TI ENI A++ EI EAAK ANA FI GY
Sbjct 1095 TIYENIKYGNENASEAEIMEAAKAANAHEFIIKMEEGY 1132
> ath:AT3G28390 PGP18; PGP18 (P-GLYCOPROTEIN 18); ATPase, coupled
to transmembrane movement of substances
Length=1225
Score = 129 bits (323), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 71/180 (39%), Positives = 104/180 (57%), Gaps = 28/180 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I+R ID + G++L E +GE++ + + FTYP+RP+ ++ +L + G+ VALV
Sbjct 324 INRVPGIDSDNLEGQIL--EKTRGEVEFNHVKFTYPSRPETPIFDDLCLRVPSGKTVALV 381
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L++RFYD P++ G IL+DG + +
Sbjct 382 GGSGSGKSTVISLLQRFYD--------PIA------------------GEILIDGLPINK 415
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
L V +R Q GLVSQEP+LF +I+ENI K A+ +E+ EAAK +NA FIS FP Y
Sbjct 416 LQVKWLRSQMGLVSQEPVLFATSIKENILFGKEDASMDEVVEAAKASNAHSFISQFPNSY 475
Score = 113 bits (283), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 72/184 (39%), Positives = 101/184 (54%), Gaps = 34/184 (18%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+DR + I+ + G V + KG+I ++F YPTRPD +++N + I++G++ A+V
Sbjct 957 LDRNTTIEPENPDGYV--PKKVKGQISFSNVDFAYPTRPDVIIFQNFSIDIEDGKSTAIV 1014
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKSTI+ L+ERFYD P K G + +DG D+R
Sbjct 1015 GPSGSGKSTIISLIERFYD-----------------PLK---------GIVKIDGRDIRS 1048
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGAT----QEEIREAAKLANAAGFISTF 176
++ S+RQ LVSQEP LF TI ENI GA+ + EI EAAK ANA FI++
Sbjct 1049 CHLRSLRQHIALVSQEPTLFAGTIRENIMYG--GASNKIDESEIIEAAKAANAHDFITSL 1106
Query 177 PAGY 180
GY
Sbjct 1107 SNGY 1110
> ath:AT3G55320 PGP20; PGP20 (P-GLYCOPROTEIN 20); ATPase, coupled
to transmembrane movement of substances
Length=1408
Score = 129 bits (323), Expect = 8e-30, Method: Compositional matrix adjust.
Identities = 72/180 (40%), Positives = 101/180 (56%), Gaps = 28/180 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+DR I+ D+ L G I++ ++F YPTRP+ V N + I G+ VA+V
Sbjct 1136 VDRVPTIEPDDNSA--LKPPNVYGSIELKNVDFCYPTRPEILVLSNFSLKISGGQTVAVV 1193
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKSTI+ LVER+YD PV+ G++LLDG DL+
Sbjct 1194 GVSGSGKSTIISLVERYYD--------PVA------------------GQVLLDGRDLKL 1227
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
N+ +R GLV QEPI+F TI ENI ++ A++ E++EAA++ANA FIS+ P GY
Sbjct 1228 YNLRWLRSHMGLVQQEPIIFSTTIRENIIYARHNASEAEMKEAARIANAHHFISSLPHGY 1287
Score = 98.6 bits (244), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 62/173 (35%), Positives = 95/173 (54%), Gaps = 29/173 (16%)
Query 8 DVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGK 67
V + G VL+ + +G I+ + F+Y +RP+ + ++ +AVALVG +G GK
Sbjct 398 SVANQEGAVLA--SVQGNIEFRNVYFSYLSRPEIPILSGFYLTVPAKKAVALVGRNGSGK 455
Query 68 STIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVR 127
S+I+ L+ERFYD P + G +LLDG++++ L + +R
Sbjct 456 SSIIPLMERFYD--------P------------------TLGEVLLDGENIKNLKLEWLR 489
Query 128 QQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
Q GLV+QEP L ++I ENIA + AT ++I EAAK A+A FIS+ GY
Sbjct 490 SQIGLVTQEPALLSLSIRENIAYGR-DATLDQIEEAAKNAHAHTFISSLEKGY 541
> hsa:5243 ABCB1, ABC20, CD243, CLCS, GP170, MDR1, MGC163296,
P-GP, PGY1; ATP-binding cassette, sub-family B (MDR/TAP), member
1 (EC:3.6.3.44); K05658 ATP-binding cassette, subfamily
B (MDR/TAP), member 1
Length=1280
Score = 128 bits (321), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 75/182 (41%), Positives = 102/182 (56%), Gaps = 30/182 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I++ ID + G L T +G + + F YPTRPD V + L+ +K+G+ +ALV
Sbjct 1012 IEKTPLIDSYSTEG--LMPNTLEGNVTFGEVVFNYPTRPDIPVLQGLSLEVKKGQTLALV 1069
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G+SGCGKST+VQL+ERFYD P++ G++LLDG +++
Sbjct 1070 GSSGCGKSTVVQLLERFYD--------PLA------------------GKVLLDGKEIKR 1103
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPG--ATQEEIREAAKLANAAGFISTFPA 178
LNV +R G+VSQEPILFD +I ENIA +QEEI AAK AN FI + P
Sbjct 1104 LNVQWLRAHLGIVSQEPILFDCSIAENIAYGDNSRVVSQEEIVRAAKEANIHAFIESLPN 1163
Query 179 GY 180
Y
Sbjct 1164 KY 1165
Score = 113 bits (282), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 65/177 (36%), Positives = 93/177 (52%), Gaps = 28/177 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
ID ID G + KG ++ ++F+YP+R + ++ + L ++ G+ VALV
Sbjct 369 IDNKPSIDSYSKSGH--KPDNIKGNLEFRNVHFSYPSRKEVKILKGLNLKVQSGQTVALV 426
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SGCGKST VQL++R YD P +EG + +DG D+R
Sbjct 427 GNSGCGKSTTVQLMQRLYD--------P------------------TEGMVSVDGQDIRT 460
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFP 177
+NV +R+ G+VSQEP+LF TI ENI + T +EI +A K ANA FI P
Sbjct 461 INVRFLREIIGVVSQEPVLFATTIAENIRYGRENVTMDEIEKAVKEANAYDFIMKLP 517
> mmu:27413 Abcb11, ABC16, Bsep, Lith1, PFIC2, PGY4, SPGP; ATP-binding
cassette, sub-family B (MDR/TAP), member 11; K05664
ATP-binding cassette, subfamily B (MDR/TAP), member 11
Length=1321
Score = 127 bits (320), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 73/177 (41%), Positives = 96/177 (54%), Gaps = 28/177 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
IDR +D G L + KGEI+ + F YP+RP+ ++ NL+ IK GE A V
Sbjct 397 IDRQPVMDCMSGDGYKL--DRIKGEIEFHNVTFHYPSRPEVKILNNLSMVIKPGETTAFV 454
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G+SG GKST +QL++RFYD P EG + LDG D+R
Sbjct 455 GSSGAGKSTALQLIQRFYD--------P------------------CEGMVTLDGHDIRS 488
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFP 177
LN+ +R Q G+V QEP+LF TI ENI + + AT E+I +AAK ANA FI P
Sbjct 489 LNIRWLRDQIGIVEQEPVLFSTTIAENIRLGREEATMEDIVQAAKDANAYNFIMALP 545
Score = 114 bits (284), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 69/185 (37%), Positives = 99/185 (53%), Gaps = 36/185 (19%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+DR IDV G+ + F+G+ID FTYP+RPD +V L+ S+ G+ +A V
Sbjct 1055 LDRKPPIDVYSGAGE--KWDNFQGKIDFIDCKFTYPSRPDIQVLNGLSVSVDPGQTLAFV 1112
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G+SGCGKST +QL+ERFYD P+ +G +++DG D ++
Sbjct 1113 GSSGCGKSTSIQLLERFYD-----------------PD---------QGTVMIDGHDSKK 1146
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIR-----EAAKLANAAGFIST 175
+NV +R G+VSQEP+LFD +I +NI K G +EI AAK A F+ +
Sbjct 1147 VNVQFLRSNIGIVSQEPVLFDCSIMDNI---KYGDNTKEISVERAIAAAKQAQLHDFVMS 1203
Query 176 FPAGY 180
P Y
Sbjct 1204 LPEKY 1208
> ath:AT3G28415 P-glycoprotein, putative
Length=1221
Score = 127 bits (320), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 74/180 (41%), Positives = 101/180 (56%), Gaps = 28/180 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I R +ID + G+VL E KGE+ + F Y +RP+ ++ +L I G++VALV
Sbjct 315 IKRVPDIDSDNPRGQVL--ENIKGEVQFKHVKFMYSSRPETPIFDDLCLRIPSGKSVALV 372
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L++RFYD P+ G IL+DG +++
Sbjct 373 GGSGSGKSTVISLLQRFYD--------PIV------------------GEILIDGVSIKK 406
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
L V +R Q GLVSQEP LF +IEENI K A+ +E+ EAAK +NA FIS FP GY
Sbjct 407 LQVKWLRSQMGLVSQEPALFATSIEENILFGKEDASFDEVVEAAKSSNAHDFISQFPLGY 466
Score = 116 bits (291), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 76/184 (41%), Positives = 100/184 (54%), Gaps = 34/184 (18%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+DR + I+ G V + KG+I ++F YPTRPD +++N + I EG++ A+V
Sbjct 953 LDRYTNIEPEKPDGFV--PQNIKGQIKFVNVDFAYPTRPDVIIFKNFSIDIDEGKSTAIV 1010
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKSTI+ L+ERFYD P K G + +DG D+R
Sbjct 1011 GPSGSGKSTIIGLIERFYD-----------------PLK---------GIVKIDGRDIRS 1044
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGAT----QEEIREAAKLANAAGFISTF 176
++ S+RQ GLVSQEPILF TI ENI GA+ + EI EAAK ANA FI T
Sbjct 1045 YHLRSLRQHIGLVSQEPILFAGTIRENIMYG--GASDKIDESEIIEAAKAANAHDFIVTL 1102
Query 177 PAGY 180
GY
Sbjct 1103 SDGY 1106
> ath:AT4G01820 PGP3; PGP3 (P-GLYCOPROTEIN 3); ATPase, coupled
to transmembrane movement of substances
Length=1229
Score = 127 bits (319), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 84/182 (46%), Positives = 110/182 (60%), Gaps = 30/182 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
ID S ID RD G VL E KG+I++ I+FTY TRPD +++R+L F+I+ G+ VALV
Sbjct 961 IDGKSMIDSRDESGLVL--ENVKGDIELCHISFTYQTRPDVQIFRDLCFAIRAGQTVALV 1018
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L++RFYD P+ G I LD +L++
Sbjct 1019 GESGSGKSTVISLLQRFYD-----------------PD---------SGHITLDRVELKK 1052
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPG--ATQEEIREAAKLANAAGFISTFPA 178
L + VRQQ GLV QEP+LF+ TI NIA K G A++ EI AA+LANA GFIS+
Sbjct 1053 LQLKWVRQQMGLVGQEPVLFNDTIRSNIAYGKGGDEASEAEIIAAAELANAHGFISSIQQ 1112
Query 179 GY 180
GY
Sbjct 1113 GY 1114
Score = 112 bits (281), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 76/179 (42%), Positives = 98/179 (54%), Gaps = 28/179 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I+R ID D GKVL E +GEI++ + F+YP RP V+ + I G ALV
Sbjct 325 IERKPSIDAFDLNGKVL--EDIRGEIELRDVCFSYPARPMEEVFGGFSLLIPSGATAALV 382
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKS+++ L+ERFYD P S G +L+DG +L+E
Sbjct 383 GESGSGKSSVISLIERFYD--------P------------------SSGSVLIDGVNLKE 416
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAG 179
+ +R + GLVSQEP+LF +I ENI K AT EEI+ AAKLANAA FI P G
Sbjct 417 FQLKWIRGKIGLVSQEPVLFSSSIMENIGYGKENATVEEIQAAAKLANAANFIDKLPRG 475
> ath:AT3G28360 PGP16; PGP16 (P-GLYCOPROTEIN 16); ATPase, coupled
to transmembrane movement of substances
Length=1228
Score = 126 bits (317), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 71/180 (39%), Positives = 100/180 (55%), Gaps = 28/180 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I R +ID + G +L ET +GE++ + + YP+RP+ ++ +L I G+ VALV
Sbjct 323 IKRVPDIDSDNLNGHIL--ETIRGEVEFNNVKCKYPSRPETLIFDDLCLKIPSGKTVALV 380
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L++RFYD PN EG IL+D +
Sbjct 381 GGSGSGKSTVISLLQRFYD-----------------PN---------EGDILIDSVSINN 414
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
+ V +R Q G+VSQEP LF +I+ENI K A+ +E+ EAAK +NA FIS FP GY
Sbjct 415 MQVKWLRSQMGMVSQEPSLFATSIKENILFGKEDASFDEVVEAAKASNAHNFISQFPHGY 474
Score = 110 bits (274), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 67/182 (36%), Positives = 97/182 (53%), Gaps = 30/182 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+DR + I+ + G +L E KG+I ++F YPTRP+ ++ N + I EG++ A+V
Sbjct 958 LDRRTTIEPENPDGYIL--EKIKGQITFLNVDFAYPTRPNMVIFNNFSIEIHEGKSTAIV 1015
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G S GKST++ L+ERFYD P+ +G + +DG D+R
Sbjct 1016 GPSRSGKSTVIGLIERFYD--------PL------------------QGIVKIDGRDIRS 1049
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGAT--QEEIREAAKLANAAGFISTFPA 178
++ S+RQ LVSQEP LF TI ENI + + EI EA K ANA FI++
Sbjct 1050 YHLRSLRQHMSLVSQEPTLFAGTIRENIMYGRASNKIDESEIIEAGKTANAHEFITSLSD 1109
Query 179 GY 180
GY
Sbjct 1110 GY 1111
> hsa:5244 ABCB4, ABC21, GBD1, MDR2, MDR2/3, MDR3, PFIC-3, PGY3;
ATP-binding cassette, sub-family B (MDR/TAP), member 4 (EC:3.6.3.44);
K05659 ATP-binding cassette, subfamily B (MDR/TAP),
member 4
Length=1279
Score = 125 bits (314), Expect = 9e-29, Method: Compositional matrix adjust.
Identities = 71/166 (42%), Positives = 96/166 (57%), Gaps = 28/166 (16%)
Query 17 LSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVER 76
L + F+G I + + F YPTR + V + L+ +K+G+ +ALVG+SGCGKST+VQL+ER
Sbjct 1025 LKPDKFEGNITFNEVVFNYPTRANVPVLQGLSLEVKKGQTLALVGSSGCGKSTVVQLLER 1084
Query 77 FYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQE 136
FYD P++ G +LLDG + ++LNV +R Q G+VSQE
Sbjct 1085 FYD--------PLA------------------GTVLLDGQEAKKLNVQWLRAQLGIVSQE 1118
Query 137 PILFDMTIEENIAISKPG--ATQEEIREAAKLANAAGFISTFPAGY 180
PILFD +I ENIA +Q+EI AAK AN FI T P Y
Sbjct 1119 PILFDCSIAENIAYGDNSRVVSQDEIVSAAKAANIHPFIETLPHKY 1164
Score = 118 bits (295), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 61/161 (37%), Positives = 92/161 (57%), Gaps = 26/161 (16%)
Query 20 ETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYD 79
++ KG ++ + ++F+YP+R + ++ + L ++ G+ VALVG+SGCGKST VQL++R YD
Sbjct 388 DSIKGNLEFNDVHFSYPSRANVKILKGLNLKVQSGQTVALVGSSGCGKSTTVQLIQRLYD 447
Query 80 LKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPIL 139
P+ EG I +DG D+R NV +R+ G+VSQEP+L
Sbjct 448 -----------------PD---------EGTINIDGQDIRNFNVNYLREIIGVVSQEPVL 481
Query 140 FDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
F TI ENI + T +EI++A K ANA FI P +
Sbjct 482 FSTTIAENICYGRGNVTMDEIKKAVKEANAYEFIMKLPQKF 522
> xla:397812 abcb1, xemdr; ATP-binding cassette, sub-family B
(MDR/TAP), member 1; K05658 ATP-binding cassette, subfamily
B (MDR/TAP), member 1
Length=1287
Score = 124 bits (312), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 75/177 (42%), Positives = 97/177 (54%), Gaps = 28/177 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
ID +ID G L + KG+I+ + FTYP+R D +V + L +I G+ VALV
Sbjct 379 IDNQPKIDSFSKEG--LKPDKIKGDIEFKNVIFTYPSRKDIQVLKGLNLNIPSGKTVALV 436
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G+SGCGKST VQL++RFYD P +G I LDG D+R
Sbjct 437 GSSGCGKSTTVQLIQRFYD-----------------PE---------DGVITLDGQDIRS 470
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFP 177
LN+ +R+ G+VSQEPILFD TI +NI + T+EEI A K ANA FI P
Sbjct 471 LNIRYLREIIGVVSQEPILFDTTIADNIRYGREDVTKEEIERATKEANAYDFIMKLP 527
Score = 123 bits (309), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 72/159 (45%), Positives = 89/159 (55%), Gaps = 28/159 (17%)
Query 24 GEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYDLKST 83
G + +NF YPTRPD V + L S+K+GE +ALVG+SGCGKST V L+ERFYD
Sbjct 1042 GNVVFKGVNFNYPTRPDITVLQGLDISVKQGETLALVGSSGCGKSTTVSLLERFYD---- 1097
Query 84 YKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPILFDMT 143
P EG +L+DG +R LN+ VR Q G+VSQEPILFD +
Sbjct 1098 ----PF------------------EGEVLVDGLSVRNLNIQWVRAQMGIVSQEPILFDCS 1135
Query 144 IEENIAI--SKPGATQEEIREAAKLANAAGFISTFPAGY 180
I +NIA + TQEEI AAK AN FI + Y
Sbjct 1136 IGDNIAYGDNNRKVTQEEIETAAKEANIHSFIESLTDKY 1174
> mmu:18669 Abcb1b, Abcb1, Mdr1, Mdr1b, Pgy-1, Pgy1, mdr; ATP-binding
cassette, sub-family B (MDR/TAP), member 1B (EC:3.6.3.44);
K05658 ATP-binding cassette, subfamily B (MDR/TAP), member
1
Length=1276
Score = 124 bits (312), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 72/182 (39%), Positives = 102/182 (56%), Gaps = 30/182 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I++ EID + G L +G + + + F YPTRP+ V + L+ +K+G+ +ALV
Sbjct 1010 IEKTPEIDSYSTEG--LKPTLLEGNVKFNGVQFNYPTRPNIPVLQGLSLEVKKGQTLALV 1067
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G+SGCGKST+VQL+ERFYD P++ G + LDG ++++
Sbjct 1068 GSSGCGKSTVVQLLERFYD--------PMA------------------GSVFLDGKEIKQ 1101
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAI--SKPGATQEEIREAAKLANAAGFISTFPA 178
LNV +R G+VSQEPILFD +I ENIA + + EEI AAK AN FI + P
Sbjct 1102 LNVQWLRAHLGIVSQEPILFDCSIAENIAYGDNSRAVSHEEIVRAAKEANIHQFIDSLPD 1161
Query 179 GY 180
Y
Sbjct 1162 KY 1163
Score = 111 bits (277), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 65/177 (36%), Positives = 93/177 (52%), Gaps = 28/177 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
ID ID + G ++ G ++ ++F YP+R + ++ + L +K G+ VALV
Sbjct 368 IDNEPSIDSFSTKG--YKPDSIMGNLEFKNVHFNYPSRSEVQILKGLNLKVKSGQTVALV 425
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SGCGKST VQL++R YD P+ EG + +DG D+R
Sbjct 426 GNSGCGKSTTVQLMQRLYD--------PL------------------EGVVSIDGQDIRT 459
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFP 177
+NV +R+ G+VSQEP+LF TI ENI + T +EI +A K ANA FI P
Sbjct 460 INVRYLREIIGVVSQEPVLFATTIAENIRYGREDVTMDEIEKAVKEANAYDFIMKLP 516
> ath:AT1G10680 PGP10; PGP10 (P-GLYCOPROTEIN 10); ATPase, coupled
to transmembrane movement of substances
Length=1227
Score = 124 bits (311), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 70/180 (38%), Positives = 107/180 (59%), Gaps = 30/180 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+DR +++ V D+G ++ + E G I++ ++F+YP+RPD ++ + + G+++ALV
Sbjct 961 LDRRTQV-VGDTGEELSNVE---GTIELKGVHFSYPSRPDVTIFSDFNLLVPSGKSMALV 1016
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKS+++ LV RFYD P + G I++DG D+++
Sbjct 1017 GQSGSGKSSVLSLVLRFYD-----------------P---------TAGIIMIDGQDIKK 1050
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
L + S+R+ GLV QEP LF TI ENI K GA++ E+ EAAKLANA FIS+ P GY
Sbjct 1051 LKLKSLRRHIGLVQQEPALFATTIYENILYGKEGASESEVMEAAKLANAHSFISSLPEGY 1110
Score = 124 bits (311), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 67/157 (42%), Positives = 90/157 (57%), Gaps = 26/157 (16%)
Query 24 GEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYDLKST 83
G+I + FTYP+RPD ++ L F I G+ VALVG SG GKST++ L+ERFY
Sbjct 359 GDILFKDVTFTYPSRPDVVIFDKLNFVIPAGKVVALVGGSGSGKSTMISLIERFY----- 413
Query 84 YKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPILFDMT 143
EP ++G ++LDG+D+R L++ +R GLV+QEP+LF T
Sbjct 414 ------------EP---------TDGAVMLDGNDIRYLDLKWLRGHIGLVNQEPVLFATT 452
Query 144 IEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
I ENI K AT EEI AAKL+ A FI+ P G+
Sbjct 453 IRENIMYGKDDATSEEITNAAKLSEAISFINNLPEGF 489
> cel:C47A10.1 pgp-9; P-GlycoProtein related family member (pgp-9)
Length=1294
Score = 124 bits (311), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 71/179 (39%), Positives = 102/179 (56%), Gaps = 28/179 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
IDR EID + G+ S G I V+++ FTYPTR D ++ + ++ + G+ VALV
Sbjct 360 IDRIPEIDAYSTEGQTPSK--ISGRISVNKVEFTYPTRADVKILKGVSLDAQPGQTVALV 417
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G+SGCGKSTI+QL++RFY+ P+ G+IL+D + +
Sbjct 418 GSSGCGKSTIIQLLQRFYN-----------------PDA---------GQILIDDIPIED 451
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAG 179
N+ +RQ G+VSQEP LF+ +IE+NI + + E+I A K ANAA FI TFP G
Sbjct 452 FNIKYLRQLVGVVSQEPNLFNTSIEQNIRYGRSDVSDEDIARALKEANAADFIKTFPEG 510
Score = 109 bits (273), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 66/176 (37%), Positives = 92/176 (52%), Gaps = 31/176 (17%)
Query 7 IDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCG 66
ID S G ++ GE+ ++++ F YP RP + + L +K G+ +ALVG SGCG
Sbjct 1014 IDGMTSSG---TYPQLSGEVKLNKVFFRYPERPAVPILQGLNVHVKPGQTLALVGPSGCG 1070
Query 67 KSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSV 126
KST++ L+ER YD P+ EG + +D +DLR++N +
Sbjct 1071 KSTVISLLERLYD--------PL------------------EGAVTVDNNDLRQMNPKHL 1104
Query 127 RQQEGLVSQEPILFDMTIEENIAIS-KPG-ATQEEIREAAKLANAAGFISTFPAGY 180
R+ LVSQEPILFD +I ENI +PG T E+I A AN FI P GY
Sbjct 1105 RKHIALVSQEPILFDTSIRENIVYGLQPGEYTHEQIETACSKANIHKFIDELPDGY 1160
> ath:AT1G28010 PGP14; PGP14 (P-GLYCOPROTEIN 14); ATPase, coupled
to transmembrane movement of substances
Length=1247
Score = 124 bits (310), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 67/157 (42%), Positives = 91/157 (57%), Gaps = 27/157 (17%)
Query 24 GEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYDLKST 83
G+I+ ++F YP+RP N V+ NL+F+I G+ A VG SG GKSTI+ +V+RFY+ +S
Sbjct 371 GKIEFCGVSFAYPSRP-NMVFENLSFTIHSGKTFAFVGPSGSGKSTIISMVQRFYEPRS- 428
Query 84 YKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPILFDMT 143
G ILLDG+D++ L + +R+Q GLVSQEP LF T
Sbjct 429 -------------------------GEILLDGNDIKNLKLKWLREQMGLVSQEPALFATT 463
Query 144 IEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
I NI + K A ++I EAAK ANA FI + P GY
Sbjct 464 IASNILLGKEKANMDQIIEAAKAANADSFIKSLPNGY 500
Score = 102 bits (254), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 65/180 (36%), Positives = 99/180 (55%), Gaps = 28/180 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+ R +EI ++++H KG+I+ ++F YPTRP+ +++NL + G+++A+V
Sbjct 983 LHRETEIPPDQPNSRLVTH--IKGDIEFRNVSFAYPTRPEIAIFKNLNLRVSAGKSLAVV 1040
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L+ RFYD P S G + +DG D++
Sbjct 1041 GPSGSGKSTVIGLIMRFYD--------P------------------SNGNLCIDGHDIKS 1074
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
+N+ S+R++ LV QEP LF +I ENI A++ EI EAAK ANA FIS GY
Sbjct 1075 VNLRSLRKKLALVQQEPALFSTSIHENIKYGNENASEAEIIEAAKAANAHEFISRMEEGY 1134
> hsa:8647 ABCB11, ABC16, BRIC2, BSEP, PFIC-2, PFIC2, PGY4, SPGP;
ATP-binding cassette, sub-family B (MDR/TAP), member 11;
K05664 ATP-binding cassette, subfamily B (MDR/TAP), member
11
Length=1321
Score = 123 bits (309), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 73/177 (41%), Positives = 94/177 (53%), Gaps = 28/177 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
IDR ID G L + KGEI+ + F YP+RP+ ++ +L IK GE ALV
Sbjct 397 IDRKPIIDCMSEDGYKL--DRIKGEIEFHNVTFHYPSRPEVKILNDLNMVIKPGEMTALV 454
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST +QL++RFYD P EG + +DG D+R
Sbjct 455 GPSGAGKSTALQLIQRFYD--------P------------------CEGMVTVDGHDIRS 488
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFP 177
LN+ +R Q G+V QEP+LF TI ENI + AT E+I +AAK ANA FI P
Sbjct 489 LNIQWLRDQIGIVEQEPVLFSTTIAENIRYGREDATMEDIVQAAKEANAYNFIMDLP 545
Score = 112 bits (281), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 68/185 (36%), Positives = 101/185 (54%), Gaps = 36/185 (19%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+DR I V ++ G+ + F+G+ID FTYP+RPD++V L+ SI G+ +A V
Sbjct 1055 LDRQPPISVYNTAGE--KWDNFQGKIDFVDCKFTYPSRPDSQVLNGLSVSISPGQTLAFV 1112
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G+SGCGKST +QL+ERFYD P+ +G++++DG D ++
Sbjct 1113 GSSGCGKSTSIQLLERFYD-----------------PD---------QGKVMIDGHDSKK 1146
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIR-----EAAKLANAAGFIST 175
+NV +R G+VSQEP+LF +I +NI K G +EI AAK A F+ +
Sbjct 1147 VNVQFLRSNIGIVSQEPVLFACSIMDNI---KYGDNTKEIPMERVIAAAKQAQLHDFVMS 1203
Query 176 FPAGY 180
P Y
Sbjct 1204 LPEKY 1208
> cel:T21E8.1 pgp-6; P-GlycoProtein related family member (pgp-6);
K02021 putative ABC transport system ATP-binding protein
Length=1263
Score = 123 bits (309), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 62/156 (39%), Positives = 89/156 (57%), Gaps = 26/156 (16%)
Query 24 GEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYDLKST 83
G I + F+YPTRPD +V + ++F ++ GE +ALVGASG GKST+VQL+ +Y++ S
Sbjct 379 GNISFKNVKFSYPTRPDAQVLKGISFDVQNGECIALVGASGSGKSTVVQLLLHYYNIDS- 437
Query 84 YKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPILFDMT 143
G I +DG DL ++N+ +R+ G+VSQEP+LF+ T
Sbjct 438 -------------------------GNIFIDGMDLNDMNIKRLRRVIGVVSQEPVLFNTT 472
Query 144 IEENIAISKPGATQEEIREAAKLANAAGFISTFPAG 179
IEENI P + EI A + ANA F+ +FP G
Sbjct 473 IEENIRFGNPNVSLPEIYGALRKANAYDFVCSFPKG 508
Score = 95.1 bits (235), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 53/158 (33%), Positives = 80/158 (50%), Gaps = 27/158 (17%)
Query 23 KGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYDLKS 82
KGEI + + F Y RPD + + + G+ +ALVG SGCGKSTI+ L+ERFY
Sbjct 1013 KGEIIGENVQFHYDQRPDRMILNGVNLKVDPGKTLALVGPSGCGKSTIISLLERFY---- 1068
Query 83 TYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPILFDM 142
H +G + +D +++ ++N+ +R LVSQEP LF+
Sbjct 1069 ----------------------HAVDGEVKIDSENVEDINLNHLRSNLALVSQEPTLFNC 1106
Query 143 TIEENIAISKPGAT-QEEIREAAKLANAAGFISTFPAG 179
+I EN+ + Q E+ +A + ANA F+ FP G
Sbjct 1107 SIRENLLYGLTRSVPQLELEKALQTANAFNFVFQFPQG 1144
> cel:T21E8.2 pgp-7; P-GlycoProtein related family member (pgp-7);
K02021 putative ABC transport system ATP-binding protein
Length=1263
Score = 123 bits (309), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 62/156 (39%), Positives = 89/156 (57%), Gaps = 26/156 (16%)
Query 24 GEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYDLKST 83
G I + F+YPTRPD +V + ++F ++ GE +ALVGASG GKST+VQL+ +Y++ S
Sbjct 379 GNISFKNVKFSYPTRPDAQVLKGISFDVQNGECIALVGASGSGKSTVVQLLLHYYNIDS- 437
Query 84 YKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPILFDMT 143
G I +DG DL ++N+ +R+ G+VSQEP+LF+ T
Sbjct 438 -------------------------GNIFIDGMDLNDMNIKRLRRVIGVVSQEPVLFNTT 472
Query 144 IEENIAISKPGATQEEIREAAKLANAAGFISTFPAG 179
IEENI P + EI A + ANA F+ +FP G
Sbjct 473 IEENIRFGNPNVSLPEIYGALRKANAYDFVCSFPKG 508
Score = 95.1 bits (235), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 53/158 (33%), Positives = 80/158 (50%), Gaps = 27/158 (17%)
Query 23 KGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYDLKS 82
KGEI + + F Y RPD + + + G+ +ALVG SGCGKSTI+ L+ERFY
Sbjct 1013 KGEIIGENVQFHYDQRPDRMILNGVNLKVDPGKTLALVGPSGCGKSTIISLLERFY---- 1068
Query 83 TYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPILFDM 142
H +G + +D +++ ++N+ +R LVSQEP LF+
Sbjct 1069 ----------------------HAVDGEVKIDSENVEDINLNHLRSNLALVSQEPTLFNC 1106
Query 143 TIEENIAISKPGAT-QEEIREAAKLANAAGFISTFPAG 179
+I EN+ + Q E+ +A + ANA F+ FP G
Sbjct 1107 SIRENLLYGLTRSVPQLELEKALQTANAFNFVFQFPQG 1144
> mmu:18671 Abcb1a, Abcb4, Evi32, Mdr1a, Mdr3, P-gp, Pgp, Pgy-3,
Pgy3, mdr-3; ATP-binding cassette, sub-family B (MDR/TAP),
member 1A (EC:3.6.3.44); K05658 ATP-binding cassette, subfamily
B (MDR/TAP), member 1
Length=1276
Score = 123 bits (308), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 73/182 (40%), Positives = 100/182 (54%), Gaps = 30/182 (16%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I++ EID + G L +G + + F YPTRP V + L+ +K+G+ +ALV
Sbjct 1008 IEKTPEIDSYSTQG--LKPNMLEGNVQFSGVVFNYPTRPSIPVLQGLSLEVKKGQTLALV 1065
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G+SGCGKST+VQL+ERFYD P++ G + LDG ++++
Sbjct 1066 GSSGCGKSTVVQLLERFYD--------PMA------------------GSVFLDGKEIKQ 1099
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPG--ATQEEIREAAKLANAAGFISTFPA 178
LNV +R Q G+VSQEPILFD +I ENIA + EEI AAK AN FI + P
Sbjct 1100 LNVQWLRAQLGIVSQEPILFDCSIAENIAYGDNSRVVSYEEIVRAAKEANIHQFIDSLPD 1159
Query 179 GY 180
Y
Sbjct 1160 KY 1161
Score = 112 bits (280), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 65/176 (36%), Positives = 95/176 (53%), Gaps = 29/176 (16%)
Query 2 DRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVG 61
++PS SG K + +G ++ I+F+YP+R + ++ + L +K G+ VALVG
Sbjct 367 NKPSIDSFSKSGHK---PDNIQGNLEFKNIHFSYPSRKEVQILKGLNLKVKSGQTVALVG 423
Query 62 ASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLREL 121
SGCGKST VQL++R YD P+ +G + +DG D+R +
Sbjct 424 NSGCGKSTTVQLMQRLYD--------PL------------------DGMVSIDGQDIRTI 457
Query 122 NVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFP 177
NV +R+ G+VSQEP+LF TI ENI + T +EI +A K ANA FI P
Sbjct 458 NVRYLREIIGVVSQEPVLFATTIAENIRYGREDVTMDEIEKAVKEANAYDFIMKLP 513
> cel:F22E10.1 pgp-12; P-GlycoProtein related family member (pgp-12);
K02021 putative ABC transport system ATP-binding protein
Length=1318
Score = 122 bits (306), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 68/180 (37%), Positives = 99/180 (55%), Gaps = 28/180 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
IDR +IDV + G+ +++ G++ + ++F YPTR +V L ++K G +VALV
Sbjct 394 IDRTPDIDVYSTEGQKMTNVV--GKVVFEEVHFRYPTRKKVKVLNGLNLTVKPGTSVALV 451
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SGCGKST V L+ R Y EP G++++DG D+R
Sbjct 452 GHSGCGKSTSVGLLTRLY-----------------EPEG---------GKVMIDGQDVRS 485
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
LN+ +R+ G+V QEPILF+ TI N+ I P AT+E++ K+ANA FI P GY
Sbjct 486 LNIDWLRKTVGIVQQEPILFNDTIHNNLLIGNPSATREDMIRVCKMANAHDFIQKMPNGY 545
Score = 103 bits (257), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 55/157 (35%), Positives = 84/157 (53%), Gaps = 26/157 (16%)
Query 23 KGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYDLKS 82
+G + + + F+YP RP V +L FS G+ VALVG SG GKST + ++ERFYD
Sbjct 1074 RGNVLFESVKFSYPQRPMQPVMTDLHFSAHSGQTVALVGPSGTGKSTCIAMLERFYD--- 1130
Query 83 TYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPILFDM 142
+S G + +DG +++ L++ +R Q LV QEP LF
Sbjct 1131 -----------------------VSGGALRIDGQNIKSLSLHHLRTQMALVGQEPRLFAG 1167
Query 143 TIEENIAISKPGATQEEIREAAKLANAAGFISTFPAG 179
TI+EN+ + E++ +A +LANA+ F++ PAG
Sbjct 1168 TIKENVCFGLKDVSVEKVHQALELANASRFLANLPAG 1204
> mmu:56199 Abcb10, AU016331, Abc-me, Abcb12, C76534; ATP-binding
cassette, sub-family B (MDR/TAP), member 10; K05657 ATP-binding
cassette, subfamily B (MDR/TAP), member 10
Length=715
Score = 122 bits (306), Expect = 7e-28, Method: Composition-based stats.
Identities = 67/170 (39%), Positives = 95/170 (55%), Gaps = 29/170 (17%)
Query 14 GKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQL 73
G VL +TF+G ++ ++FTYP RP+ V+++ + SI G ALVG SG GKST+V L
Sbjct 445 GMVLDEKTFQGALEFRNVHFTYPARPEVSVFQDFSLSIPSGSVTALVGPSGSGKSTVVSL 504
Query 74 VERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLV 133
+ R YD PN G + LDG D+R+LN V +R + G V
Sbjct 505 LLRLYD-----------------PN---------SGTVSLDGHDIRQLNPVWLRSKIGTV 538
Query 134 SQEPILFDMTIEENIAISK---PGATQEEIREAAKLANAAGFISTFPAGY 180
SQEP+LF ++ ENIA T +++ AA++ANAA FI +FP G+
Sbjct 539 SQEPVLFSCSVAENIAYGADNLSSVTAQQVERAAEVANAAEFIRSFPQGF 588
> dre:100136865 abcb4, zgc:172149; ATP-binding cassette, sub-family
B (MDR/TAP), member 4
Length=650
Score = 122 bits (305), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 65/155 (41%), Positives = 87/155 (56%), Gaps = 26/155 (16%)
Query 23 KGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYDLKS 82
KG I+ INF YP+R D +V + + G+ +ALVG+SGCGKST +QL++RFYD
Sbjct 394 KGNIEFKNINFRYPSRDDVKVLNGMNLKVMSGQTIALVGSSGCGKSTTIQLLQRFYD--- 450
Query 83 TYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPILFDM 142
P EG + +DG D+R LNV +R+ G+VSQEP+LF
Sbjct 451 --------------PQ---------EGSVSIDGHDIRSLNVRGLRELIGVVSQEPVLFAT 487
Query 143 TIEENIAISKPGATQEEIREAAKLANAAGFISTFP 177
TI ENI + TQ+EI +AA+ ANA FI P
Sbjct 488 TIAENIRYGRQDVTQDEIEQAAREANAYNFIMKLP 522
> pfa:PFE1150w PfMDR1, Pgh-1; multidrug resistance protein
Length=1419
Score = 122 bits (305), Expect = 9e-28, Method: Composition-based stats.
Identities = 68/199 (34%), Positives = 107/199 (53%), Gaps = 21/199 (10%)
Query 3 RPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGA 62
R S IDVRD GG ++ KG++D+ +NF Y +RP+ +Y+NL+F+ + A+VG
Sbjct 1103 RKSNIDVRDDGGIRINKNLIKGKVDIKDVNFRYISRPNVPIYKNLSFTCDSKKTTAIVGE 1162
Query 63 SGCGKSTIVQLVERFYDLKSTYKV---------------------KPVSQSEQGEPNKEA 101
+G GKST + L+ RFYDLK+ + + K V++ + E
Sbjct 1163 TGSGKSTFMNLLLRFYDLKNDHIILKNDMTNFQDYQNNNNNSLVLKNVNEFSNQSGSAED 1222
Query 102 ERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIR 161
V + G ILLD ++ + N+ +R +VSQEP+LF+M+I ENI + AT E+++
Sbjct 1223 YTVFNNNGEILLDDINICDYNLRDLRNLFSIVSQEPMLFNMSIYENIKFGREDATLEDVK 1282
Query 162 EAAKLANAAGFISTFPAGY 180
+K A FI + P Y
Sbjct 1283 RVSKFAAIDEFIESLPNKY 1301
Score = 84.3 bits (207), Expect = 2e-16, Method: Composition-based stats.
Identities = 46/128 (35%), Positives = 73/128 (57%), Gaps = 27/128 (21%)
Query 25 EIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYDLKSTY 84
+I+ + F Y TR D +Y++L+F++KEG+ A VG SGCGKSTI++L+ER YD
Sbjct 377 KIEFKNVRFHYDTRKDVEIYKDLSFTLKEGKTYAFVGESGCGKSTILKLIERLYD----- 431
Query 85 KVKPVSQSEQGEPNKEAERVHISEGRILL-DGDDLRELNVVSVRQQEGLVSQEPILFDMT 143
+EG I++ D +L+++N+ R + G+VSQ+P+LF +
Sbjct 432 ---------------------PTEGDIIVNDSHNLKDINLKWWRSKIGVVSQDPLLFSNS 470
Query 144 IEENIAIS 151
I+ NI S
Sbjct 471 IKNNIKYS 478
> mmu:77706 Abcb5, 9230106F14Rik; ATP-binding cassette, sub-family
B (MDR/TAP), member 5; K05660 ATP-binding cassette, subfamily
B (MDR/TAP), member 5
Length=1255
Score = 121 bits (303), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 71/181 (39%), Positives = 100/181 (55%), Gaps = 31/181 (17%)
Query 2 DRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVG 61
++P+ SG K +T +G ++ ++F YP RP+ V +N++ SI++G+ VA VG
Sbjct 995 NKPTINSCSQSGEK---PDTCEGNLEFREVSFVYPCRPEVPVLQNMSLSIEKGKTVAFVG 1051
Query 62 ASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLREL 121
+SGCGKST VQL++RFYD P K G++LLDG D++EL
Sbjct 1052 SSGCGKSTCVQLLQRFYD-----------------PMK---------GQVLLDGVDVKEL 1085
Query 122 NVVSVRQQEGLVSQEPILFDMTIEENIAISKPG--ATQEEIREAAKLANAAGFISTFPAG 179
NV +R Q +VSQEP+LF+ +I ENIA EEI+E A AN FI P
Sbjct 1086 NVQWLRSQTAIVSQEPVLFNCSIAENIAYGDNSRMVPLEEIKEVADAANIHSFIEGLPRK 1145
Query 180 Y 180
Y
Sbjct 1146 Y 1146
Score = 115 bits (288), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 69/180 (38%), Positives = 95/180 (52%), Gaps = 28/180 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
ID+ ID + G V E +G I+ ++F+YP+RP +V + L IK GE VALV
Sbjct 364 IDKKPNIDNFSTAGFV--PECIEGNIEFKNVSFSYPSRPSAKVLKGLNLKIKAGETVALV 421
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST VQL++R YD P +G I +D +D+R
Sbjct 422 GPSGSGKSTTVQLLQRLYD-----------------PE---------DGCITVDENDIRA 455
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
NV R+Q G+V QEP+LF TI NI + G ++E+ +AA+ ANA FI FP +
Sbjct 456 QNVRHYREQIGVVRQEPVLFGTTIGNNIKFGREGVGEKEMEQAAREANAYDFIMAFPKKF 515
> mmu:18670 Abcb4, Mdr2, Pgy-2, Pgy2, mdr-2; ATP-binding cassette,
sub-family B (MDR/TAP), member 4 (EC:3.6.3.44); K05659
ATP-binding cassette, subfamily B (MDR/TAP), member 4
Length=1276
Score = 121 bits (303), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 70/169 (41%), Positives = 96/169 (56%), Gaps = 28/169 (16%)
Query 14 GKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQL 73
G+ L + F+G + + + F YPTR + V + L+ +K+G+ +ALVG+SGCGKST+VQL
Sbjct 1019 GEGLWPDKFEGSVTFNEVVFNYPTRANVPVLQGLSLEVKKGQTLALVGSSGCGKSTVVQL 1078
Query 74 VERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLV 133
+ERFYD P++ G +LLDG + ++LNV +R Q G+V
Sbjct 1079 LERFYD--------PMA------------------GSVLLDGQEAKKLNVQWLRAQLGIV 1112
Query 134 SQEPILFDMTIEENIAISKPGAT--QEEIREAAKLANAAGFISTFPAGY 180
SQEPILFD +I ENIA +EI AAK AN FI T P Y
Sbjct 1113 SQEPILFDCSIAENIAYGDNSRVVPHDEIVRAAKEANIHPFIETLPQKY 1161
Score = 117 bits (292), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 62/161 (38%), Positives = 89/161 (55%), Gaps = 26/161 (16%)
Query 20 ETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYD 79
+ KG ++ ++F+YP+R + ++ + L +K G+ VALVG SGCGKST VQL++R YD
Sbjct 385 DNIKGNLEFSDVHFSYPSRANIKILKGLNLKVKSGQTVALVGNSGCGKSTTVQLLQRLYD 444
Query 80 LKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPIL 139
P +EG+I +DG D+R NV +R+ G+VSQEP+L
Sbjct 445 --------P------------------TEGKISIDGQDIRNFNVRCLREIIGVVSQEPVL 478
Query 140 FDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
F TI ENI + T +EI +A K ANA FI P +
Sbjct 479 FSTTIAENIRYGRGNVTMDEIEKAVKEANAYDFIMKLPQKF 519
> cel:F22E10.3 pgp-14; P-GlycoProtein related family member (pgp-14);
K02021 putative ABC transport system ATP-binding protein
Length=1327
Score = 120 bits (302), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 69/180 (38%), Positives = 94/180 (52%), Gaps = 28/180 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
IDR +ID GK L + G + + ++F YP+R D ++ L ++ G +VALV
Sbjct 406 IDRVPKIDPYSKAGKRL--QNVVGRVKFENVHFRYPSRKDAKILNGLNLVVEPGTSVALV 463
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SGCGKST V L+ R Y EP G + +DG D+RE
Sbjct 464 GHSGCGKSTSVGLLTRLY-----------------EP---------EAGNVTIDGTDVRE 497
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
LN+ +R G+V QEPILF+ TI N+ I PG+T+E + E K+ANA FI P GY
Sbjct 498 LNIEWLRNTVGIVQQEPILFNDTIHNNLLIGNPGSTRETMIEVCKMANAHDFIEKMPKGY 557
Score = 101 bits (251), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 55/157 (35%), Positives = 83/157 (52%), Gaps = 26/157 (16%)
Query 23 KGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYDLKS 82
+G I + + F+YP RP + + L ++ G+ VALVG SG GKST + ++ERFYD
Sbjct 1083 RGNILFENVKFSYPQRPLQPIMKGLQWTALRGQTVALVGPSGSGKSTNIGMLERFYD--- 1139
Query 83 TYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPILFDM 142
++ G + +DG D+R+L++ +R Q LV QEP LF
Sbjct 1140 -----------------------VTGGALRIDGQDIRKLSLFHLRTQMALVGQEPRLFAG 1176
Query 143 TIEENIAISKPGATQEEIREAAKLANAAGFISTFPAG 179
TI EN+ + E+I +A +LANA F++ PAG
Sbjct 1177 TIRENVCLGLKDVPLEKINQALELANANRFLANLPAG 1213
> cel:T21E8.3 pgp-8; P-GlycoProtein related family member (pgp-8);
K02021 putative ABC transport system ATP-binding protein
Length=1243
Score = 120 bits (300), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 63/162 (38%), Positives = 90/162 (55%), Gaps = 26/162 (16%)
Query 18 SHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERF 77
S++T +G + + + F YP+RPD + R ++F +K+GE +ALVGASG GKSTIVQL+ F
Sbjct 372 SNKTIQGMLSFNNVKFAYPSRPDVDILRGISFDVKQGECIALVGASGSGKSTIVQLLLHF 431
Query 78 YDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEP 137
Y+++S G I + L ++N+ +R G+VSQEP
Sbjct 432 YNIQS--------------------------GTIKIGDSHLHDINLKQLRNAIGVVSQEP 465
Query 138 ILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAG 179
+LF+ TIEENI P AT EI EA + ANA F+ G
Sbjct 466 VLFNTTIEENIRFGNPNATSSEIYEALRKANAYDFVCNIKDG 507
Score = 88.2 bits (217), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 53/170 (31%), Positives = 85/170 (50%), Gaps = 31/170 (18%)
Query 11 DSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTI 70
D+G + ++ KGE++ + + F Y RPD V ++ ++ G+ +A+VG SG GKSTI
Sbjct 991 DTGSRSIT----KGEVNGENVKFHYHQRPDYTVLDSVNLKVEAGKTLAIVGPSGSGKSTI 1046
Query 71 VQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQE 130
+ L+E FY +G I +D D++ +N+ +R
Sbjct 1047 ISLLEMFY--------------------------RADQGFIKIDNDNVENINLDHLRSNL 1080
Query 131 GLVSQEPILFDMTIEENIAISKP-GATQEEIREAAKLANAAGFISTFPAG 179
GLVSQ P+LF+ +I +NI +Q EI A ++ANA F+ P G
Sbjct 1081 GLVSQGPVLFNCSIRDNILYGLTRNISQTEIENALQIANAFNFVFQLPQG 1130
> cel:DH11.3 pgp-11; P-GlycoProtein related family member (pgp-11);
K02021 putative ABC transport system ATP-binding protein
Length=1270
Score = 120 bits (300), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 63/164 (38%), Positives = 91/164 (55%), Gaps = 26/164 (15%)
Query 17 LSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVER 76
LS G+++V + F YP+ P V + +I +G ++ALVGASGCGKSTI+ ++ER
Sbjct 1021 LSKPEINGKVEVSDVTFAYPSTPHRNVCEGFSLNIPKGHSIALVGASGCGKSTIISMLER 1080
Query 77 FYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQE 136
FY K+ GRI +D +D+ ++V +R +V QE
Sbjct 1081 FYSAKA--------------------------GRISVDDNDIDGIDVNHLRNNISVVGQE 1114
Query 137 PILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
P+LF+ TI ENI I + EE+++A K ANAAGFI +FP GY
Sbjct 1115 PVLFNATIRENITIGIDEVSVEEVQKACKAANAAGFIESFPLGY 1158
Score = 99.0 bits (245), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 61/158 (38%), Positives = 83/158 (52%), Gaps = 28/158 (17%)
Query 23 KGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYDLKS 82
+G I+ + F Y TR DN V + L+ + G+ VALVG SGCGKST + L+ + Y
Sbjct 397 RGNIEFRDVRFKYFTR-DNEVLQGLSLQVLPGQTVALVGTSGCGKSTSIGLLTKLY---- 451
Query 83 TYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPILFDM 142
SEG IL+DG ++ L+ S+RQQ G+V QEP LFD
Sbjct 452 ----------------------RASEGEILIDGKNIDMLDAKSLRQQIGIVQQEPKLFDG 489
Query 143 TIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
TI ENI + + +E I+ AA +ANA+ FI GY
Sbjct 490 TIMENIKLGR-NVDEETIKTAADIANASSFIEKLENGY 526
> ath:AT4G25960 PGP2; PGP2 (P-GLYCOPROTEIN 2); ATPase, coupled
to transmembrane movement of substances
Length=1273
Score = 119 bits (299), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 70/167 (41%), Positives = 92/167 (55%), Gaps = 32/167 (19%)
Query 14 GKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQL 73
GKV H FK F+YP+RPD ++ L +I G+ VALVG SG GKST++ L
Sbjct 395 GKVDGHIQFKDA------TFSYPSRPDVVIFDRLNLAIPAGKIVALVGGSGSGKSTVISL 448
Query 74 VERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLV 133
+ERFY+ P+S G +LLDG+++ EL++ +R Q GLV
Sbjct 449 IERFYE--------PIS------------------GAVLLDGNNISELDIKWLRGQIGLV 482
Query 134 SQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
+QEP LF TI ENI K AT EEI AAKL+ A FI+ P G+
Sbjct 483 NQEPALFATTIRENILYGKDDATAEEITRAAKLSEAISFINNLPEGF 529
Score = 119 bits (298), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 58/158 (36%), Positives = 96/158 (60%), Gaps = 26/158 (16%)
Query 23 KGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALVGASGCGKSTIVQLVERFYDLKS 82
+G I++ ++F+YP+RPD ++R+ ++ G+++ALVG SG GKS+++ L+ RFYD
Sbjct 1027 EGTIELKGVHFSYPSRPDVVIFRDFDLIVRAGKSMALVGQSGSGKSSVISLILRFYD--- 1083
Query 83 TYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRELNVVSVRQQEGLVSQEPILFDM 142
P + G+++++G D+++L++ ++R+ GLV QEP LF
Sbjct 1084 -----P------------------TAGKVMIEGKDIKKLDLKALRKHIGLVQQEPALFAT 1120
Query 143 TIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
TI ENI GA+Q E+ E+A LANA FI++ P GY
Sbjct 1121 TIYENILYGNEGASQSEVVESAMLANAHSFITSLPEGY 1158
> ath:AT3G28345 ABC transporter family protein
Length=1240
Score = 119 bits (297), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 68/180 (37%), Positives = 98/180 (54%), Gaps = 28/180 (15%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
I+R +ID + G L E +GE++ + F YP+R + ++ + + G+ VALV
Sbjct 336 INRVPKIDSDNPDGHKL--EKIRGEVEFKNVKFVYPSRLETSIFDDFCLRVPSGKTVALV 393
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKST++ L++RFYD P++ G IL+DG + +
Sbjct 394 GGSGSGKSTVISLLQRFYD--------PLA------------------GEILIDGVSIDK 427
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIREAAKLANAAGFISTFPAGY 180
L V +R Q GLVSQEP LF TI+ENI K A+ +++ EAAK +NA FIS P GY
Sbjct 428 LQVKWLRSQMGLVSQEPALFATTIKENILFGKEDASMDDVVEAAKASNAHNFISQLPNGY 487
Score = 99.8 bits (247), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 70/185 (37%), Positives = 99/185 (53%), Gaps = 36/185 (19%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+DR + ID D G E G+++ ++F+YPTRPD +++N + I+EG++ A+V
Sbjct 972 LDRYTSIDPEDPDG--YETERITGQVEFLDVDFSYPTRPDVIIFKNFSIKIEEGKSTAIV 1029
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G SG GKSTI+ L+ERFYD P K G + +DG D+R
Sbjct 1030 GPSGSGKSTIIGLIERFYD-----------------PLK---------GIVKIDGRDIRS 1063
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEI-----REAAKLANAAGFIST 175
++ S+R+ LVSQEP LF TI ENI G ++I EAAK ANA FI++
Sbjct 1064 YHLRSLRRHIALVSQEPTLFAGTIRENIIY---GGVSDKIDEAEIIEAAKAANAHDFITS 1120
Query 176 FPAGY 180
GY
Sbjct 1121 LTEGY 1125
> dre:571189 abcb11b; ATP-binding cassette, sub-family B (MDR/TAP),
member 11b; K05664 ATP-binding cassette, subfamily B (MDR/TAP),
member 11
Length=1297
Score = 119 bits (297), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 73/185 (39%), Positives = 99/185 (53%), Gaps = 36/185 (19%)
Query 1 IDRPSEIDVRDSGGKVLSHETFKGEIDVDRINFTYPTRPDNRVYRNLTFSIKEGEAVALV 60
+DR +I V G+ + FKG+I+ FTYP+RPD +V L S+K G+ +A V
Sbjct 1031 LDRIPKISVYSKDGQ--KWDNFKGDIEFIDCKFTYPSRPDIQVLNGLNVSVKPGQTLAFV 1088
Query 61 GASGCGKSTIVQLVERFYDLKSTYKVKPVSQSEQGEPNKEAERVHISEGRILLDGDDLRE 120
G+SGCGKST VQL+ERFYD PN GR+L+DG + +
Sbjct 1089 GSSGCGKSTSVQLLERFYD-----------------PN---------SGRVLIDGRESSQ 1122
Query 121 LNVVSVRQQEGLVSQEPILFDMTIEENIAISKPGATQEEIR-----EAAKLANAAGFIST 175
+NV +R + G+VSQEPILFD +I ENI + G Q E+ AAK A F+ +
Sbjct 1123 INVAYLRSKIGIVSQEPILFDCSIAENI---RYGDNQRELSMNDVISAAKKAQLHDFVMS 1179
Query 176 FPAGY 180
P Y
Sbjct 1180 LPEKY 1184
Lambda K H
0.314 0.134 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4859948100
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40