bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1434_orf1
Length=211
Score E
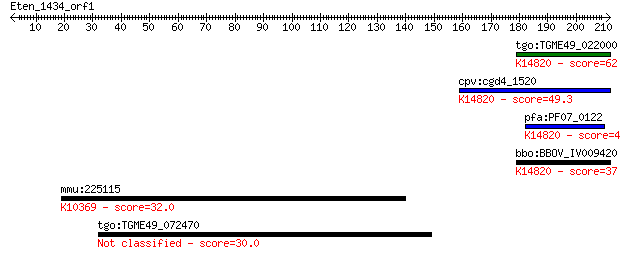
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_022000 ribosome biogenesis protein Brix, putative ;... 62.8 1e-09
cpv:cgd4_1520 Brx1p nucleolar protein required for biogenesis ... 49.3 9e-06
pfa:PF07_0122 nucleolus BRIX protein, putative; K14820 ribosom... 41.2 0.003
bbo:BBOV_IV009420 23.m05729; ribosome biogenesis Brix protein;... 37.4 0.038
mmu:225115 Svil, AU024053, B430302E16Rik; supervillin; K10369 ... 32.0 1.5
tgo:TGME49_072470 protein kinase, putative (EC:2.7.11.1 3.4.21... 30.0 6.0
> tgo:TGME49_022000 ribosome biogenesis protein Brix, putative
; K14820 ribosome biogenesis protein BRX1
Length=517
Score = 62.8 bits (151), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 27/33 (81%), Positives = 30/33 (90%), Gaps = 0/33 (0%)
Query 179 AAADPYLLEDAEYLRRETRWLNRQRVLVLGSRG 211
A ADPYLL +A+YLRRE RW+NRQRVLVLGSRG
Sbjct 209 APADPYLLAEADYLRREKRWVNRQRVLVLGSRG 241
> cpv:cgd4_1520 Brx1p nucleolar protein required for biogenesis
of the 60S ribosomal subunit ; K14820 ribosome biogenesis
protein BRX1
Length=353
Score = 49.3 bits (116), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 35/56 (62%), Gaps = 3/56 (5%)
Query 159 SAENDAAADAAD---SAAAIDAAAAADPYLLEDAEYLRRETRWLNRQRVLVLGSRG 211
S +N+ D + ++ + DPY+++ AEYL +E+RW N+QRVLVL +RG
Sbjct 22 SEDNEIKMDIEEGHITSNNNNNNINTDPYMMKGAEYLNKESRWKNKQRVLVLSTRG 77
> pfa:PF07_0122 nucleolus BRIX protein, putative; K14820 ribosome
biogenesis protein BRX1
Length=416
Score = 41.2 bits (95), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 16/28 (57%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 182 DPYLLEDAEYLRRETRWLNRQRVLVLGS 209
DPYLL+DA Y+++ W NRQRVL++ S
Sbjct 144 DPYLLKDASYIKKNELWKNRQRVLIVRS 171
> bbo:BBOV_IV009420 23.m05729; ribosome biogenesis Brix protein;
K14820 ribosome biogenesis protein BRX1
Length=281
Score = 37.4 bits (85), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 24/33 (72%), Gaps = 0/33 (0%)
Query 179 AAADPYLLEDAEYLRRETRWLNRQRVLVLGSRG 211
DPY L+DA Y+++ +++ N+++ +VL SRG
Sbjct 71 VNLDPYRLKDAYYIKQNSKYTNKKKCMVLASRG 103
> mmu:225115 Svil, AU024053, B430302E16Rik; supervillin; K10369
supervillin
Length=2170
Score = 32.0 bits (71), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 42/144 (29%), Positives = 61/144 (42%), Gaps = 25/144 (17%)
Query 19 EVAAAAQKAAGATPEAAAAAIQEATKKAPKALGKRP-ASSAKGECGAPATTSFRKRPRKQ 77
EVAA ATP A QE++++ + L K P A A GE P SF P K
Sbjct 1024 EVAARKASVQMATP--GAWKQQESSEQLAEKLFKNPCAMFASGEVKVPVGDSFLDSPSKT 1081
Query 78 GRLRQ-----EKQQPEQQPPRAD----------GVHRQEDEHGNKKRRRQQQQQHQVH-Q 121
+++ +K E R G+H QE E KK+R + ++ Q+ +
Sbjct 1082 MSIKERLALLKKSGEEDWKNRLIRKQEYGKATGGLHTQEVEQSLKKKRVTESRESQMTIE 1141
Query 122 DRNQTTDM------GKGEGAASDS 139
+R + KG GAA+DS
Sbjct 1142 ERKHLITVREEAWKTKGRGAANDS 1165
> tgo:TGME49_072470 protein kinase, putative (EC:2.7.11.1 3.4.21.10)
Length=1734
Score = 30.0 bits (66), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 33/129 (25%), Positives = 52/129 (40%), Gaps = 14/129 (10%)
Query 32 PEAAAAAIQEA----TKKAPKALGKRPASSAKGECGAPATTSFRKR--------PRKQGR 79
P AA A+ +E TKK + G R S+ +C P TS R+R P QG
Sbjct 659 PSAAVASFEEILLSFTKKGQRRDGSRDPDSSFRDCSTPPATSSRRRQNGALPGPPMWQGD 718
Query 80 LRQEKQQPEQQPPRADGVHRQEDEHGNKKRRRQQQQQHQVHQDRNQTTDMGKGEGAASDS 139
++++ E + GV R G + R+ D + D+ + E AS
Sbjct 719 FVRQRENSESR--VHHGVSRTSSMSGKRGTHRRGPDDTSTSHDWRDSGDVRQNETVASRG 776
Query 140 LGQSEKTET 148
L + E + +
Sbjct 777 LVKREASNS 785
Lambda K H
0.307 0.123 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6623499460
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40