bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1442_orf1
Length=160
Score E
Sequences producing significant alignments: (Bits) Value
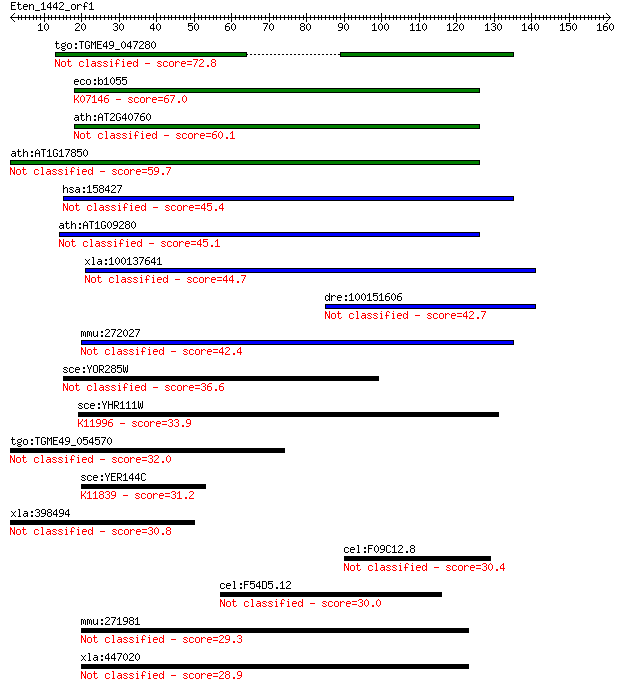
tgo:TGME49_047280 hypothetical protein 72.8 4e-13
eco:b1055 yceA, ECK1040, JW1042; predicted rhodanese-related s... 67.0 3e-11
ath:AT2G40760 rhodanese-like domain-containing protein 60.1 3e-09
ath:AT1G17850 hypothetical protein 59.7 4e-09
hsa:158427 TSTD2, C9orf97, FLJ36724; thiosulfate sulfurtransfe... 45.4 9e-05
ath:AT1G09280 hypothetical protein 45.1 1e-04
xla:100137641 tstd2; thiosulfate sulfurtransferase (rhodanese)... 44.7 1e-04
dre:100151606 hypothetical LOC100151606 42.7 5e-04
mmu:272027 Tstd2, 3010020C06; thiosulfate sulfurtransferase (r... 42.4 7e-04
sce:YOR285W RDL1; Protein of unknown function containing a rho... 36.6 0.032
sce:YHR111W UBA4, YHR1; Protein that activates Urm1p before it... 33.9 0.23
tgo:TGME49_054570 hypothetical protein 32.0 1.0
sce:YER144C UBP5; Putative ubiquitin-specific protease, closes... 31.2 1.5
xla:398494 hnrnpu; heterogeneous nuclear ribonucleoprotein U (... 30.8 2.1
cel:F09C12.8 hypothetical protein 30.4 2.7
cel:F54D5.12 hypothetical protein 30.0 3.3
mmu:271981 Tbck, 1700120J03Rik, 9430001M19, A630047E20Rik, Tbc... 29.3 6.4
xla:447020 tbck, MGC82809, hspc302, tbckl; TBC1 domain contain... 28.9 7.3
> tgo:TGME49_047280 hypothetical protein
Length=2373
Score = 72.8 bits (177), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 30/46 (65%), Positives = 38/46 (82%), Gaps = 0/46 (0%)
Query 89 IMMYCTGGIRCVKAGAFVKQILGFPRVTRLKGGILAYKNFIESFKK 134
+MMYCTGGIRCVKAGA+VKQ LG P V RLKGG+L Y ++++ +K
Sbjct 1222 VMMYCTGGIRCVKAGAYVKQRLGIPCVFRLKGGVLRYVEYLQTLEK 1267
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/51 (43%), Positives = 33/51 (64%), Gaps = 0/51 (0%)
Query 13 QKQRTPKVVLLDMRNASEYAVGHFKGSQCVDTLTFADSFTSGGPLEEALAR 63
+++R ++VLLD RN E +G FKG+ ++T F++SF S G L LAR
Sbjct 1082 EQRRRKRIVLLDCRNMYESDIGFFKGAVPLNTNIFSESFGSDGALRTVLAR 1132
> eco:b1055 yceA, ECK1040, JW1042; predicted rhodanese-related
sulfurtransferase; K07146 UPF0176 protein
Length=350
Score = 67.0 bits (162), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 40/108 (37%), Positives = 57/108 (52%), Gaps = 24/108 (22%)
Query 18 PKVVLLDMRNASEYAVGHFKGSQCVDTLTFADSFTSGGPLEEALARVGIQLPSREATSVS 77
P + +DMRN EY VGHF+ + + TF + QLP +A +
Sbjct 148 PDALFIDMRNHYEYEVGHFENALEIPADTFRE-----------------QLP--KAVEMM 188
Query 78 SKGSCSRKDVEIMMYCTGGIRCVKAGAFVKQILGFPRVTRLKGGILAY 125
+ KD +I+MYCTGGIRC KA A++K GF +V ++GGI+ Y
Sbjct 189 Q----AHKDKKIVMYCTGGIRCEKASAWMKHN-GFNKVWHIEGGIIEY 231
> ath:AT2G40760 rhodanese-like domain-containing protein
Length=474
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 44/116 (37%), Positives = 59/116 (50%), Gaps = 11/116 (9%)
Query 18 PKVVLLDMRNASEYAVGHFKGSQCVDTLTFA-DSFTSGGPLEEALARVGIQLPSR----- 71
P+ V++D+RN E +G FKG+ VD T A +F S + AL + G + ++
Sbjct 247 PETVVIDVRNTYETRIGKFKGA--VDPCTTAFRNFPSWVENQFALKQEGNETQAKVEKED 304
Query 72 --EATSVSSKGSCSRKDVEIMMYCTGGIRCVKAGAFVKQILGFPRVTRLKGGILAY 125
E T K + I MYCTGGIRC KA + + GF V LKGGIL Y
Sbjct 305 FSEITHKEDKAEKPKTLPRIAMYCTGGIRCEKASSLLLS-QGFEEVYHLKGGILKY 359
> ath:AT1G17850 hypothetical protein
Length=446
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 43/126 (34%), Positives = 63/126 (50%), Gaps = 16/126 (12%)
Query 1 RKLVALLQEQQLQKQRTPK-VVLLDMRNASEYAVGHFKGSQCVDTLTFADSFTSGGPLEE 59
RKL L + + + K +LLD+RN E+ VGHF+G+ + F + TS G +E
Sbjct 202 RKLKDLTDDDEASPSNSGKSYILLDVRNGYEWDVGHFRGAHRPEVDCFRN--TSFGLSDE 259
Query 60 ALARVGIQLPSREATSVSSKGSCSRKDVEIMMYCTGGIRCVKAGAFVKQILGFPRVTRLK 119
A PS +V ++ +I+MYCTGGIRC ++Q GF + LK
Sbjct 260 KEA------PSDPLINVD------KEKTDILMYCTGGIRCDVYSTVLRQ-RGFKNLYTLK 306
Query 120 GGILAY 125
GG+ Y
Sbjct 307 GGVSHY 312
> hsa:158427 TSTD2, C9orf97, FLJ36724; thiosulfate sulfurtransferase
(rhodanese)-like domain containing 2
Length=516
Score = 45.4 bits (106), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 35/121 (28%), Positives = 51/121 (42%), Gaps = 24/121 (19%)
Query 15 QRTPKVVLLDMRNASEYAVGHFKGSQCVDTLTFADSFTSGGPLEEALARVGIQLPSREAT 74
Q +LLD RN E +G F+G D F+ PS
Sbjct 300 QEQSDTILLDCRNFYESKIGRFQGCLAPDIRKFS------------------YFPSYVDK 341
Query 75 SVSSKGSCSRKDVEIMMYCTGGIRCVKAGAFVKQILGFPRVTRLKGGILAY-KNFIESFK 133
++ ++ ++MYCTGGIRC + A++K V +LKGGI Y + F + F
Sbjct 342 NLEL-----FREKRVLMYCTGGIRCERGSAYLKAKGVCKEVFQLKGGIHKYLEEFPDGFY 396
Query 134 K 134
K
Sbjct 397 K 397
> ath:AT1G09280 hypothetical protein
Length=581
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 37/114 (32%), Positives = 54/114 (47%), Gaps = 22/114 (19%)
Query 14 KQRTPKVVLLDMRNASEYAVGHFKGSQCVDTLTFADSFTSGGPLEEALARVGIQLPSR-E 72
K ++VLLD RN E +G F+ S+ V+TL + R LP+ +
Sbjct 156 KSENKELVLLDARNLYETRIGKFE-SENVETL-------------DPEIRQYSDLPTWID 201
Query 73 ATSVSSKGSCSRKDVEIMMYCTGGIRCVKAGAFVK-QILGFPRVTRLKGGILAY 125
+ KG ++MYCTGGIRC A A+++ + GF +L GGI Y
Sbjct 202 QNAEKMKGK------NVLMYCTGGIRCEMASAYIRSKGAGFENTFQLYGGIQRY 249
> xla:100137641 tstd2; thiosulfate sulfurtransferase (rhodanese)-like
domain containing 2
Length=482
Score = 44.7 bits (104), Expect = 1e-04, Method: Composition-based stats.
Identities = 35/120 (29%), Positives = 50/120 (41%), Gaps = 26/120 (21%)
Query 21 VLLDMRNASEYAVGHFKGSQCVDTLTFADSFTSGGPLEEALARVGIQLPSREATSVSSKG 80
+LLD RN E +G F+ D F+ PS ++
Sbjct 295 ILLDCRNFYESKIGKFQNCLAPDIRKFS------------------YFPSYVDQNLEL-- 334
Query 81 SCSRKDVEIMMYCTGGIRCVKAGAFVKQILGFPRVTRLKGGILAYKNFIESFKKRPNRGE 140
KD ++MYCTGGIRC + A+++ V +LKGGI Y +E F RG+
Sbjct 335 ---FKDKTVLMYCTGGIRCERGSAYLRSKSVCKEVYQLKGGIHKY---LEQFPDGYFRGK 388
> dre:100151606 hypothetical LOC100151606
Length=324
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 32/56 (57%), Gaps = 3/56 (5%)
Query 85 KDVEIMMYCTGGIRCVKAGAFVKQILGFPRVTRLKGGILAYKNFIESFKKRPNRGE 140
+D ++MYCTGGIRC + A+++ V +LKGG+ Y +E F RG+
Sbjct 244 RDKRVLMYCTGGIRCERGSAYLRSKRVCKEVYQLKGGVHKY---LEQFPDGFYRGK 296
> mmu:272027 Tstd2, 3010020C06; thiosulfate sulfurtransferase
(rhodanese)-like domain containing 2
Length=495
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 32/116 (27%), Positives = 49/116 (42%), Gaps = 24/116 (20%)
Query 20 VVLLDMRNASEYAVGHFKGSQCVDTLTFADSFTSGGPLEEALARVGIQLPSREATSVSSK 79
++LD RN E +G F+G D F+ PS ++
Sbjct 304 TIILDCRNFYESKIGRFQGCLAPDIRKFS------------------YFPSYVDKNLDI- 344
Query 80 GSCSRKDVEIMMYCTGGIRCVKAGAFVKQILGFPRVTRLKGGILAY-KNFIESFKK 134
+ ++MYCTGGIRC + A+++ V +LKGGI Y + F + F K
Sbjct 345 ----FRQKRVLMYCTGGIRCERGSAYLRAKGVCKEVFQLKGGIHKYLEEFPDGFYK 396
> sce:YOR285W RDL1; Protein of unknown function containing a rhodanese-like
domain; localized to the mitochondrial outer membrane
(EC:2.8.1.1)
Length=139
Score = 36.6 bits (83), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 27/84 (32%), Positives = 39/84 (46%), Gaps = 16/84 (19%)
Query 15 QRTPKVVLLDMRNASEYAVGHFKGSQCVDTLTFADSFTSGGPLEEALARVGIQLPSREAT 74
+ P VVL+D+R SEY++ H S V + D+F PLE ++GI P
Sbjct 36 KHDPNVVLVDVREPSEYSIVHIPASINVPYRSHPDAFAL-DPLEFE-KQIGIPKPD---- 89
Query 75 SVSSKGSCSRKDVEIMMYCTGGIR 98
S+K E++ YC G R
Sbjct 90 --SAK--------ELIFYCASGKR 103
> sce:YHR111W UBA4, YHR1; Protein that activates Urm1p before
its conjugation to proteins (urmylation); one target is the
thioredoxin peroxidase Ahp1p, suggesting a role of urmylation
in the oxidative stress response; K11996 adenylyltransferase
and sulfurtransferase
Length=440
Score = 33.9 bits (76), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 27/112 (24%), Positives = 44/112 (39%), Gaps = 19/112 (16%)
Query 19 KVVLLDMRNASEYAVGHFKGSQCVDTLTFADSFTSGGPLEEALARVGIQLPSREATSVSS 78
K + LD+R + Y + HF + + D + L ++ +LPS E
Sbjct 342 KHIFLDVRPSHHYEISHFPEAVNIPIKNLRD-------MNGDLKKLQEKLPSVE------ 388
Query 79 KGSCSRKDVEIMMYCTGGIRCVKAGAFVKQILGFPRVTRLKGGILAYKNFIE 130
KD I++ C G A +K GF V ++GG Y + I+
Sbjct 389 ------KDSNIVILCRYGNDSQLATRLLKDKFGFSNVRDVRGGYFKYIDDID 434
> tgo:TGME49_054570 hypothetical protein
Length=1972
Score = 32.0 bits (71), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 33/73 (45%), Gaps = 7/73 (9%)
Query 1 RKLVALLQEQQLQKQRTPKVVLLDMRNASEYAVGHFKGSQCVDTLTFADSFTSGGPLEEA 60
+KLVA LQ +R R E V + C D+ + DS +GG LEE
Sbjct 958 KKLVAFLQ------RRASSSAFGTERREGESEVSAHTEASCTDSRSCTDSRAAGGSLEEC 1011
Query 61 LARVGIQLPSREA 73
L ++ QL REA
Sbjct 1012 L-KLQTQLAQREA 1023
> sce:YER144C UBP5; Putative ubiquitin-specific protease, closest
paralog of Doa4p but has no functional overlap; concentrates
at the bud neck (EC:3.1.2.15); K11839 ubiquitin carboxyl-terminal
hydrolase 8 [EC:3.1.2.15]
Length=805
Score = 31.2 bits (69), Expect = 1.5, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 21/35 (60%), Gaps = 2/35 (5%)
Query 20 VVLLDMRNASEYAVGHFKGSQ--CVDTLTFADSFT 52
++L+D+R SE+ H K C+D +F DSFT
Sbjct 163 LLLIDVRPRSEFVRAHIKCKNIICIDPASFKDSFT 197
> xla:398494 hnrnpu; heterogeneous nuclear ribonucleoprotein U
(scaffold attachment factor A)
Length=797
Score = 30.8 bits (68), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 29/56 (51%), Gaps = 7/56 (12%)
Query 1 RKLVALLQEQQLQKQRTPKVVLLDMRNASEYAVGHFKG-------SQCVDTLTFAD 49
RK V + +L K+RT K ++ ++ E+A+ KG S+C D +T+ D
Sbjct 549 RKAVVVCPTDELYKERTQKKSEVEGKDLPEHAILKMKGNFTLPEASECFDEITYVD 604
> cel:F09C12.8 hypothetical protein
Length=306
Score = 30.4 bits (67), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 10/39 (25%), Positives = 26/39 (66%), Gaps = 2/39 (5%)
Query 90 MMYCTGGIRCVKAGAFVKQILGFPRVTRLKGGILAYKNF 128
++YC+ +RC++ +V+++ G + R++ G+ ++NF
Sbjct 124 VIYCSPSLRCIQTATWVREMSGSKALLRVEPGL--FENF 160
> cel:F54D5.12 hypothetical protein
Length=487
Score = 30.0 bits (66), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 29/59 (49%), Gaps = 6/59 (10%)
Query 57 LEEALARVGIQLPSREATSVSSKGSCSRKDVEIMMYCTGGIRCVKAGAFVKQILGFPRV 115
L+ LA++G +P + +KGSC + C GGIR ++ G+ +LG V
Sbjct 144 LDNKLAKLGYMMP----FDLGAKGSCQIGGN--IATCAGGIRLIRYGSLHAHLLGLTVV 196
> mmu:271981 Tbck, 1700120J03Rik, 9430001M19, A630047E20Rik, Tbckl;
TBC1 domain containing kinase
Length=893
Score = 29.3 bits (64), Expect = 6.4, Method: Composition-based stats.
Identities = 25/103 (24%), Positives = 46/103 (44%), Gaps = 19/103 (18%)
Query 20 VVLLDMRNASEYAVGHFKGSQCVDTLTFADSFTSGGPLEEALARVGIQLPSREATSVSSK 79
++++D+RN+ ++ GH GS + F+ +FT+ G L + L + + +
Sbjct 794 LLVVDIRNSEDFVRGHIAGSI---NIPFSAAFTAEGELSQ--GPYTTMLHNFKGKVIVVV 848
Query 80 GSCSRKDVEIMMYCTGGIRCVKAGAFVKQILGFPRVTRLKGGI 122
G +++ E A VK + +PRV L GGI
Sbjct 849 GHVAKQTAEF------------AAHLVK--MKYPRVCILDGGI 877
> xla:447020 tbck, MGC82809, hspc302, tbckl; TBC1 domain containing
kinase
Length=893
Score = 28.9 bits (63), Expect = 7.3, Method: Composition-based stats.
Identities = 25/103 (24%), Positives = 47/103 (45%), Gaps = 19/103 (18%)
Query 20 VVLLDMRNASEYAVGHFKGSQCVDTLTFADSFTSGGPLEEALARVGIQLPSREATSVSSK 79
++++D+R++ ++ GH GS + F +FT+ G L A +Q + + V +
Sbjct 794 LLVVDIRHSEDFNRGHIPGSI---NMPFGSAFTTEGDLAPCPASTTLQ--NFKGKVVVTV 848
Query 80 GSCSRKDVEIMMYCTGGIRCVKAGAFVKQILGFPRVTRLKGGI 122
G ++ + ++ VK L +PRV L GGI
Sbjct 849 GHAAQNAAKFAVH------------LVK--LAYPRVCILDGGI 877
Lambda K H
0.318 0.134 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3712313100
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40