bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1474_orf3
Length=235
Score E
Sequences producing significant alignments: (Bits) Value
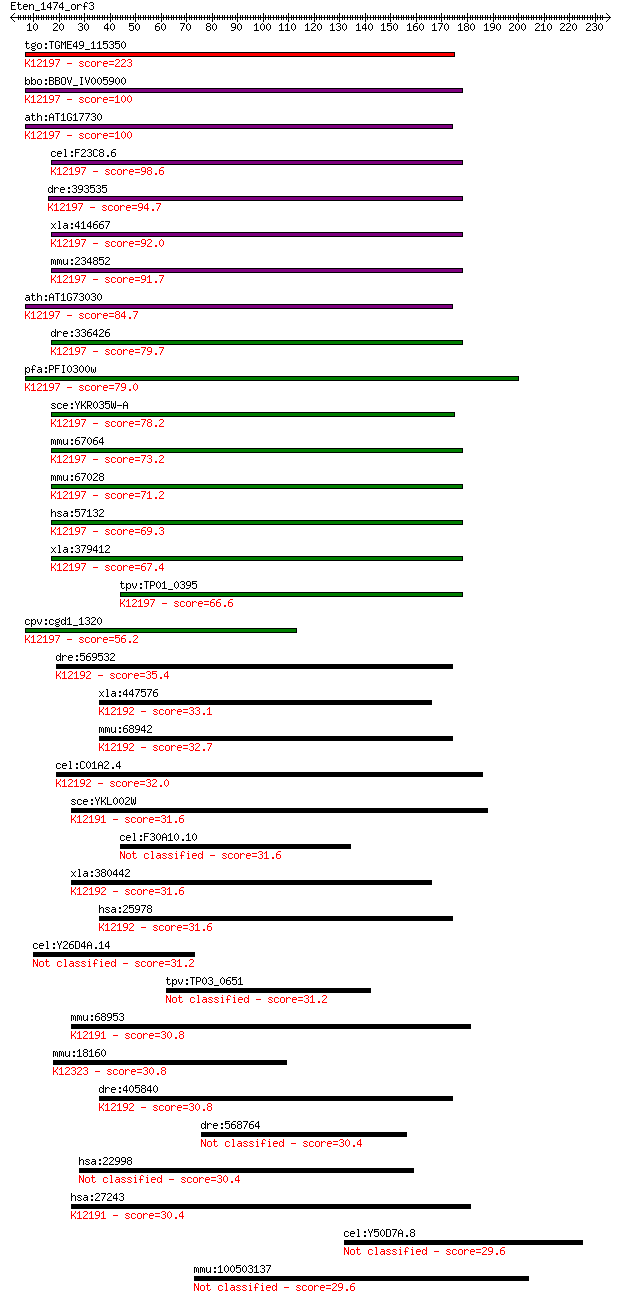
tgo:TGME49_115350 hypothetical protein ; K12197 charged multiv... 223 6e-58
bbo:BBOV_IV005900 23.m06461; hypothetical protein; K12197 char... 100 4e-21
ath:AT1G17730 VPS46.1 (VACUOLAR PROTEIN SORTING 46.1); K12197 ... 100 5e-21
cel:F23C8.6 hypothetical protein; K12197 charged multivesicula... 98.6 2e-20
dre:393535 chmp1a, KM0004, MGC65914, pcoln3, wu:fb50a12, zgc:6... 94.7 2e-19
xla:414667 chmp1a; chromatin modifying protein 1A; K12197 char... 92.0 2e-18
mmu:234852 Chmp1a, 2900018H07Rik, Pcoln3; chromatin modifying ... 91.7 2e-18
ath:AT1G73030 VPS46.2; K12197 charged multivesicular body prot... 84.7 3e-16
dre:336426 chmp1b, Chmp1.5, fb10c03, wu:fb10c03, zgc:56134; ch... 79.7 9e-15
pfa:PFI0300w developmental protein, putative; K12197 charged m... 79.0 1e-14
sce:YKR035W-A DID2, CHM1, FTI1, VPL30, VPS46; Class E protein ... 78.2 2e-14
mmu:67064 Chmp1b, 2810405I11Rik, Chmp1b1; chromatin modifying ... 73.2 8e-13
mmu:67028 2610002M06Rik, 8430408J07Rik, AI876413, Chmp1b2; RIK... 71.2 3e-12
hsa:57132 CHMP1B, C10orf2, C18-ORF2, C18orf2, CHMP1.5, Vps46-2... 69.3 1e-11
xla:379412 chmp1b, MGC64275; chromatin modifying protein 1B; K... 67.4 4e-11
tpv:TP01_0395 hypothetical protein; K12197 charged multivesicu... 66.6 6e-11
cpv:cgd1_1320 developmental protein ; K12197 charged multivesi... 56.2 1e-07
dre:569532 novel protein similar to H.sapiens CHMP2B, chromati... 35.4 0.17
xla:447576 chmp2b-b, MGC84310, chmp2.5, dmt1, vps2-2, vps2b; c... 33.1 0.77
mmu:68942 Chmp2b, 1190006E07Rik; chromatin modifying protein 2... 32.7 1.3
cel:C01A2.4 hypothetical protein; K12192 charged multivesicula... 32.0 1.9
sce:YKL002W DID4, CHM2, GRD7, REN1, VPL2, VPS14, VPS2, VPT14; ... 31.6 2.2
cel:F30A10.10 hypothetical protein 31.6 2.5
xla:380442 chmp2b-a, MGC64560, cg4618, chmp2.5, dmt1, vps2-2, ... 31.6 2.5
hsa:25978 CHMP2B, CHMP2.5, DKFZp564O123, DMT1, VPS2-2, VPS2B; ... 31.6 2.9
cel:Y26D4A.14 hypothetical protein 31.2 3.3
tpv:TP03_0651 hypothetical protein 31.2 3.5
mmu:68953 Chmp2a, 1500016L11Rik; chromatin modifying protein 2... 30.8 4.0
mmu:18160 Npr1, AI893888, GC-A, MGC117511, NPR-A, NPRA, Pndr; ... 30.8 4.2
dre:405840 chmp2bl, MGC77025, chmp2b, zgc:77025; chromatin mod... 30.8 4.7
dre:568764 mKIAA0969 protein-like 30.4 5.1
hsa:22998 LIMCH1, DKFZp434I0312, DKFZp686A01247, DKFZp686B2470... 30.4 5.5
hsa:27243 CHMP2A, BC-2, BC2, CHMP2, VPS2, VPS2A; chromatin mod... 30.4 5.6
cel:Y50D7A.8 hypothetical protein 29.6 9.0
mmu:100503137 putative uncharacterized protein C8orf73-like 29.6 9.5
> tgo:TGME49_115350 hypothetical protein ; K12197 charged multivesicular
body protein 1
Length=199
Score = 223 bits (567), Expect = 6e-58, Method: Compositional matrix adjust.
Identities = 107/168 (63%), Positives = 138/168 (82%), Gaps = 0/168 (0%)
Query 7 MGNHHSVLPVILDLRIKARELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVR 66
MGN HSV+P+I D++IKARELKRQS+RCYRES E+ K++ AL + N EGAR+FAQN++R
Sbjct 1 MGNKHSVIPLIFDMKIKARELKRQSERCYRESEGEKGKVKAALQKNNSEGARVFAQNFIR 60
Query 67 KKQEGLNSLHMSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIEL 126
K+QEGL+ L +SSKL+AVASRLD AHRS Q++ ++ S A GLS ALR LD +SLR++E
Sbjct 61 KQQEGLHCLQLSSKLDAVASRLDAAHRSQQMSKQMRSAAYGLSNALRTLDGGTSLRQMEE 120
Query 127 FSKLFDDLDVRSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLE 174
F+KLFDDLDVRSDSVS+LLDASTS+++P +QVD VL VA+ I +E
Sbjct 121 FAKLFDDLDVRSDSVSTLLDASTSTSIPLEQVDDVLKHVATAAKIHME 168
> bbo:BBOV_IV005900 23.m06461; hypothetical protein; K12197 charged
multivesicular body protein 1
Length=190
Score = 100 bits (250), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 57/171 (33%), Positives = 96/171 (56%), Gaps = 1/171 (0%)
Query 7 MGNHHSVLPVILDLRIKARELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVR 66
MG + S +P LDLR++ARE + +C RE + E+EK++R+L + N E ARI A N +R
Sbjct 1 MGINQSSMPTSLDLRLQAREALKLQSQCEREEVNEKEKVKRSLEKGNMEAARIHAGNAIR 60
Query 67 KKQEGLNSLHMSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIEL 126
K E L L SKLE + S+++ A R++ L ++ LS + K S+ + ++
Sbjct 61 KHNEALRYLQYHSKLEILRSQVESAERTNALNEQLKEALPRLS-KMTKYKSNGNTNLMQE 119
Query 127 FSKLFDDLDVRSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLEDQL 177
K+FDDLDV +++ + P +VD ++++VA + + + D L
Sbjct 120 LEKIFDDLDVSEAYGDMTMNSVNAHLAPQNEVDNLISKVADEYALDIGDML 170
> ath:AT1G17730 VPS46.1 (VACUOLAR PROTEIN SORTING 46.1); K12197
charged multivesicular body protein 1
Length=203
Score = 100 bits (249), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 48/167 (28%), Positives = 101/167 (60%), Gaps = 0/167 (0%)
Query 7 MGNHHSVLPVILDLRIKARELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVR 66
MGN ++ I +L+ ++ L+RQ+ +C +E E+ K+++A+ + N +GARI+A+N +R
Sbjct 1 MGNTDKLMNQIFELKFTSKSLQRQARKCEKEERSEKLKVKKAIEKGNMDGARIYAENAIR 60
Query 67 KKQEGLNSLHMSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIEL 126
K+ E +N L +SS+L+AV +RLD + +T + ++ L +L + ++
Sbjct 61 KRSEQMNYLRLSSRLDAVVARLDTQAKMATITKSMTNIVKSLESSLTTGNLQKMSETMDS 120
Query 127 FSKLFDDLDVRSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISL 173
F K F +++V+++ + + + STS + P +V+ ++ +VA +G+ +
Sbjct 121 FEKQFVNMEVQAEFMDNAMAGSTSLSTPEGEVNSLMQQVADDYGLEV 167
> cel:F23C8.6 hypothetical protein; K12197 charged multivesicular
body protein 1
Length=205
Score = 98.6 bits (244), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 46/161 (28%), Positives = 95/161 (59%), Gaps = 0/161 (0%)
Query 17 ILDLRIKARELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLH 76
+ DL+ A++L++ + RC ++ E++K+ A+ + N+E A++ A+N +RKK E +N +
Sbjct 14 LFDLKFAAKQLEKNAQRCEKDEKVEKDKLTAAIKKGNKEVAQVHAENAIRKKNEAVNYIK 73
Query 77 MSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDV 136
M+++++AVA+R+ A ++T + V + A++ ++ + ++ F + F+DLDV
Sbjct 74 MAARIDAVAARVQTAATQKRVTASMSGVVKAMESAMKSMNLEKVQQLMDRFERDFEDLDV 133
Query 137 RSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLEDQL 177
+ ++ +D +T P QVD ++ E A GI L +L
Sbjct 134 TTKTMEKTMDGTTVLNAPKSQVDALIAEAADKAGIELNQEL 174
> dre:393535 chmp1a, KM0004, MGC65914, pcoln3, wu:fb50a12, zgc:65914,
zgc:85863; chromatin modifying protein 1A; K12197 charged
multivesicular body protein 1
Length=198
Score = 94.7 bits (234), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 50/162 (30%), Positives = 95/162 (58%), Gaps = 0/162 (0%)
Query 16 VILDLRIKARELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVRKKQEGLNSL 75
+ L+ +++L+R + + ++S E+ K+++AL ++N E AR++A+N +RKK EGLN L
Sbjct 4 TLFQLKFTSKQLERLAKKAEKDSKSEQAKVKKALQQKNVECARVYAENAIRKKNEGLNWL 63
Query 76 HMSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLD 135
M+S+++AVAS++ A + + V L AL +D ++ F +LD
Sbjct 64 RMASRVDAVASKVQTALTMKGVAKNMTQVTKALDKALSSMDLQKVSAVMDKFETQVQNLD 123
Query 136 VRSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLEDQL 177
V + + + ++T+ + P QQVD ++ ++A G+ +EDQL
Sbjct 124 VHTSVMEDSMSSATTLSTPQQQVDDLILQIAEESGLEVEDQL 165
> xla:414667 chmp1a; chromatin modifying protein 1A; K12197 charged
multivesicular body protein 1
Length=196
Score = 92.0 bits (227), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 53/165 (32%), Positives = 97/165 (58%), Gaps = 8/165 (4%)
Query 17 ILDLRIKARELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLH 76
+ L+ A++L++ + + ++S E+ K+++AL ++N E AR++A+N +RKK EGLN L
Sbjct 5 LFQLKFTAKQLEKLAKKAEKDSNTEQAKVKKALQQKNVEVARVYAENAIRKKNEGLNWLR 64
Query 77 MSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDV 136
M+S+++AVAS++ A +T + V L AL +D ++ F + +LDV
Sbjct 65 MASRVDAVASKVQTAVTMKGVTKNMAQVTKALDKALSSMDLQKVSAVMDKFDQQVQNLDV 124
Query 137 RS----DSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLEDQL 177
+ DS+SS + +T P +QVD ++ ++A G+ + DQL
Sbjct 125 HTSVMEDSMSSAMTLTT----PQEQVDNLIVQIAEENGLEVMDQL 165
> mmu:234852 Chmp1a, 2900018H07Rik, Pcoln3; chromatin modifying
protein 1A; K12197 charged multivesicular body protein 1
Length=196
Score = 91.7 bits (226), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 48/161 (29%), Positives = 95/161 (59%), Gaps = 0/161 (0%)
Query 17 ILDLRIKARELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLH 76
+ L+ A++L++ + + ++S E+ K+++AL ++N E AR++A+N +RKK EG+N L
Sbjct 5 LFQLKFTAKQLEKLAKKAEKDSKAEQAKVKKALQQKNVECARVYAENAIRKKNEGVNWLR 64
Query 77 MSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDV 136
M+S+++AVAS++ A +T + V L AL +D ++ F + +LDV
Sbjct 65 MASRVDAVASKVQTAVTMKGVTKNMAQVTKALDKALSAMDLQKVSAVMDRFEQQVQNLDV 124
Query 137 RSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLEDQL 177
+ + + ++T+ P +QVD ++ ++A G+ + DQL
Sbjct 125 HTSVMEDSVSSATTLTTPQEQVDSLIVQIAEENGLEVLDQL 165
> ath:AT1G73030 VPS46.2; K12197 charged multivesicular body protein
1
Length=203
Score = 84.7 bits (208), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 49/167 (29%), Positives = 101/167 (60%), Gaps = 0/167 (0%)
Query 7 MGNHHSVLPVILDLRIKARELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVR 66
MGN ++ I DL+ ++ L+RQS +C +E E+ K+++A+ + N +GARI+A+N +R
Sbjct 1 MGNTDKLMNQIFDLKFTSKSLQRQSRKCEKEEKAEKLKVKKAIEKGNMDGARIYAENAIR 60
Query 67 KKQEGLNSLHMSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIEL 126
K+ E +N L ++S+L+AV +RLD + +T + ++ L +L + ++
Sbjct 61 KRSEQMNYLRLASRLDAVVARLDTQAKMTTITKSMTNIVKSLESSLATGNLQKMSETMDS 120
Query 127 FSKLFDDLDVRSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISL 173
F K F +++V+++ + + + STS + P +V+ ++ +VA +G+ +
Sbjct 121 FEKQFVNMEVQAEFMENAMAGSTSLSTPEGEVNSLMQQVADDYGLEV 167
> dre:336426 chmp1b, Chmp1.5, fb10c03, wu:fb10c03, zgc:56134;
chromatin modifying protein 1B; K12197 charged multivesicular
body protein 1
Length=199
Score = 79.7 bits (195), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 50/161 (31%), Positives = 96/161 (59%), Gaps = 0/161 (0%)
Query 17 ILDLRIKARELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLH 76
+ +L+ A+EL+R S +C +E E+ K+++A+ + N E ARI A+N +R+K + +N L
Sbjct 8 LFNLKFAAKELQRNSKKCDKEEKAEKVKVKKAIQKGNMEVARIHAENAIRQKNQSVNFLR 67
Query 77 MSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDV 136
MS++++AVA+R+ A +Q+T + V G+ L+ ++ +E F + F+ LDV
Sbjct 68 MSARVDAVAARVQTAVTMNQVTKSMAGVVKGMDATLKSMNLEKISGLMEKFERQFETLDV 127
Query 137 RSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLEDQL 177
++ + + ++T+ P QVD ++ E+A G+ L +L
Sbjct 128 QTAQMEDSMSSTTTLTTPQGQVDTLMMEMADEAGLDLNMEL 168
> pfa:PFI0300w developmental protein, putative; K12197 charged
multivesicular body protein 1
Length=195
Score = 79.0 bits (193), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 49/193 (25%), Positives = 102/193 (52%), Gaps = 4/193 (2%)
Query 7 MGNHHSVLPVILDLRIKARELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVR 66
MGN S I L++K +EL++ S R E + +++A+ E AR++A+ +R
Sbjct 1 MGNKISTEDHIFRLKLKTKELEKLSQRSELEEKKLIGDVKKAIQAGKIELARLYAEKCIR 60
Query 67 KKQEGLNSLHMSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIEL 126
KK E +N L++S+KL+ + SRL+GAHR L + + + + ++ ++
Sbjct 61 KKNEKVNYLNLSNKLDVLVSRLEGAHRCASLVKDVGVMIPLIQKINAETNAIKIGNDVTK 120
Query 127 FSKLFDDLDVRSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLEDQLEGFEGVDTP 186
+FD++ + S+ ++ + S++ + P ++VD++++++A + L+ Q+ G P
Sbjct 121 LENIFDEISISSELINDTVQTSSAISAPTEEVDELISKIADEHALKLDGQI----GSVHP 176
Query 187 QKEHCERQRAMGD 199
+H E M +
Sbjct 177 INKHLEEISNMSE 189
> sce:YKR035W-A DID2, CHM1, FTI1, VPL30, VPS46; Class E protein
of the vacuolar protein-sorting (Vps) pathway; binds Vps4p
and directs it to dissociate ESCRT-III complexes; forms a functional
and physical complex with Ist1p; human ortholog may
be altered in breast tumors; K12197 charged multivesicular
body protein 1
Length=204
Score = 78.2 bits (191), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 45/158 (28%), Positives = 93/158 (58%), Gaps = 1/158 (0%)
Query 17 ILDLRIKARELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLH 76
+ L+ +++L++Q+++ +E QE K++RAL +N++ +RI+A N +RKK E L L
Sbjct 13 LFQLKFTSKQLQKQANKASKEEKQETNKLKRAL-NENEDISRIYASNAIRKKNERLQLLK 71
Query 77 MSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDV 136
++S++++VASR+ A Q++ + V G+ AL+ ++ ++ F + F+DLD
Sbjct 72 LASRVDSVASRVQTAVTMRQVSASMGQVCKGMDKALQNMNLQQITMIMDKFEQQFEDLDT 131
Query 137 RSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLE 174
+ + S + V +VD+++++VA G+ L+
Sbjct 132 SVNVYEDMGVNSDAMLVDNDKVDELMSKVADENGMELK 169
> mmu:67064 Chmp1b, 2810405I11Rik, Chmp1b1; chromatin modifying
protein 1B; K12197 charged multivesicular body protein 1
Length=199
Score = 73.2 bits (178), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 50/161 (31%), Positives = 93/161 (57%), Gaps = 0/161 (0%)
Query 17 ILDLRIKARELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLH 76
+ +L+ A+EL R S +C +E E+ KI++A+ + N E ARI A+N +R+K +G+N L
Sbjct 8 LFNLKFAAKELNRSSKKCDKEEKAEKAKIKKAIQKGNMEVARIHAENAIRQKNQGVNFLR 67
Query 77 MSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDV 136
MS++++AVA+R+ A ++T + V + L+ ++ ++ F F+ LDV
Sbjct 68 MSARVDAVAARVQTAVTMGKVTKSMAGVVKSMDATLKSMNLEKISALMDKFEHQFETLDV 127
Query 137 RSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLEDQL 177
++ + + ++T+ P QVD +L E+A G+ L +L
Sbjct 128 QTQQMEDTMSSTTTLTTPQNQVDMLLQEMADEAGLDLNMEL 168
> mmu:67028 2610002M06Rik, 8430408J07Rik, AI876413, Chmp1b2; RIKEN
cDNA 2610002M06 gene; K12197 charged multivesicular body
protein 1
Length=199
Score = 71.2 bits (173), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 49/161 (30%), Positives = 92/161 (57%), Gaps = 0/161 (0%)
Query 17 ILDLRIKARELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLH 76
+ +L+ A+EL R + +C +E E+ KI++A+ + N E ARI A+N +R+K + +N L
Sbjct 8 LFNLKFAAKELNRNAKKCDKEEKAEKAKIKKAIQKGNTEVARIHAENAIRQKNQAINFLR 67
Query 77 MSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDV 136
MS++++AVA+R+ A ++T + V + LR ++ ++ F F+ LDV
Sbjct 68 MSARVDAVAARVQTAVTMGKVTKSMAGVVKSMDATLRSMNLEKISALMDKFEHQFETLDV 127
Query 137 RSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLEDQL 177
++ + + ++T+ P QVD +L E+A G+ L +L
Sbjct 128 QTQQMEDTMSSTTTLTTPQNQVDMLLQEMADEAGLDLNMEL 168
> hsa:57132 CHMP1B, C10orf2, C18-ORF2, C18orf2, CHMP1.5, Vps46-2,
Vps46B; chromatin modifying protein 1B; K12197 charged multivesicular
body protein 1
Length=199
Score = 69.3 bits (168), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 48/161 (29%), Positives = 92/161 (57%), Gaps = 0/161 (0%)
Query 17 ILDLRIKARELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLH 76
+ +L+ A+EL R + +C +E E+ KI++A+ + N E ARI A+N +R+K + +N L
Sbjct 8 LFNLKFAAKELSRSAKKCDKEEKAEKAKIKKAIQKGNMEVARIHAENAIRQKNQAVNFLR 67
Query 77 MSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDV 136
MS++++AVA+R+ A ++T + V + L+ ++ ++ F F+ LDV
Sbjct 68 MSARVDAVAARVQTAVTMGKVTKSMAGVVKSMDATLKTMNLEKISALMDKFEHQFETLDV 127
Query 137 RSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLEDQL 177
++ + + ++T+ P QVD +L E+A G+ L +L
Sbjct 128 QTQQMEDTMSSTTTLTTPQNQVDMLLQEMADEAGLDLNMEL 168
> xla:379412 chmp1b, MGC64275; chromatin modifying protein 1B;
K12197 charged multivesicular body protein 1
Length=199
Score = 67.4 bits (163), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 49/161 (30%), Positives = 92/161 (57%), Gaps = 0/161 (0%)
Query 17 ILDLRIKARELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLH 76
+ +L+ A+EL R + +C +E E+ KI++A+ + N E ARI A+N +R+K +G+N L
Sbjct 8 LFNLKFAAKELNRNAKKCEKEEKTEKAKIKKAIQKGNTEIARIHAENAIRQKNQGINFLR 67
Query 77 MSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDV 136
MS++++AVA+R+ A ++T + V + L+ ++ ++ F F+ LDV
Sbjct 68 MSARVDAVAARVQTAVTMGKVTKSMAGVVKSMDATLKSMNLEKISALMDKFEHQFETLDV 127
Query 137 RSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLEDQL 177
++ + + +T+ P QVD +L E+A G+ L +L
Sbjct 128 QTQQMEDTMSNTTTLTTPQNQVDNLLHEMADEAGLDLSMEL 168
> tpv:TP01_0395 hypothetical protein; K12197 charged multivesicular
body protein 1
Length=154
Score = 66.6 bits (161), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 39/136 (28%), Positives = 75/136 (55%), Gaps = 5/136 (3%)
Query 44 KIRRALLRQNQEGARIFAQNYVRKKQEGLNSLHMSSKLEAVASRLDGAHRSHQLTTRIHS 103
K++ A+ +N E ARI A N + K+ E + L + S+L+ + +++D A R+ +LT +
Sbjct 2 KVKSAIESENPETARIHAANSIFKRNEAMRFLELKSRLDILTAQVDSAIRTQRLTEELKK 61
Query 104 VASGLSGALR--KLDSSSSLREIELFSKLFDDLDVRSDSVSSLLDASTSSAVPAQQVDKV 161
V L+ LR KLDS + + F K+F+++DV +S + + P ++VD +
Sbjct 62 VLPQLNRFLRYKKLDSVDVITD---FKKIFENMDVNESQMSENMSKDSPFKPPQREVDAL 118
Query 162 LTEVASTFGISLEDQL 177
++ VA + + + + L
Sbjct 119 ISRVAQEYELDMTEML 134
> cpv:cgd1_1320 developmental protein ; K12197 charged multivesicular
body protein 1
Length=188
Score = 56.2 bits (134), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 64/106 (60%), Gaps = 0/106 (0%)
Query 7 MGNHHSVLPVILDLRIKARELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVR 66
MGN S+ I +L+++ +EL+RQ ++ R+S ER+K + A++ + ++I+A+N +R
Sbjct 1 MGNRISIDDSIFELKLQKKELERQYNKRDRDSKSERKKAKDAIISGKADLSKIYAENSLR 60
Query 67 KKQEGLNSLHMSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGAL 112
++E L M SKL+ + +RL+ A +++T++ + L L
Sbjct 61 MQEECKELLIMISKLDRLCARLEKASSKEKISTQLLDIMPKLKKQL 106
> dre:569532 novel protein similar to H.sapiens CHMP2B, chromatin
modifying protein 2B (CHMP2B); K12192 charged multivesicular
body protein 2B
Length=216
Score = 35.4 bits (80), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 27/155 (17%), Positives = 73/155 (47%), Gaps = 0/155 (0%)
Query 19 DLRIKARELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLHMS 78
+LR R++ R R+ Q +I++ N+E +I A+ V+ +++ + +S
Sbjct 20 ELRGTQRQITRDRAALERQERQMEMEIKKMAKTGNREACKILAKQLVQLRKQKNRTYAVS 79
Query 79 SKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDVRS 138
SK+ +++++ + ++ + + A + +++D +L+ ++ F K + +
Sbjct 80 SKVTSMSTQTKVMNSQMKMAGAMSTTAKTMQTVNKRMDPKKTLQTMQDFQKENMKMGMTE 139
Query 139 DSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISL 173
D ++S LD + ++ ++ +V GI +
Sbjct 140 DMLNSTLDEIFDESADEEECQDIVNQVLDEIGIEI 174
> xla:447576 chmp2b-b, MGC84310, chmp2.5, dmt1, vps2-2, vps2b;
chromatin modifying protein 2B; K12192 charged multivesicular
body protein 2B
Length=214
Score = 33.1 bits (74), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 27/141 (19%), Positives = 70/141 (49%), Gaps = 13/141 (9%)
Query 36 RESLQEREK-----IRRALLRQNQEGARIFAQNYVRKKQEGLNSLHMSSKLEAVASRLDG 90
R +L+++EK I++ N++ R+ A+ V+ +++ + +SSK+ +++++
Sbjct 32 RAALEKQEKQLEMEIKKMAKTGNKDACRVLAKQLVQLRKQKTRTYAVSSKVTSMSTQTKV 91
Query 91 AHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSK------LFDDLDVRSDSVSSL 144
++ + + A + +K+D +L+ ++ F K + D++ +D++ +
Sbjct 92 MSSQMKMAGAMSTTAKTMQAVNKKMDPQKTLQTMQNFQKENMKMEMTDEM--INDTLDDI 149
Query 145 LDASTSSAVPAQQVDKVLTEV 165
DAS V++VL E+
Sbjct 150 FDASEDEEESQDIVNQVLDEI 170
> mmu:68942 Chmp2b, 1190006E07Rik; chromatin modifying protein
2B; K12192 charged multivesicular body protein 2B
Length=213
Score = 32.7 bits (73), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 24/143 (16%), Positives = 69/143 (48%), Gaps = 5/143 (3%)
Query 36 RESLQEREK-----IRRALLRQNQEGARIFAQNYVRKKQEGLNSLHMSSKLEAVASRLDG 90
R +L+++EK I++ N+E R+ A+ V +++ + +SSK+ +++++
Sbjct 32 RAALEKQEKQLELEIKKMAKIGNKEACRVLAKQLVHLRKQKTRTFAVSSKVTSMSTQTKV 91
Query 91 AHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDVRSDSVSSLLDASTS 150
+ ++ + + A + +K+D +L+ ++ F K +++ + ++ LD
Sbjct 92 MNSQMKMAGAMSTTAKTMQAVNKKMDPQKTLQTMQNFQKENMKMEMTEEMINDTLDDIFD 151
Query 151 SAVPAQQVDKVLTEVASTFGISL 173
+ ++ ++ +V GI +
Sbjct 152 GSDDEEESQDIVNQVLDEIGIEI 174
> cel:C01A2.4 hypothetical protein; K12192 charged multivesicular
body protein 2B
Length=207
Score = 32.0 bits (71), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 40/179 (22%), Positives = 84/179 (46%), Gaps = 24/179 (13%)
Query 19 DLRIKAREL---KRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVRKKQEGLNSL 75
+LR R+L +RQ DR +E QE I++ + + + AR A+ V+ + + S+
Sbjct 19 ELRKTNRDLESDRRQMDRREKELEQE---IKKLAAKGHNDAARHLAKQLVQLRNQKTKSI 75
Query 76 HMSSKLEAVASRLDGAHRSH-QLTTRIHSVASGLSGALRKLDS-------SSSLREIELF 127
MS+++ V A SH Q ++ S ++ +++ ++++RE ++
Sbjct 76 GMSARISGVQ-----AQNSHMQSMAKMGSAMGTTVKTMKAMNAQMPLEKVAANMREFQMA 130
Query 128 -SKLFDDLDVRSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLEDQLEGFEGVDT 185
K+ ++ +D++ S+LDA A + D ++ +V GI + +L + T
Sbjct 131 QEKMGLTEEMMNDTLDSILDAPGD----ADEQDAIVNQVLDEIGIEMNSKLANVPALPT 185
> sce:YKL002W DID4, CHM2, GRD7, REN1, VPL2, VPS14, VPS2, VPT14;
Class E Vps protein of the ESCRT-III complex, required for
sorting of integral membrane proteins into lumenal vesicles
of multivesicular bodies, and for delivery of newly synthesized
vacuolar enzymes to the vacuole, involved in endocytosis;
K12191 charged multivesicular body protein 2A
Length=232
Score = 31.6 bits (70), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 41/169 (24%), Positives = 78/169 (46%), Gaps = 16/169 (9%)
Query 25 RELKRQSDRCYRESLQEREKIRRALLRQNQEGARIFAQNYVRKKQEGLNSLHMSSKLEAV 84
REL+R+ + + + +I+++ A++ A++ VR + +M ++L+A+
Sbjct 30 RELEREKRKLELQDKKLVSEIKKSAKNGQVAAAKVQAKDLVRTRNYIQKFDNMKAQLQAI 89
Query 85 ASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIEL-FSKLFDDLDVRSDSVSS 143
+ R+ S Q+ TR S A+GL + + + L+ I + F K D + R + +
Sbjct 90 SLRIQAVRSSDQM-TRSMSEATGLLAGMNRTMNLPQLQRISMEFEKQSDLMGQRQEFMDE 148
Query 144 LLDASTSSAVPAQQ-----VDKVLTEVASTFGISLEDQLEGFEGVDTPQ 187
+D V + V+KVL E+ G+ L QL+ TPQ
Sbjct 149 AIDNVMGDEVDEDEEADEIVNKVLDEI----GVDLNSQLQS-----TPQ 188
> cel:F30A10.10 hypothetical protein
Length=1262
Score = 31.6 bits (70), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 24/96 (25%), Positives = 47/96 (48%), Gaps = 6/96 (6%)
Query 44 KIRRALLRQNQEGARIFAQNYVRKKQEGLNSLHMSSKLEAVASRLDGAHRSHQLTTRIH- 102
++++ +QN E +++Q RK G N + S + + V+SR+ SH++ T H
Sbjct 637 ELKKRTRQQNGEVKYVYSQRTPRKSHNGTNGTNSSPQKQPVSSRVAALLSSHEIPTCGHG 696
Query 103 --SVASGLSGALRKLDSSSS---LREIELFSKLFDD 133
S+ L G ++ + + + LRE + K+ D
Sbjct 697 KMSIDPILYGDVKAVSRAPAIALLREYDFRVKIVYD 732
> xla:380442 chmp2b-a, MGC64560, cg4618, chmp2.5, dmt1, vps2-2,
vps2b; chromatin modifying protein 2B; K12192 charged multivesicular
body protein 2B
Length=214
Score = 31.6 bits (70), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 29/152 (19%), Positives = 75/152 (49%), Gaps = 11/152 (7%)
Query 25 RELKRQSDRCYRE--SLQEREK-----IRRALLRQNQEGARIFAQNYVRKKQEGLNSLHM 77
+ELK Q R+ +L+++EK I++ N++ ++ A+ V+ +++ + +
Sbjct 19 KELKGQQRAITRDRTALEKQEKQLEMEIKKMAKAGNKDACKVLAKQLVQLRKQKTRTYAV 78
Query 78 SSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDVR 137
SSK+ +++++ ++ + + A + +K+D +L+ ++ F K +++
Sbjct 79 SSKVTSMSTQTKVMSSQMKMAGAMSTTAKTMQAVNKKMDPQKTLQTMQNFQKENMKMEMT 138
Query 138 ----SDSVSSLLDASTSSAVPAQQVDKVLTEV 165
+D++ + DAS V++VL E+
Sbjct 139 EEMINDTLDDIFDASEDEEESQDIVNQVLDEI 170
> hsa:25978 CHMP2B, CHMP2.5, DKFZp564O123, DMT1, VPS2-2, VPS2B;
chromatin modifying protein 2B; K12192 charged multivesicular
body protein 2B
Length=213
Score = 31.6 bits (70), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 23/143 (16%), Positives = 69/143 (48%), Gaps = 5/143 (3%)
Query 36 RESLQEREK-----IRRALLRQNQEGARIFAQNYVRKKQEGLNSLHMSSKLEAVASRLDG 90
R +L+++EK I++ N+E ++ A+ V +++ + +SSK+ +++++
Sbjct 32 RAALEKQEKQLELEIKKMAKIGNKEACKVLAKQLVHLRKQKTRTFAVSSKVTSMSTQTKV 91
Query 91 AHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDVRSDSVSSLLDASTS 150
+ ++ + + A + +K+D +L+ ++ F K +++ + ++ LD
Sbjct 92 MNSQMKMAGAMSTTAKTMQAVNKKMDPQKTLQTMQNFQKENMKMEMTEEMINDTLDDIFD 151
Query 151 SAVPAQQVDKVLTEVASTFGISL 173
+ ++ ++ +V GI +
Sbjct 152 GSDDEEESQDIVNQVLDEIGIEI 174
> cel:Y26D4A.14 hypothetical protein
Length=952
Score = 31.2 bits (69), Expect = 3.3, Method: Composition-based stats.
Identities = 21/70 (30%), Positives = 36/70 (51%), Gaps = 9/70 (12%)
Query 10 HHSVLPVILDLRIKARELKRQSDRCYRESLQEREKIRRALLR-------QNQEGARIFAQ 62
+HS P + + I RE +Q +++ L+E EK + +LR QNQE +
Sbjct 182 NHSRFPKQIPVAISTREELQQQR--HKQQLEEYEKRNQEILRREEEIKQQNQEREKQIND 239
Query 63 NYVRKKQEGL 72
++ ++KQE L
Sbjct 240 DFEKRKQEKL 249
> tpv:TP03_0651 hypothetical protein
Length=2053
Score = 31.2 bits (69), Expect = 3.5, Method: Composition-based stats.
Identities = 29/88 (32%), Positives = 42/88 (47%), Gaps = 8/88 (9%)
Query 62 QNYVRKKQEGLNSLHMSSKLEAVASRLDGAHRSHQL-TTRIHSVASGL------SGALR- 113
+N+V + + L + +AS + +H L TTRIH V SG +G+L
Sbjct 1493 RNFVPTSRNNMLELVGNLVFHNMASFIRFSHSVDSLDTTRIHPVESGFIAQKLCNGSLDD 1552
Query 114 KLDSSSSLREIELFSKLFDDLDVRSDSV 141
KLD S+ EI D+LD+ S SV
Sbjct 1553 KLDGEDSIHEILANPAKLDELDLESYSV 1580
> mmu:68953 Chmp2a, 1500016L11Rik; chromatin modifying protein
2A; K12191 charged multivesicular body protein 2A
Length=222
Score = 30.8 bits (68), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 32/167 (19%), Positives = 78/167 (46%), Gaps = 11/167 (6%)
Query 25 RELKRQSDRCYRESLQEREKIR----------RALLRQNQ-EGARIFAQNYVRKKQEGLN 73
R+ +R +R RE +ER+K+ + + +Q Q + RI A++ VR ++
Sbjct 16 RQNQRALNRAMRELDRERQKLETQEKKIIADIKKMAKQGQMDAVRIMAKDLVRTRRYVRK 75
Query 74 SLHMSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDD 133
+ M + ++AV+ ++ ++ + + V + R+L + + F + +
Sbjct 76 FVLMRANIQAVSLKIQTLKSNNSMAQAMKGVTKAMGTMNRQLKLPQIQKIMMEFERQAEI 135
Query 134 LDVRSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLEDQLEGF 180
+D++ + ++ +D + ++ D V+++V G+SL D+L
Sbjct 136 MDMKEEMMNDAIDDAMGDEEDEEESDAVVSQVLDELGLSLTDELSNL 182
> mmu:18160 Npr1, AI893888, GC-A, MGC117511, NPR-A, NPRA, Pndr;
natriuretic peptide receptor 1 (EC:4.6.1.2); K12323 atrial
natriuretic peptide receptor A [EC:4.6.1.2]
Length=1057
Score = 30.8 bits (68), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 28/96 (29%), Positives = 48/96 (50%), Gaps = 6/96 (6%)
Query 18 LDLRIKARELKRQSDRCYRESLQER---EKIRRALLRQNQEGARIFAQNYVRKKQEGLNS 74
+DL+ EL + RC+ E QER ++IR AL + N+E + N + + ++ N+
Sbjct 763 MDLQSHLEELGQLMQRCWAEDPQERPPFQQIRLALRKFNKENSSNILDNLLSRMEQYANN 822
Query 75 LHMSSKLEAVASRLDGAHRSHQLTTRI--HSVASGL 108
L + E + L+ ++ L +I HSVA L
Sbjct 823 LEELVE-ERTQAYLEEKRKAEALLYQILPHSVAEQL 857
> dre:405840 chmp2bl, MGC77025, chmp2b, zgc:77025; chromatin modifying
protein 2B, like; K12192 charged multivesicular body
protein 2B
Length=214
Score = 30.8 bits (68), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 21/143 (14%), Positives = 69/143 (48%), Gaps = 5/143 (3%)
Query 36 RESLQEREK-----IRRALLRQNQEGARIFAQNYVRKKQEGLNSLHMSSKLEAVASRLDG 90
R +L+++EK I++ N++ ++ A+ V+ +++ + +SSK+ +++++
Sbjct 32 RTALEKQEKQLEMEIKKMAKTGNRDACKVLAKQLVQVRKQKTRTYAVSSKVTSMSTQTKL 91
Query 91 AHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDVRSDSVSSLLDASTS 150
+ ++ + + + +K+D +++ ++ F K +D+ + ++ LD
Sbjct 92 MNSQMKMAGAMATTTKTMQAVNKKMDPKKTMQTLQNFQKETAKMDMTEEMMNDTLDEIFE 151
Query 151 SAVPAQQVDKVLTEVASTFGISL 173
+ ++ ++ +V GI +
Sbjct 152 DSGDEEESQDIVNQVLDEIGIEI 174
> dre:568764 mKIAA0969 protein-like
Length=1086
Score = 30.4 bits (67), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 34/83 (40%), Gaps = 3/83 (3%)
Query 76 HMSSKLEAVASRLDGAHRSHQ---LTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFD 132
H+ + V L+G H+++ T H VASG SG+ +S L + L S L
Sbjct 673 HIQKEFWRVQDVLEGLHKNNPSRGTDTAKHRVASGASGSFSTNSPASPLSSVSLTSPLSP 732
Query 133 DLDVRSDSVSSLLDASTSSAVPA 155
V S +T A+P
Sbjct 733 FSPVPGSQPSPTKHLATEEAIPP 755
> hsa:22998 LIMCH1, DKFZp434I0312, DKFZp686A01247, DKFZp686B2470,
DKFZp686G18243, DKFZp686G2094, DKFZp781C1754, DKFZp781I1455,
LIMCH1A, LMO7B, MGC72127; LIM and calponin homology domains
1
Length=1057
Score = 30.4 bits (67), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 39/138 (28%), Positives = 61/138 (44%), Gaps = 21/138 (15%)
Query 28 KRQSDRCYRESLQEREKIRRALLR-----QNQEGARIFAQNYVRKKQEGLNSLHMSSKLE 82
+R+S + YRE +QE+E+ R L ++QE A Q Y+ + +S E
Sbjct 405 RRKSIKTYREIVQEKERRERELHEAYKNARSQEEAEGILQQYIER-------FTIS---E 454
Query 83 AVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDDLDVRSDSVS 142
AV RL+ ++ R HS LS L + LR+ L F + + +
Sbjct 455 AVLERLEMP----KILERSHSTEPNLSSFLNDPNPMKYLRQQSLPPPKFTATVETTIARA 510
Query 143 SLLDASTS--SAVPAQQV 158
S+LD S S S P++ V
Sbjct 511 SVLDTSMSAGSGSPSKTV 528
> hsa:27243 CHMP2A, BC-2, BC2, CHMP2, VPS2, VPS2A; chromatin modifying
protein 2A; K12191 charged multivesicular body protein
2A
Length=222
Score = 30.4 bits (67), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 32/167 (19%), Positives = 78/167 (46%), Gaps = 11/167 (6%)
Query 25 RELKRQSDRCYRESLQEREKIR----------RALLRQNQ-EGARIFAQNYVRKKQEGLN 73
R+ +R +R RE +ER+K+ + + +Q Q + RI A++ VR ++
Sbjct 16 RQNQRALNRAMRELDRERQKLETQEKKIIADIKKMAKQGQMDAVRIMAKDLVRTRRYVRK 75
Query 74 SLHMSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLREIELFSKLFDD 133
+ M + ++AV+ ++ ++ + + V + R+L + + F + +
Sbjct 76 FVLMRANIQAVSLKIQTLKSNNSMAQAMKGVTKAMGTMNRQLKLPQIQKIMMEFERQAEI 135
Query 134 LDVRSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLEDQLEGF 180
+D++ + ++ +D + ++ D V+++V G+SL D+L
Sbjct 136 MDMKEEMMNDAIDDAMGDEEDEEESDAVVSQVLDELGLSLTDELSNL 182
> cel:Y50D7A.8 hypothetical protein
Length=569
Score = 29.6 bits (65), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 30/96 (31%), Positives = 45/96 (46%), Gaps = 11/96 (11%)
Query 132 DDLDVRSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLEDQLE---GFEGVDTPQK 188
DD++ D+ L+D+S + + QQ K+L +++S+ I QL E ++ P
Sbjct 294 DDMEHYGDNEEHLMDSSGTDFLDEQQEVKLLEDISSSNTIVPHHQLHHPAAAEVMEAPNN 353
Query 189 EHCERQRAMGDISGVYTLQANAPNRLLGKGIEGRKE 224
E C Q M D V A+ PN L GI R E
Sbjct 354 EIC--QEHMLD---VINQVAHPPNEL---GIFTRME 381
> mmu:100503137 putative uncharacterized protein C8orf73-like
Length=722
Score = 29.6 bits (65), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 34/132 (25%), Positives = 55/132 (41%), Gaps = 12/132 (9%)
Query 73 NSLHMSSKLEAVASRLDGAHRSHQLTTRIHSVASGLSGALRKLDSSSSLRE-IELFSKLF 131
+L +SS LEA R +L ++H + GL + L R + + S L
Sbjct 132 TALTLSSALEA---------RGQRLEDQVHGLVRGLLAQVPSLAEGRPRRAALRVLSAL- 181
Query 132 DDLDVRSDSVSSLLDASTSSAVPAQQVDKVLTEVASTFGISLEDQLEGFEGVDTPQKEHC 191
L+ D V +LL +S A ++ + L+ G L L +GV P+ E
Sbjct 182 -ALEHAQDVVCALLPSSLPPDRAASELWRSLSRNQRVNGQVLVQLLWALKGVAAPESEAL 240
Query 192 ERQRAMGDISGV 203
RA+G++ V
Sbjct 241 AATRALGEMLAV 252
Lambda K H
0.315 0.129 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8081806824
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40