bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1504_orf2
Length=63
Score E
Sequences producing significant alignments: (Bits) Value
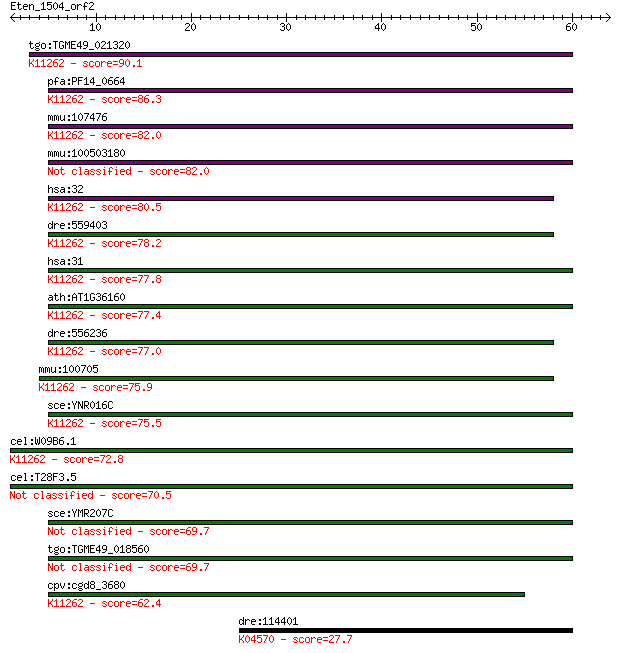
tgo:TGME49_021320 acetyl-CoA carboxylase, putative (EC:6.3.4.1... 90.1 1e-18
pfa:PF14_0664 ACC1; biotin carboxylase subunit of acetyl CoA c... 86.3 2e-17
mmu:107476 Acaca, A530025K05Rik, Acac, Acc1, Gm738; acetyl-Coe... 82.0 4e-16
mmu:100503180 acetyl-CoA carboxylase 1-like 82.0 4e-16
hsa:32 ACACB, ACC2, ACCB, HACC275; acetyl-CoA carboxylase beta... 80.5 1e-15
dre:559403 acaca, im:7138837, si:ch211-199d18.1, wu:fj43d01; a... 78.2 6e-15
hsa:31 ACACA, ACAC, ACC, ACC1, ACCA; acetyl-CoA carboxylase al... 77.8 8e-15
ath:AT1G36160 ACC1; ACC1 (ACETYL-COENZYME A CARBOXYLASE 1); ac... 77.4 1e-14
dre:556236 acetyl-CoA carboxylase 1-like; K11262 acetyl-CoA ca... 77.0 1e-14
mmu:100705 Acacb, AI597064, AW743042, Acc2, Accb; acetyl-Coenz... 75.9 3e-14
sce:YNR016C ACC1, ABP2, FAS3, MTR7; Acc1p (EC:6.4.1.2 6.3.4.14... 75.5 4e-14
cel:W09B6.1 pod-2; Polarity and Osmotic sensitivity Defect fam... 72.8 3e-13
cel:T28F3.5 hypothetical protein 70.5 1e-12
sce:YMR207C HFA1; Hfa1p (EC:6.4.1.2 6.3.4.14) 69.7 2e-12
tgo:TGME49_018560 acetyl-coA carboxylase (EC:6.3.4.14 2.3.1.12... 69.7 2e-12
cpv:cgd8_3680 acetyl-CoA carboxylase like biotin dependent car... 62.4 4e-10
dre:114401 bcl2l1, bcl2l, blp1, cb1021, zfBLP1; bcl2-like 1; K... 27.7 8.7
> tgo:TGME49_021320 acetyl-CoA carboxylase, putative (EC:6.3.4.14
2.3.1.12 6.4.1.3); K11262 acetyl-CoA carboxylase / biotin
carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2599
Score = 90.1 bits (222), Expect = 1e-18, Method: Composition-based stats.
Identities = 44/57 (77%), Positives = 47/57 (82%), Gaps = 0/57 (0%)
Query 3 QHLGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
+ +GVENLRGSG IAGETS AYDE FTLS V+GRSVGIGAYIVRLAQR IQ PL
Sbjct 2084 KFIGVENLRGSGTIAGETSRAYDETFTLSYVTGRSVGIGAYIVRLAQRTIQMVRGPL 2140
> pfa:PF14_0664 ACC1; biotin carboxylase subunit of acetyl CoA
carboxylase, putative (EC:6.4.1.2); K11262 acetyl-CoA carboxylase
/ biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=3367
Score = 86.3 bits (212), Expect = 2e-17, Method: Composition-based stats.
Identities = 41/55 (74%), Positives = 48/55 (87%), Gaps = 0/55 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
+GVENLRGSG IAGETS AY+E+FTLS V+GRSVGIGAY+VRL +R IQ+K S L
Sbjct 2735 VGVENLRGSGLIAGETSKAYEEIFTLSYVTGRSVGIGAYLVRLGKRTIQKKGSSL 2789
> mmu:107476 Acaca, A530025K05Rik, Acac, Acc1, Gm738; acetyl-Coenzyme
A carboxylase alpha (EC:6.4.1.2 6.3.4.14); K11262 acetyl-CoA
carboxylase / biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2345
Score = 82.0 bits (201), Expect = 4e-16, Method: Composition-based stats.
Identities = 39/55 (70%), Positives = 46/55 (83%), Gaps = 0/55 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
LG ENLRGSG IAGE+SLAYDEV T+SLV+ R++GIGAY+VRL QR IQ + S L
Sbjct 1791 LGAENLRGSGMIAGESSLAYDEVITISLVTCRAIGIGAYLVRLGQRTIQVENSHL 1845
> mmu:100503180 acetyl-CoA carboxylase 1-like
Length=580
Score = 82.0 bits (201), Expect = 4e-16, Method: Composition-based stats.
Identities = 39/55 (70%), Positives = 46/55 (83%), Gaps = 0/55 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
LG ENLRGSG IAGE+SLAYDEV T+SLV+ R++GIGAY+VRL QR IQ + S L
Sbjct 46 LGAENLRGSGMIAGESSLAYDEVITISLVTCRAIGIGAYLVRLGQRTIQVENSHL 100
> hsa:32 ACACB, ACC2, ACCB, HACC275; acetyl-CoA carboxylase beta
(EC:6.4.1.2 6.3.4.14); K11262 acetyl-CoA carboxylase / biotin
carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2458
Score = 80.5 bits (197), Expect = 1e-15, Method: Composition-based stats.
Identities = 37/53 (69%), Positives = 46/53 (86%), Gaps = 0/53 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTS 57
LGVENLRGSG IAGE+SLAY+E+ T+SLV+ R++GIGAY+VRL QR IQ + S
Sbjct 1903 LGVENLRGSGMIAGESSLAYEEIVTISLVTCRAIGIGAYLVRLGQRVIQVENS 1955
> dre:559403 acaca, im:7138837, si:ch211-199d18.1, wu:fj43d01;
acetyl-Coenzyme A carboxylase alpha; K11262 acetyl-CoA carboxylase
/ biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2349
Score = 78.2 bits (191), Expect = 6e-15, Method: Composition-based stats.
Identities = 35/53 (66%), Positives = 46/53 (86%), Gaps = 0/53 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTS 57
LGVENLRGSG IAGE+SLAY+++ T++LV+ R++GIGAY+VRL QR IQ + S
Sbjct 1794 LGVENLRGSGMIAGESSLAYEDIITMNLVTCRAIGIGAYLVRLGQRTIQVENS 1846
> hsa:31 ACACA, ACAC, ACC, ACC1, ACCA; acetyl-CoA carboxylase
alpha (EC:6.4.1.2 6.3.4.14); K11262 acetyl-CoA carboxylase /
biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2383
Score = 77.8 bits (190), Expect = 8e-15, Method: Composition-based stats.
Identities = 36/55 (65%), Positives = 46/55 (83%), Gaps = 0/55 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
+G ENLRGSG IAGE+SLAY+E+ T+SLV+ R++GIGAY+VRL QR IQ + S L
Sbjct 1829 IGPENLRGSGMIAGESSLAYNEIITISLVTCRAIGIGAYLVRLGQRTIQVENSHL 1883
> ath:AT1G36160 ACC1; ACC1 (ACETYL-COENZYME A CARBOXYLASE 1);
acetyl-CoA carboxylase; K11262 acetyl-CoA carboxylase / biotin
carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2254
Score = 77.4 bits (189), Expect = 1e-14, Method: Composition-based stats.
Identities = 35/55 (63%), Positives = 42/55 (76%), Gaps = 0/55 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
+GVENL GSG IAG S AY+E FTL+ VSGR+VGIGAY+ RL RCIQ+ P+
Sbjct 1709 IGVENLTGSGAIAGAYSKAYNETFTLTFVSGRTVGIGAYLARLGMRCIQRLDQPI 1763
> dre:556236 acetyl-CoA carboxylase 1-like; K11262 acetyl-CoA
carboxylase / biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2240
Score = 77.0 bits (188), Expect = 1e-14, Method: Composition-based stats.
Identities = 35/53 (66%), Positives = 44/53 (83%), Gaps = 0/53 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTS 57
+GVENLRGSG IAGETS AY E+ T+S+V+ R++GIGAY+VRL QR IQ + S
Sbjct 1685 IGVENLRGSGTIAGETSQAYKEIITISMVTCRAIGIGAYLVRLGQRVIQVENS 1737
> mmu:100705 Acacb, AI597064, AW743042, Acc2, Accb; acetyl-Coenzyme
A carboxylase beta (EC:6.4.1.2); K11262 acetyl-CoA carboxylase
/ biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2448
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 35/54 (64%), Positives = 45/54 (83%), Gaps = 0/54 (0%)
Query 4 HLGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTS 57
+LGVENLRGSG IAGE SLAY++ T+S+V+ R++GIGAY+VRL QR IQ + S
Sbjct 1892 NLGVENLRGSGMIAGEASLAYEKTVTISMVTCRALGIGAYLVRLGQRVIQVENS 1945
> sce:YNR016C ACC1, ABP2, FAS3, MTR7; Acc1p (EC:6.4.1.2 6.3.4.14);
K11262 acetyl-CoA carboxylase / biotin carboxylase [EC:6.4.1.2
6.3.4.14]
Length=2233
Score = 75.5 bits (184), Expect = 4e-14, Method: Composition-based stats.
Identities = 36/55 (65%), Positives = 44/55 (80%), Gaps = 0/55 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
LGVE LRGSG IAG TS AY ++FT++LV+ RSVGIGAY+VRL QR IQ + P+
Sbjct 1700 LGVECLRGSGLIAGATSRAYHDIFTITLVTCRSVGIGAYLVRLGQRAIQVEGQPI 1754
> cel:W09B6.1 pod-2; Polarity and Osmotic sensitivity Defect family
member (pod-2); K11262 acetyl-CoA carboxylase / biotin
carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2054
Score = 72.8 bits (177), Expect = 3e-13, Method: Composition-based stats.
Identities = 36/59 (61%), Positives = 43/59 (72%), Gaps = 0/59 (0%)
Query 1 EGQHLGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
+ + +GVENL+GSG IAGET+ AY EV T V+GRSVGIGAY RLA R +Q K S L
Sbjct 1543 KNEKIGVENLQGSGLIAGETARAYAEVPTYCYVTGRSVGIGAYTARLAHRIVQHKQSHL 1601
> cel:T28F3.5 hypothetical protein
Length=1679
Score = 70.5 bits (171), Expect = 1e-12, Method: Composition-based stats.
Identities = 34/59 (57%), Positives = 43/59 (72%), Gaps = 0/59 (0%)
Query 1 EGQHLGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
+ +++GVENL GSG I GETS AY E+ T V+GRSVGIGAY RLA+R IQ + + L
Sbjct 1223 KNEYIGVENLMGSGAIGGETSRAYREIPTYCYVTGRSVGIGAYTARLARRIIQHEKAHL 1281
> sce:YMR207C HFA1; Hfa1p (EC:6.4.1.2 6.3.4.14)
Length=2123
Score = 69.7 bits (169), Expect = 2e-12, Method: Composition-based stats.
Identities = 33/55 (60%), Positives = 43/55 (78%), Gaps = 0/55 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
LGVE L+GSG IAG TS AY ++FT++ V+ RSVGIG+Y+VRL QR IQ + P+
Sbjct 1595 LGVECLQGSGLIAGATSKAYRDIFTITAVTCRSVGIGSYLVRLGQRTIQVEDKPI 1649
> tgo:TGME49_018560 acetyl-coA carboxylase (EC:6.3.4.14 2.3.1.12
6.4.1.3 3.2.1.17)
Length=3399
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 36/56 (64%), Positives = 42/56 (75%), Gaps = 1/56 (1%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQK-TSPL 59
LGVENL GSG IAGET+ AY FT++ SGRSVGIGAY+ RL QR IQ+ +PL
Sbjct 2819 LGVENLCGSGAIAGETARAYKSTFTITFCSGRSVGIGAYLTRLGQRVIQKSVNAPL 2874
> cpv:cgd8_3680 acetyl-CoA carboxylase like biotin dependent carboxylase
involved in fatty acid biosynthesis ; K11262 acetyl-CoA
carboxylase / biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=2733
Score = 62.4 bits (150), Expect = 4e-10, Method: Composition-based stats.
Identities = 29/50 (58%), Positives = 36/50 (72%), Gaps = 0/50 (0%)
Query 5 LGVENLRGSGKIAGETSLAYDEVFTLSLVSGRSVGIGAYIVRLAQRCIQQ 54
LGVENL GSG IAGETS A + T++ + R+VGIGAY+ RL R IQ+
Sbjct 2232 LGVENLSGSGGIAGETSKASKSIMTITYATTRTVGIGAYLARLGHRVIQK 2281
> dre:114401 bcl2l1, bcl2l, blp1, cb1021, zfBLP1; bcl2-like 1;
K04570 BCL2-like 1 (apoptosis regulator Bcl-X)
Length=238
Score = 27.7 bits (60), Expect = 8.7, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 25 DEVFTLSLVSGRSVGIGAYIVRLAQRCIQQKTSPL 59
DEVF + GR VG+ A+ L C++++ SPL
Sbjct 131 DEVFRDGVNWGRIVGLFAFGGALCVECVEKEMSPL 165
Lambda K H
0.321 0.136 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2056037364
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40