bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1589_orf2
Length=123
Score E
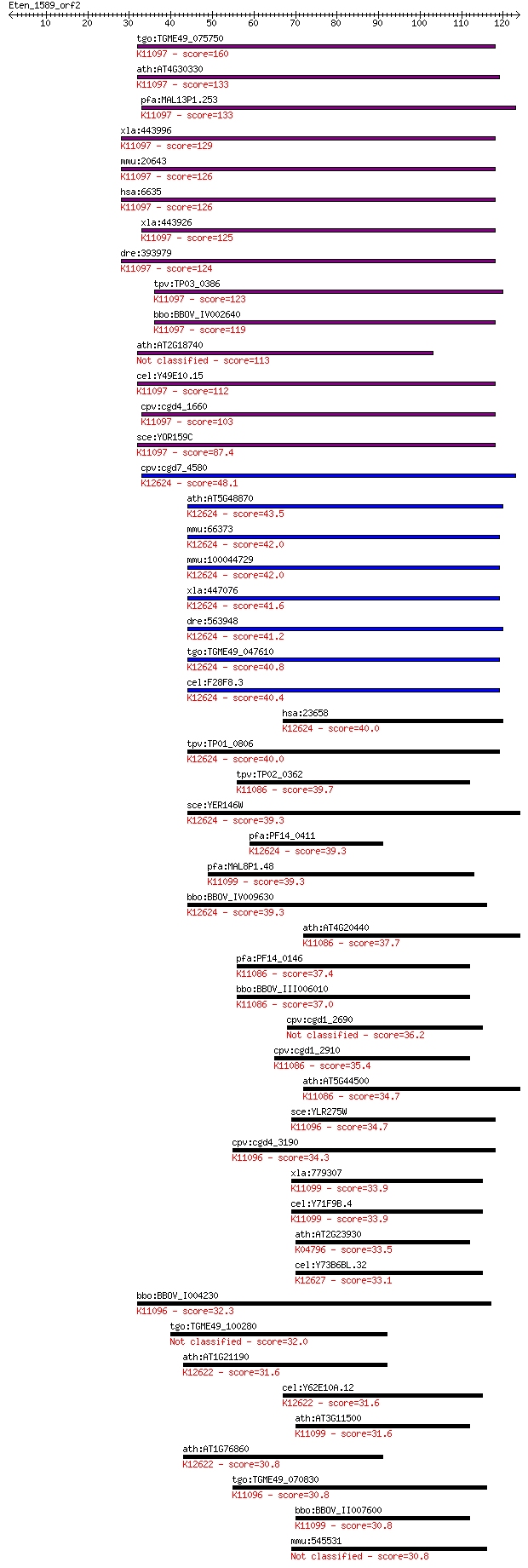
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_075750 small nuclear ribonucleoprotein E, putative ... 160 7e-40
ath:AT4G30330 small nuclear ribonucleoprotein E, putative / sn... 133 1e-31
pfa:MAL13P1.253 small nuclear ribonucleoprotein, putative; K11... 133 1e-31
xla:443996 snrpe; small nuclear ribonucleoprotein polypeptide ... 129 2e-30
mmu:20643 Snrpe, AL022645, C76690, SME; small nuclear ribonucl... 126 1e-29
hsa:6635 SNRPE, B-raf, SME, Sm-E; small nuclear ribonucleoprot... 126 1e-29
xla:443926 MGC80249 protein; K11097 small nuclear ribonucleopr... 125 2e-29
dre:393979 snrpe, MGC56682, zgc:56682; small nuclear ribonucle... 124 7e-29
tpv:TP03_0386 small nuclear ribonucleoprotein; K11097 small nu... 123 1e-28
bbo:BBOV_IV002640 21.m02844; small nuclear ribonucleoprotein E... 119 2e-27
ath:AT2G18740 small nuclear ribonucleoprotein E, putative / sn... 113 1e-25
cel:Y49E10.15 snr-6; Small Nuclear Ribonucleoprotein family me... 112 2e-25
cpv:cgd4_1660 small nuclear ribonucleoprotein ; K11097 small n... 103 1e-22
sce:YOR159C SME1; Sm E; SmE; K11097 small nuclear ribonucleopr... 87.4 1e-17
cpv:cgd7_4580 U6 snRNA-associated Sm-like protein LSm5. SM dom... 48.1 7e-06
ath:AT5G48870 SAD1; SAD1 (SUPERSENSITIVE TO ABA AND DROUGHT 1)... 43.5 1e-04
mmu:66373 Lsm5, 2010208O10Rik, 2310034K10Rik; LSM5 homolog, U6... 42.0 5e-04
mmu:100044729 u6 snRNA-associated Sm-like protein LSm5-like; K... 42.0 5e-04
xla:447076 lsm5; LSM5 homolog, U6 small nuclear RNA associated... 41.6 6e-04
dre:563948 zgc:171959; K12624 U6 snRNA-associated Sm-like prot... 41.2 8e-04
tgo:TGME49_047610 snRNP protein Lsm5, putative ; K12624 U6 snR... 40.8 0.001
cel:F28F8.3 lsm-5; LSM Sm-like protein family member (lsm-5); ... 40.4 0.001
hsa:23658 LSM5, FLJ12710, YER146W; LSM5 homolog, U6 small nucl... 40.0 0.002
tpv:TP01_0806 U6 small nuclear ribonucleoprotein E; K12624 U6 ... 40.0 0.002
tpv:TP02_0362 small nuclear ribonucleoprotein B; K11086 small ... 39.7 0.002
sce:YER146W LSM5; Lsm (Like Sm) protein; part of heteroheptame... 39.3 0.003
pfa:PF14_0411 small nuclear ribonucleoprotein, putative; K1262... 39.3 0.003
pfa:MAL8P1.48 small nuclear ribonucleoprotein, putative; K1109... 39.3 0.003
bbo:BBOV_IV009630 23.m05873; snRNP Sm-like protein; K12624 U6 ... 39.3 0.003
ath:AT4G20440 smB; smB (small nuclear ribonucleoprotein associ... 37.7 0.008
pfa:PF14_0146 ribonucleoprotein, putative; K11086 small nuclea... 37.4 0.012
bbo:BBOV_III006010 17.m07530; Sm domain containing protein; K1... 37.0 0.015
cpv:cgd1_2690 hypothetical protein 36.2 0.028
cpv:cgd1_2910 hypothetical protein ; K11086 small nuclear ribo... 35.4 0.042
ath:AT5G44500 small nuclear ribonucleoprotein associated prote... 34.7 0.081
sce:YLR275W SMD2; Sm D2; K11096 small nuclear ribonucleoprotei... 34.7 0.081
cpv:cgd4_3190 small nuclear ribonucleoprotein ; K11096 small n... 34.3 0.095
xla:779307 snrpg, MGC154809; small nuclear ribonucleoprotein p... 33.9 0.11
cel:Y71F9B.4 snr-7; Small Nuclear Ribonucleoprotein family mem... 33.9 0.13
ath:AT2G23930 SNRNP-G (PROBABLE SMALL NUCLEAR RIBONUCLEOPROTEI... 33.5 0.17
cel:Y73B6BL.32 lsm-8; LSM Sm-like protein family member (lsm-8... 33.1 0.24
bbo:BBOV_I004230 19.m02306; small nuclear ribonucleoprotein; K... 32.3 0.38
tgo:TGME49_100280 small nuclear ribonucleoprotein, putative 32.0 0.48
ath:AT1G21190 small nuclear ribonucleoprotein, putative / snRN... 31.6 0.59
cel:Y62E10A.12 lsm-3; LSM Sm-like protein family member (lsm-3... 31.6 0.62
ath:AT3G11500 small nuclear ribonucleoprotein G, putative / sn... 31.6 0.72
ath:AT1G76860 small nuclear ribonucleoprotein, putative / snRN... 30.8 0.94
tgo:TGME49_070830 small nuclear ribonucleoprotein Sm D2, putat... 30.8 0.94
bbo:BBOV_II007600 18.m06632; small nuclear ribonucleoprotein G... 30.8 1.0
mmu:545531 Gm5848, EG545531; predicted pseudogene 5848 30.8
> tgo:TGME49_075750 small nuclear ribonucleoprotein E, putative
; K11097 small nuclear ribonucleoprotein E
Length=91
Score = 160 bits (406), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 73/86 (84%), Positives = 82/86 (95%), Gaps = 0/86 (0%)
Query 32 MSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEI 91
MSNKKLQKIMTQPINLIFRFFT + RVQ+WLYEQPD+RIEGRIMGFDEYMNMVLDDAEE+
Sbjct 6 MSNKKLQKIMTQPINLIFRFFTQRMRVQIWLYEQPDMRIEGRIMGFDEYMNMVLDDAEEV 65
Query 92 HLKKNTRVFVGRILLKGENISLIMST 117
+LKK TR+ VGRILLKGEN++LIM+T
Sbjct 66 YLKKRTRIPVGRILLKGENVTLIMAT 91
> ath:AT4G30330 small nuclear ribonucleoprotein E, putative /
snRNP-E, putative / Sm protein E, putative; K11097 small nuclear
ribonucleoprotein E
Length=88
Score = 133 bits (335), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 59/87 (67%), Positives = 77/87 (88%), Gaps = 0/87 (0%)
Query 32 MSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEI 91
M++ K+Q+IMTQPINLIFRF +K+R+Q+WL+EQ DLRIEGRI GFDEYMN+VLD+AEE+
Sbjct 1 MASTKVQRIMTQPINLIFRFLQSKARIQIWLFEQKDLRIEGRITGFDEYMNLVLDEAEEV 60
Query 92 HLKKNTRVFVGRILLKGENISLIMSTG 118
+KK TR +GRILLKG+NI+L+M+ G
Sbjct 61 SIKKKTRKPLGRILLKGDNITLMMNAG 87
> pfa:MAL13P1.253 small nuclear ribonucleoprotein, putative; K11097
small nuclear ribonucleoprotein E
Length=93
Score = 133 bits (335), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 61/90 (67%), Positives = 76/90 (84%), Gaps = 0/90 (0%)
Query 33 SNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIH 92
+NKKLQKIMTQPIN IFRFFTNK+ VQ+WLY++PD+RIEG I+GFDEYMNMVLD +EI
Sbjct 4 TNKKLQKIMTQPINQIFRFFTNKTVVQIWLYDKPDMRIEGIILGFDEYMNMVLDQTKEIS 63
Query 93 LKKNTRVFVGRILLKGENISLIMSTGSTNT 122
+KKNT+ +G+ILLKG+ I+LIM + T
Sbjct 64 VKKNTKKELGKILLKGDTITLIMEVKNEET 93
> xla:443996 snrpe; small nuclear ribonucleoprotein polypeptide
E; K11097 small nuclear ribonucleoprotein E
Length=92
Score = 129 bits (324), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 57/90 (63%), Positives = 75/90 (83%), Gaps = 0/90 (0%)
Query 28 MTHQMSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDD 87
M ++ +K+QK+M QPINLIFR+ N+SRVQ+WLYEQ ++RIEG I+GFDEYMN+VLDD
Sbjct 1 MAYRGQGQKVQKVMVQPINLIFRYLQNRSRVQVWLYEQVNMRIEGCIIGFDEYMNIVLDD 60
Query 88 AEEIHLKKNTRVFVGRILLKGENISLIMST 117
AEEIHLK +R +GRI+LKG+NI+L+ S
Sbjct 61 AEEIHLKTKSRKQLGRIMLKGDNITLLQSV 90
> mmu:20643 Snrpe, AL022645, C76690, SME; small nuclear ribonucleoprotein
E; K11097 small nuclear ribonucleoprotein E
Length=92
Score = 126 bits (317), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 55/90 (61%), Positives = 74/90 (82%), Gaps = 0/90 (0%)
Query 28 MTHQMSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDD 87
M ++ +K+QK+M QPINLIFR+ N+SR+Q+WLYEQ ++RIEG I+GFDEYMN+VLDD
Sbjct 1 MAYRGQGQKVQKVMVQPINLIFRYLQNRSRIQVWLYEQVNMRIEGCIIGFDEYMNLVLDD 60
Query 88 AEEIHLKKNTRVFVGRILLKGENISLIMST 117
AEEIH K +R +GRI+LKG+NI+L+ S
Sbjct 61 AEEIHSKTKSRKQLGRIMLKGDNITLLQSV 90
> hsa:6635 SNRPE, B-raf, SME, Sm-E; small nuclear ribonucleoprotein
polypeptide E; K11097 small nuclear ribonucleoprotein
E
Length=92
Score = 126 bits (317), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 55/90 (61%), Positives = 74/90 (82%), Gaps = 0/90 (0%)
Query 28 MTHQMSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDD 87
M ++ +K+QK+M QPINLIFR+ N+SR+Q+WLYEQ ++RIEG I+GFDEYMN+VLDD
Sbjct 1 MAYRGQGQKVQKVMVQPINLIFRYLQNRSRIQVWLYEQVNMRIEGCIIGFDEYMNLVLDD 60
Query 88 AEEIHLKKNTRVFVGRILLKGENISLIMST 117
AEEIH K +R +GRI+LKG+NI+L+ S
Sbjct 61 AEEIHSKTKSRKQLGRIMLKGDNITLLQSV 90
> xla:443926 MGC80249 protein; K11097 small nuclear ribonucleoprotein
E
Length=93
Score = 125 bits (315), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 54/85 (63%), Positives = 72/85 (84%), Gaps = 0/85 (0%)
Query 33 SNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIH 92
+K+QK+M QPINLIFR+ N+SR+Q+WLYEQ ++RIEG I+GFDEYMN+VLDD+EEIH
Sbjct 7 GGQKVQKVMVQPINLIFRYLQNRSRIQVWLYEQVNMRIEGCIIGFDEYMNLVLDDSEEIH 66
Query 93 LKKNTRVFVGRILLKGENISLIMST 117
LK +R +GRI+LKG+NI+L+ S
Sbjct 67 LKTKSRKPIGRIMLKGDNITLLQSV 91
> dre:393979 snrpe, MGC56682, zgc:56682; small nuclear ribonucleoprotein
polypeptide E; K11097 small nuclear ribonucleoprotein
E
Length=92
Score = 124 bits (311), Expect = 7e-29, Method: Compositional matrix adjust.
Identities = 53/90 (58%), Positives = 73/90 (81%), Gaps = 0/90 (0%)
Query 28 MTHQMSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDD 87
M ++ +K+QK+M QPINLIFR+ N+SR+ +WLYEQ ++RIEG I+GFDEYMN+VLDD
Sbjct 1 MAYRGQGQKVQKVMVQPINLIFRYLQNRSRISVWLYEQVNMRIEGCIIGFDEYMNLVLDD 60
Query 88 AEEIHLKKNTRVFVGRILLKGENISLIMST 117
AEE+H+K R +GRI+LKG+NI+L+ S
Sbjct 61 AEEVHMKTKNRKPLGRIMLKGDNITLLQSV 90
> tpv:TP03_0386 small nuclear ribonucleoprotein; K11097 small
nuclear ribonucleoprotein E
Length=91
Score = 123 bits (309), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 56/84 (66%), Positives = 70/84 (83%), Gaps = 0/84 (0%)
Query 36 KLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKK 95
KLQKIMTQPIN IFRFFT +RVQ+WL++QP+ +IEG I GFDEYMNMVLD+A E+H+KK
Sbjct 7 KLQKIMTQPINQIFRFFTGGTRVQIWLFDQPNTKIEGTIRGFDEYMNMVLDNATEVHVKK 66
Query 96 NTRVFVGRILLKGENISLIMSTGS 119
N + VGRILLKG+ ++LI + S
Sbjct 67 NVKKDVGRILLKGDCMTLISAATS 90
> bbo:BBOV_IV002640 21.m02844; small nuclear ribonucleoprotein
E; K11097 small nuclear ribonucleoprotein E
Length=89
Score = 119 bits (298), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 53/82 (64%), Positives = 70/82 (85%), Gaps = 0/82 (0%)
Query 36 KLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKK 95
KLQ+IMTQPIN IFRFFT+ +RVQ+WLY+QP+L+IEG+I GFDEYMNMVL+D EE+++KK
Sbjct 7 KLQQIMTQPINQIFRFFTSGTRVQIWLYDQPNLKIEGKIRGFDEYMNMVLEDVEELYVKK 66
Query 96 NTRVFVGRILLKGENISLIMST 117
R +G ILLKG+ ++LI +
Sbjct 67 QERRALGTILLKGDAMALITAA 88
> ath:AT2G18740 small nuclear ribonucleoprotein E, putative /
snRNP-E, putative / Sm protein E, putative
Length=78
Score = 113 bits (282), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 49/71 (69%), Positives = 63/71 (88%), Gaps = 0/71 (0%)
Query 32 MSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEI 91
M++ K+Q+IMTQPINLIFRF +K+R+Q+WL+EQ DLRIEGRI GFDEYMN+VLD+AEE+
Sbjct 1 MASTKVQRIMTQPINLIFRFLQSKARIQIWLFEQKDLRIEGRITGFDEYMNLVLDEAEEV 60
Query 92 HLKKNTRVFVG 102
+KKNTR +G
Sbjct 61 SIKKNTRKPLG 71
> cel:Y49E10.15 snr-6; Small Nuclear Ribonucleoprotein family
member (snr-6); K11097 small nuclear ribonucleoprotein E
Length=90
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 49/86 (56%), Positives = 68/86 (79%), Gaps = 0/86 (0%)
Query 32 MSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEI 91
MS +KL K+M QP+NLIFR+ N++RVQ+WLYE R+EG I+GFDE+MN+V D+AEE+
Sbjct 1 MSTRKLNKVMVQPVNLIFRYLQNRTRVQIWLYEDVTHRLEGYIIGFDEFMNVVFDEAEEV 60
Query 92 HLKKNTRVFVGRILLKGENISLIMST 117
++K R +GRILLKG+NI+LI +
Sbjct 61 NMKTKGRNKIGRILLKGDNITLIHAA 86
> cpv:cgd4_1660 small nuclear ribonucleoprotein ; K11097 small
nuclear ribonucleoprotein E
Length=92
Score = 103 bits (257), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 49/87 (56%), Positives = 63/87 (72%), Gaps = 2/87 (2%)
Query 33 SNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIH 92
+ +K QK+M QPIN IF FT+K RVQ+WLY+ +L +EG I GFDEYMN+VLD A E++
Sbjct 5 TKRKTQKMMLQPINQIFHLFTSKQRVQIWLYDHKNLVLEGVIQGFDEYMNIVLDQASEVY 64
Query 93 LKKNTR--VFVGRILLKGENISLIMST 117
KK R VG++LL+GENISLI
Sbjct 65 TKKEVRRETSVGQLLLRGENISLICEC 91
> sce:YOR159C SME1; Sm E; SmE; K11097 small nuclear ribonucleoprotein
E
Length=94
Score = 87.4 bits (215), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 45/95 (47%), Positives = 63/95 (66%), Gaps = 11/95 (11%)
Query 32 MSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEI 91
MSNK K M PIN IF F ++ V +WL+EQ +RI+G+I+GFDE+MN+V+D+A EI
Sbjct 1 MSNKVKTKAMVPPINCIFNFLQQQTPVTIWLFEQIGIRIKGKIVGFDEFMNVVIDEAVEI 60
Query 92 ---------HLKKNTRVFVGRILLKGENISLIMST 117
++K T +G+ILLKG+NI+LI S
Sbjct 61 PVNSADGKEDVEKGTP--LGKILLKGDNITLITSA 93
> cpv:cgd7_4580 U6 snRNA-associated Sm-like protein LSm5. SM domain
; K12624 U6 snRNA-associated Sm-like protein LSm5
Length=121
Score = 48.1 bits (113), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 33/104 (31%), Positives = 53/104 (50%), Gaps = 19/104 (18%)
Query 33 SNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIH 92
SN+K I+ P+ LI + N+ +++ + D G + GFDEY+NMVLDD +E
Sbjct 19 SNQKGGNIIL-PLALIDKCIGNR----IYVVMKGDKEFSGVLRGFDEYVNMVLDDVQEYG 73
Query 93 LKKN-----------TRVFVGR---ILLKGENISLIMSTGSTNT 122
K + RV V R ILL G N+++++ G ++
Sbjct 74 FKADEEDISGGNKKLKRVMVNRLETILLSGNNVAMLVPGGDPDS 117
> ath:AT5G48870 SAD1; SAD1 (SUPERSENSITIVE TO ABA AND DROUGHT
1); RNA binding; K12624 U6 snRNA-associated Sm-like protein
LSm5
Length=88
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 42/78 (53%), Gaps = 6/78 (7%)
Query 44 PINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVF--V 101
P LI R +K +W+ + D + G + GFD Y+NMVL+D E + R +
Sbjct 10 PSELIDRCIGSK----IWVIMKGDKELVGILKGFDVYVNMVLEDVTEYEITAEGRRVTKL 65
Query 102 GRILLKGENISLIMSTGS 119
+ILL G NI++++ GS
Sbjct 66 DQILLNGNNIAILVPGGS 83
> mmu:66373 Lsm5, 2010208O10Rik, 2310034K10Rik; LSM5 homolog,
U6 small nuclear RNA associated (S. cerevisiae); K12624 U6 snRNA-associated
Sm-like protein LSm5
Length=91
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 44/77 (57%), Gaps = 6/77 (7%)
Query 44 PINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVF--V 101
P+ L+ + SR+ + + + D I G ++GFD+++NMVL+D E + R +
Sbjct 14 PLELVDKCI--GSRIHIVM--KSDKEIVGTLLGFDDFVNMVLEDVTEFEITPEGRRITKL 69
Query 102 GRILLKGENISLIMSTG 118
+ILL G NI++++ G
Sbjct 70 DQILLNGNNITMLVPGG 86
> mmu:100044729 u6 snRNA-associated Sm-like protein LSm5-like;
K12624 U6 snRNA-associated Sm-like protein LSm5
Length=91
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 44/77 (57%), Gaps = 6/77 (7%)
Query 44 PINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVF--V 101
P+ L+ + SR+ + + + D I G ++GFD+++NMVL+D E + R +
Sbjct 14 PLELVDKCI--GSRIHIVM--KSDKEIVGTLLGFDDFVNMVLEDVTEFEITPEGRRITKL 69
Query 102 GRILLKGENISLIMSTG 118
+ILL G NI++++ G
Sbjct 70 DQILLNGNNITMLVPGG 86
> xla:447076 lsm5; LSM5 homolog, U6 small nuclear RNA associated;
K12624 U6 snRNA-associated Sm-like protein LSm5
Length=91
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 44/77 (57%), Gaps = 6/77 (7%)
Query 44 PINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVF--V 101
P+ L+ + SR+ + + + D I G ++GFD+++NMVL+D E + R +
Sbjct 14 PLELVDKCI--GSRIHIVM--KSDKEIVGTLLGFDDFVNMVLEDVTEFEITPEGRRITKL 69
Query 102 GRILLKGENISLIMSTG 118
+ILL G NI++++ G
Sbjct 70 DQILLNGNNITMLVPGG 86
> dre:563948 zgc:171959; K12624 U6 snRNA-associated Sm-like protein
LSm5
Length=91
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 43/78 (55%), Gaps = 6/78 (7%)
Query 44 PINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVF--V 101
P+ L+ + SR+ + + D I G ++GFD+++NMVL+D E + R +
Sbjct 14 PLELVDKCI--GSRIHIVMKN--DKEIVGTLLGFDDFVNMVLEDVTEFEITPEGRRITKL 69
Query 102 GRILLKGENISLIMSTGS 119
+ILL G NI++++ G
Sbjct 70 DQILLNGNNITMLIPGGE 87
> tgo:TGME49_047610 snRNP protein Lsm5, putative ; K12624 U6 snRNA-associated
Sm-like protein LSm5
Length=119
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 41/77 (53%), Gaps = 6/77 (7%)
Query 44 PINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKKNT--RVFV 101
P+ L+ + ++ +W+ + D + G + GFD+++NMVLDD E + +
Sbjct 26 PLALVDKCIGSR----MWIIMKGDKELAGTLRGFDDFVNMVLDDVTEYTFTPTGVKKTKL 81
Query 102 GRILLKGENISLIMSTG 118
ILL G +I++++ G
Sbjct 82 QSILLNGNSITMLVPGG 98
> cel:F28F8.3 lsm-5; LSM Sm-like protein family member (lsm-5);
K12624 U6 snRNA-associated Sm-like protein LSm5
Length=91
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 42/77 (54%), Gaps = 6/77 (7%)
Query 44 PINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVF--V 101
P+ LI + +K +W+ + D I G + GFD+Y+NMVL+D E + + +
Sbjct 14 PLELIDKCIGSK----IWVIMKNDKEIVGTLTGFDDYVNMVLEDVVEYENTADGKRMTKL 69
Query 102 GRILLKGENISLIMSTG 118
ILL G +I++++ G
Sbjct 70 DTILLNGNHITMLVPGG 86
> hsa:23658 LSM5, FLJ12710, YER146W; LSM5 homolog, U6 small nuclear
RNA associated (S. cerevisiae); K12624 U6 snRNA-associated
Sm-like protein LSm5
Length=62
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 33/55 (60%), Gaps = 2/55 (3%)
Query 67 DLRIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVF--VGRILLKGENISLIMSTGS 119
D I G ++GFD+++NMVL+D E + R + +ILL G NI++++ G
Sbjct 4 DKEIVGTLLGFDDFVNMVLEDVTEFEITPEGRRITKLDQILLNGNNITMLVPGGE 58
> tpv:TP01_0806 U6 small nuclear ribonucleoprotein E; K12624 U6
snRNA-associated Sm-like protein LSm5
Length=90
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/77 (28%), Positives = 40/77 (51%), Gaps = 6/77 (7%)
Query 44 PINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKKNT--RVFV 101
P+ LI + +K +W+ + D I G + GFD+YMNMVL+D + + +
Sbjct 12 PLALIDKCLGSK----IWIIMKNDKEITGTLRGFDDYMNMVLEDVVDYSFSPDGVKTTEL 67
Query 102 GRILLKGENISLIMSTG 118
L+ G N+++++ G
Sbjct 68 NDALVNGNNVAMLVPGG 84
> tpv:TP02_0362 small nuclear ribonucleoprotein B; K11086 small
nuclear ribonucleoprotein B and B'
Length=155
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 40/71 (56%), Gaps = 17/71 (23%)
Query 56 SRVQLWLYEQPDLRIE--------GRIMGFDEYMNMVLDDAEEIHL-----KKNTRV--F 100
+R+Q WL Q ++R+ G ++ +D+YMN+VL D EE + K T V
Sbjct 6 TRMQHWL--QYNVRVTLKDNRKFVGTLVAYDKYMNLVLSDCEEFRMTLGKDKNRTEVKRT 63
Query 101 VGRILLKGENI 111
+G +LL+GENI
Sbjct 64 LGFVLLRGENI 74
> sce:YER146W LSM5; Lsm (Like Sm) protein; part of heteroheptameric
complexes (Lsm2p-7p and either Lsm1p or 8p): cytoplasmic
Lsm1p complex involved in mRNA decay; nuclear Lsm8p complex
part of U6 snRNP and possibly involved in processing tRNA,
snoRNA, and rRNA; K12624 U6 snRNA-associated Sm-like protein
LSm5
Length=93
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 49/88 (55%), Gaps = 12/88 (13%)
Query 44 PINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLK-----KNTR 98
P+ +I + K + L Q + EG ++GFD+++N++L+DA E + +N +
Sbjct 8 PLEVIDKTINQKVLIVL----QSNREFEGTLVGFDDFVNVILEDAVEWLIDPEDESRNEK 63
Query 99 VFV--GRILLKGENISLIMSTG-STNTE 123
V GR+LL G NI++++ G T TE
Sbjct 64 VMQHHGRMLLSGNNIAILVPGGKKTPTE 91
> pfa:PF14_0411 small nuclear ribonucleoprotein, putative; K12624
U6 snRNA-associated Sm-like protein LSm5
Length=101
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 59 QLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEE 90
++W+ + D I G+++GFDEY+NMVL+D E
Sbjct 23 KIWIMMKGDKEIVGKLVGFDEYVNMVLEDVTE 54
> pfa:MAL8P1.48 small nuclear ribonucleoprotein, putative; K11099
small nuclear ribonucleoprotein G
Length=83
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 43/64 (67%), Gaps = 5/64 (7%)
Query 49 FRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVFVGRILLKG 108
FR F K R+Q++L + ++ G + G+D +MN+VLD+ EI KK+ ++ +G ++++G
Sbjct 14 FRKFMEK-RLQIYL--NGNRQVVGILRGYDTFMNLVLDNTMEI--KKDEQIDIGVVVIRG 68
Query 109 ENIS 112
+IS
Sbjct 69 NSIS 72
> bbo:BBOV_IV009630 23.m05873; snRNP Sm-like protein; K12624 U6
snRNA-associated Sm-like protein LSm5
Length=87
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 40/75 (53%), Gaps = 8/75 (10%)
Query 44 PINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVFVGR 103
P+ L+ + K +W+ + + I G + GFD+YMN+VLDD E K T V
Sbjct 12 PLALVDKCLGTK----VWIIMKGEKEITGVLRGFDDYMNVVLDDVTEYTFKP-TGVETTE 66
Query 104 I---LLKGENISLIM 115
+ L+ G NI++I+
Sbjct 67 LKDALVNGTNIAMIV 81
> ath:AT4G20440 smB; smB (small nuclear ribonucleoprotein associated
protein B); K11086 small nuclear ribonucleoprotein B
and B'
Length=257
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 31/63 (49%), Gaps = 11/63 (17%)
Query 72 GRIMGFDEYMNMVLDDAEE-----------IHLKKNTRVFVGRILLKGENISLIMSTGST 120
G+ M FD +MN+VL D EE I+ ++ R +G +LL+GE + + G
Sbjct 29 GKFMAFDRHMNLVLGDCEEFRKLPPAKGKKINEEREDRRTLGLVLLRGEEVISMTVEGPP 88
Query 121 NTE 123
E
Sbjct 89 PPE 91
> pfa:PF14_0146 ribonucleoprotein, putative; K11086 small nuclear
ribonucleoprotein B and B'
Length=201
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 27/70 (38%), Positives = 38/70 (54%), Gaps = 15/70 (21%)
Query 56 SRVQLWLYEQPDLRIE------GRIMGFDEYMNMVLDDAEEIH--------LKKNTRVFV 101
SR++ WL + + I G + +D +MN+VL DAEE LK+ RV V
Sbjct 5 SRLETWLQYRVRVTISDTRYFVGTFLSYDRHMNIVLVDAEEFRKVKSQENSLKEIKRV-V 63
Query 102 GRILLKGENI 111
G IL++GENI
Sbjct 64 GLILIRGENI 73
> bbo:BBOV_III006010 17.m07530; Sm domain containing protein;
K11086 small nuclear ribonucleoprotein B and B'
Length=159
Score = 37.0 bits (84), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 37/71 (52%), Gaps = 15/71 (21%)
Query 56 SRVQLWLYEQPDLRIE------GRIMGFDEYMNMVLDDAEEIHLKKNT---------RVF 100
+R+Q WL + + I+ G + FD++MN+VL D EE + K +
Sbjct 6 TRMQQWLQYRVRVAIKDGRKFVGTFIAFDKHMNLVLADCEEFRITKGKGPDKQKVELKRT 65
Query 101 VGRILLKGENI 111
+G I+L+GENI
Sbjct 66 LGFIMLRGENI 76
> cpv:cgd1_2690 hypothetical protein
Length=67
Score = 36.2 bits (82), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 32/49 (65%), Gaps = 5/49 (10%)
Query 68 LRIEGRIMGFDEYMNMVLDDAEEI--HLKKNTRVFVGRILLKGENISLI 114
+R +G ++ FDEYMN++L+D EE + KK T +G++ ++ N+ I
Sbjct 1 MRYKGTLVSFDEYMNILLNDCEEWIDNTKKGT---LGKVFIRCNNVLYI 46
> cpv:cgd1_2910 hypothetical protein ; K11086 small nuclear ribonucleoprotein
B and B'
Length=139
Score = 35.4 bits (80), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 29/53 (54%), Gaps = 6/53 (11%)
Query 65 QPDLRIEGRIMGFDEYMNMVLDDAEEI------HLKKNTRVFVGRILLKGENI 111
Q D + G +M FD +MN+VL D +E K + +G I+L+GENI
Sbjct 14 QDDRVMVGNLMAFDRHMNLVLSDCQEYRRVKKGEESKELKRSLGLIMLRGENI 66
> ath:AT5G44500 small nuclear ribonucleoprotein associated protein
B, putative / snRNP-B, putative / Sm protein B, putative;
K11086 small nuclear ribonucleoprotein B and B'
Length=254
Score = 34.7 bits (78), Expect = 0.081, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 30/64 (46%), Gaps = 12/64 (18%)
Query 72 GRIMGFDEYMNMVLDDAEEI------------HLKKNTRVFVGRILLKGENISLIMSTGS 119
G+ M FD +MN+VL D EE + ++ R +G +LL+GE + + G
Sbjct 29 GKFMAFDRHMNLVLGDCEEFRKLPPAKGNKKTNEEREERRTLGLVLLRGEEVISMTVEGP 88
Query 120 TNTE 123
E
Sbjct 89 PPPE 92
> sce:YLR275W SMD2; Sm D2; K11096 small nuclear ribonucleoprotein
D2
Length=110
Score = 34.7 bits (78), Expect = 0.081, Method: Compositional matrix adjust.
Identities = 15/55 (27%), Positives = 33/55 (60%), Gaps = 6/55 (10%)
Query 69 RIEGRIMGFDEYMNMVLDDAEEIHLKK------NTRVFVGRILLKGENISLIMST 117
+I R+ FD + NMVL++ +E+ +K N F+ ++ L+G+++ +++ T
Sbjct 53 KIIARVKAFDRHCNMVLENVKELWTEKKGKNVINRERFISKLFLRGDSVIVVLKT 107
> cpv:cgd4_3190 small nuclear ribonucleoprotein ; K11096 small
nuclear ribonucleoprotein D2
Length=106
Score = 34.3 bits (77), Expect = 0.095, Method: Compositional matrix adjust.
Identities = 18/75 (24%), Positives = 39/75 (52%), Gaps = 12/75 (16%)
Query 55 KSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHLKK------------NTRVFVG 102
K+ Q+ + + + +I GR+ FD + N++L DA EI + N F+
Sbjct 30 KTNTQILVNCRNNRKILGRVKAFDRHCNLLLTDAREIWTESMKSGKKGSAKYINKDRFIS 89
Query 103 RILLKGENISLIMST 117
++ ++G+++ LI+ +
Sbjct 90 KLFVRGDSVILILKS 104
> xla:779307 snrpg, MGC154809; small nuclear ribonucleoprotein
polypeptide G; K11099 small nuclear ribonucleoprotein G
Length=76
Score = 33.9 bits (76), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 31/46 (67%), Gaps = 1/46 (2%)
Query 69 RIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVFVGRILLKGENISLI 114
+++G + GFD +MN+VLD++ EI N +G ++++G +I ++
Sbjct 26 QVQGILRGFDPFMNLVLDESTEISGSGNQNS-IGMVVIRGNSIIML 70
> cel:Y71F9B.4 snr-7; Small Nuclear Ribonucleoprotein family member
(snr-7); K11099 small nuclear ribonucleoprotein G
Length=77
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 30/46 (65%), Gaps = 1/46 (2%)
Query 69 RIEGRIMGFDEYMNMVLDDAEEIHLKKNTRVFVGRILLKGENISLI 114
R+ G + GFD +MNMV+D+A E + K V +G +++G ++ ++
Sbjct 26 RVSGILRGFDPFMNMVIDEAVE-YQKDGGSVNLGMTVIRGNSVVIM 70
> ath:AT2G23930 SNRNP-G (PROBABLE SMALL NUCLEAR RIBONUCLEOPROTEIN
G); K04796 small nuclear ribonucleoprotein
Length=67
Score = 33.5 bits (75), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 29/42 (69%), Gaps = 2/42 (4%)
Query 70 IEGRIMGFDEYMNMVLDDAEEIHLKKNTRVFVGRILLKGENI 111
+ G + GFD++MN+V+D+ E++ N + +G ++++G +I
Sbjct 15 VTGTLRGFDQFMNLVVDNTVEVN--GNDKTDIGMVVIRGNSI 54
> cel:Y73B6BL.32 lsm-8; LSM Sm-like protein family member (lsm-8);
K12627 U6 snRNA-associated Sm-like protein LSm8
Length=98
Score = 33.1 bits (74), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 30/48 (62%), Gaps = 3/48 (6%)
Query 70 IEGRIMGFDEYMNMVLDDAEEIHLKKNTRVF---VGRILLKGENISLI 114
I G + GFD+ +N+V++DA E + V +G +++GEN+++I
Sbjct 23 IVGLLKGFDQLINLVIEDAHERSYSETEGVLTTPLGLYIIRGENVAII 70
> bbo:BBOV_I004230 19.m02306; small nuclear ribonucleoprotein;
K11096 small nuclear ribonucleoprotein D2
Length=162
Score = 32.3 bits (72), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 19/95 (20%), Positives = 48/95 (50%), Gaps = 12/95 (12%)
Query 32 MSNKKLQKIMTQPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEI 91
MS + + + P++++ + S Q+ + + + ++ GR+ FD + NM+L D E+
Sbjct 67 MSTTEDKDCPSGPLSVLEACVRDNS--QVLINCRSNRKLLGRVKAFDRHFNMILTDVREM 124
Query 92 HLK----------KNTRVFVGRILLKGENISLIMS 116
+ N ++ R+ L+G+++ +++S
Sbjct 125 WTEVSGGGGKKKYMNKDRYISRLFLRGDSVVVVLS 159
> tgo:TGME49_100280 small nuclear ribonucleoprotein, putative
Length=236
Score = 32.0 bits (71), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 30/58 (51%), Gaps = 6/58 (10%)
Query 40 IMTQPINLIFRFFTNKSRVQLWL-YE-----QPDLRIEGRIMGFDEYMNMVLDDAEEI 91
+ T+ N + + +R+Q W+ Y Q + G ++ FD +MN+VL DAEE
Sbjct 31 VGTEERNKFSKMPSKNARLQQWINYRIRVCLQDSRMLVGSLLAFDRHMNIVLADAEEF 88
> ath:AT1G21190 small nuclear ribonucleoprotein, putative / snRNP,
putative / Sm protein, putative; K12622 U6 snRNA-associated
Sm-like protein LSm3
Length=97
Score = 31.6 bits (70), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 16/49 (32%), Positives = 31/49 (63%), Gaps = 4/49 (8%)
Query 43 QPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEI 91
+P++LI + + R+ + L +LR G++ FD+++NM+L D EE+
Sbjct 11 EPLDLIR--LSIEERIYVKLRSDRELR--GKLHAFDQHLNMILGDVEEV 55
> cel:Y62E10A.12 lsm-3; LSM Sm-like protein family member (lsm-3);
K12622 U6 snRNA-associated Sm-like protein LSm3
Length=102
Score = 31.6 bits (70), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 31/60 (51%), Gaps = 12/60 (20%)
Query 67 DLRIEGRIMGFDEYMNMVLDDAEEI------------HLKKNTRVFVGRILLKGENISLI 114
D + GR+ FD+++NMVL + EE + K T+ V + ++G+++ L+
Sbjct 36 DRELRGRLRAFDQHLNMVLSEVEETITTREVDEDTFEEIYKQTKRVVPMLFVRGDSVILV 95
> ath:AT3G11500 small nuclear ribonucleoprotein G, putative /
snRNP-G, putative / Sm protein G, putative; K11099 small nuclear
ribonucleoprotein G
Length=79
Score = 31.6 bits (70), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 13/42 (30%), Positives = 28/42 (66%), Gaps = 2/42 (4%)
Query 70 IEGRIMGFDEYMNMVLDDAEEIHLKKNTRVFVGRILLKGENI 111
+ G + GFD++MN+V+D+ E++ T +G ++++G +I
Sbjct 28 VVGTLRGFDQFMNLVVDNTVEVNGDDKTD--IGMVVIRGNSI 67
> ath:AT1G76860 small nuclear ribonucleoprotein, putative / snRNP,
putative / Sm protein, putative; K12622 U6 snRNA-associated
Sm-like protein LSm3
Length=98
Score = 30.8 bits (68), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 28/48 (58%), Gaps = 4/48 (8%)
Query 43 QPINLIFRFFTNKSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEE 90
+P++LI + V+L + D + G++ FD+++NM+L D EE
Sbjct 11 EPLDLIRLSLDERIYVKL----RSDRELRGKLHAFDQHLNMILGDVEE 54
> tgo:TGME49_070830 small nuclear ribonucleoprotein Sm D2, putative
; K11096 small nuclear ribonucleoprotein D2
Length=104
Score = 30.8 bits (68), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 36/73 (49%), Gaps = 12/73 (16%)
Query 55 KSRVQLWLYEQPDLRIEGRIMGFDEYMNMVLDDAEEIHL-------KKNTRV-----FVG 102
K Q+ + + + ++ GR+ FD + N+VL D E+ KK R F+
Sbjct 28 KDNSQVLINCRNNRKVLGRVKAFDRHCNLVLTDVREMWTETSKGGGKKKVRTVNKDRFIS 87
Query 103 RILLKGENISLIM 115
++ L+G+ + LI+
Sbjct 88 KLFLRGDAVILIL 100
> bbo:BBOV_II007600 18.m06632; small nuclear ribonucleoprotein
G; K11099 small nuclear ribonucleoprotein G
Length=80
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 28/42 (66%), Gaps = 2/42 (4%)
Query 70 IEGRIMGFDEYMNMVLDDAEEIHLKKNTRVFVGRILLKGENI 111
+ G + G+D +MN+VLD+A +I + TR +G ++++G I
Sbjct 29 VVGVLRGYDTFMNVVLDNALQIEGDEQTR--LGMVVIRGNAI 68
> mmu:545531 Gm5848, EG545531; predicted pseudogene 5848
Length=118
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 13/60 (21%), Positives = 31/60 (51%), Gaps = 13/60 (21%)
Query 69 RIEGRIMGFDEYMNMVLDDAEEI-------------HLKKNTRVFVGRILLKGENISLIM 115
++ GR+ FD + NMVL++ +E+ N ++ +I L+G+++ +++
Sbjct 51 KLLGRVKAFDRHCNMVLENVKEMWTEVPKSSKGKKKSKPVNKDCYISKIFLRGDSVIVVL 110
Lambda K H
0.319 0.131 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003197800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40