bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1610_orf4
Length=191
Score E
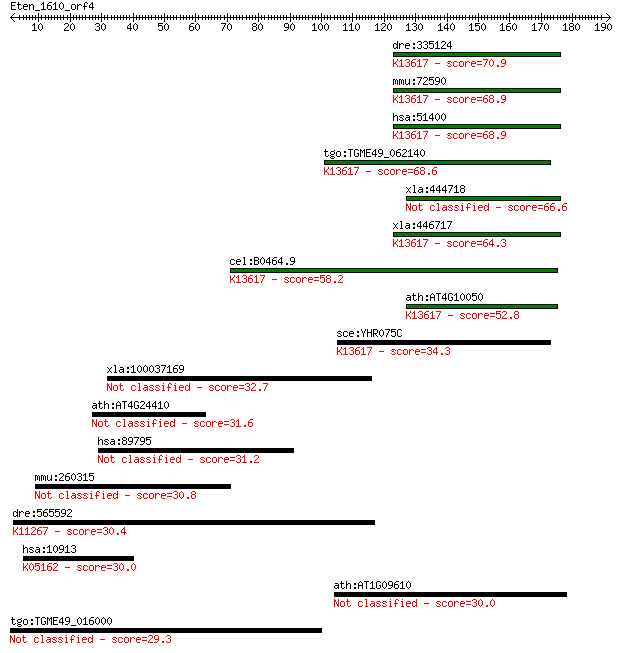
Sequences producing significant alignments: (Bits) Value
dre:335124 ppme1, fk72d03, wu:fk72d03, zgc:56239; protein phos... 70.9 3e-12
mmu:72590 Ppme1, 1110069N17Rik, 2700017M01Rik, PME-1, Pme1; pr... 68.9 9e-12
hsa:51400 PPME1, FLJ22226, PME-1; protein phosphatase methyles... 68.9 1e-11
tgo:TGME49_062140 protein phosphatase methylesterase 1, putati... 68.6 1e-11
xla:444718 MGC84679 protein 66.6 5e-11
xla:446717 ppme1, MGC84506; protein phosphatase methylesterase... 64.3 3e-10
cel:B0464.9 hypothetical protein; K13617 protein phosphatase m... 58.2 2e-08
ath:AT4G10050 hydrolase, alpha/beta fold family protein; K1361... 52.8 8e-07
sce:YHR075C PPE1; Ppe1p (EC:3.1.1.-); K13617 protein phosphata... 34.3 0.24
xla:100037169 hypothetical protein LOC100037169 32.7 0.87
ath:AT4G24410 hypothetical protein 31.6 2.0
hsa:89795 NAV3, KIAA0938, POMFIL1, STEERIN3, unc53H3; neuron n... 31.2 2.4
mmu:260315 Nav3, 4732483H20Rik, 9630020C08Rik, POMFIL1, Pomfil... 30.8 2.8
dre:565592 pds5b; PDS5, regulator of cohesion maintenance, hom... 30.4 3.6
hsa:10913 EDAR, DL, ED1R, ED3, ED5, EDA-A1R, EDA1R, EDA3, FLJ9... 30.0 4.9
ath:AT1G09610 hypothetical protein 30.0 5.4
tgo:TGME49_016000 inner membrane complex protein IMC3 (EC:1.12... 29.3 9.2
> dre:335124 ppme1, fk72d03, wu:fk72d03, zgc:56239; protein phosphatase
methylesterase 1 (EC:3.1.1.-); K13617 protein phosphatase
methylesterase 1 [EC:3.1.1.-]
Length=377
Score = 70.9 bits (172), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 33/57 (57%), Positives = 39/57 (68%), Gaps = 5/57 (8%)
Query 123 PLPTWDEFFEKCEDIRPL----ESSDTFRVYSSGSSGPLLFLLHGAGHTSLSWACFT 175
PLP W +FE ED+ S DTFR+YSSGS GP+L LLHG GH++LSWA FT
Sbjct 37 PLP-WSHYFETIEDVEVEVENDNSKDTFRIYSSGSHGPVLLLLHGGGHSALSWAVFT 92
> mmu:72590 Ppme1, 1110069N17Rik, 2700017M01Rik, PME-1, Pme1;
protein phosphatase methylesterase 1 (EC:3.1.1.-); K13617 protein
phosphatase methylesterase 1 [EC:3.1.1.-]
Length=386
Score = 68.9 bits (167), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 31/55 (56%), Positives = 39/55 (70%), Gaps = 3/55 (5%)
Query 123 PLPTWDEFFEKCEDIRPLESS--DTFRVYSSGSSGPLLFLLHGAGHTSLSWACFT 175
P+P W ++FE ED+ + DTFRVY SGS GP+L LLHG GH++LSWA FT
Sbjct 43 PVP-WSQYFESMEDVEVENETGKDTFRVYKSGSEGPVLLLLHGGGHSALSWAVFT 96
> hsa:51400 PPME1, FLJ22226, PME-1; protein phosphatase methylesterase
1 (EC:3.1.1.-); K13617 protein phosphatase methylesterase
1 [EC:3.1.1.-]
Length=386
Score = 68.9 bits (167), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 31/55 (56%), Positives = 39/55 (70%), Gaps = 3/55 (5%)
Query 123 PLPTWDEFFEKCEDIRPLESS--DTFRVYSSGSSGPLLFLLHGAGHTSLSWACFT 175
P+P W ++FE ED+ + DTFRVY SGS GP+L LLHG GH++LSWA FT
Sbjct 43 PVP-WSQYFESMEDVEVENETGKDTFRVYKSGSEGPVLLLLHGGGHSALSWAVFT 96
> tgo:TGME49_062140 protein phosphatase methylesterase 1, putative
; K13617 protein phosphatase methylesterase 1 [EC:3.1.1.-]
Length=452
Score = 68.6 bits (166), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 33/73 (45%), Positives = 48/73 (65%), Gaps = 1/73 (1%)
Query 101 RDAQTPHSGSSSRRSSCNASCGPLPTWDEFFEKCEDIRPLES-SDTFRVYSSGSSGPLLF 159
+D + ++ S + S + S PL W +++E EDI P E+ +D+FRVY SGS GP+ F
Sbjct 69 KDERAANAASRGKGRSRSRSVEPLLKWSDYWEVQEDIHPTENPNDSFRVYRSGSEGPICF 128
Query 160 LLHGAGHTSLSWA 172
LHG GHT+LSW+
Sbjct 129 FLHGGGHTALSWS 141
> xla:444718 MGC84679 protein
Length=140
Score = 66.6 bits (161), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 30/52 (57%), Positives = 38/52 (73%), Gaps = 4/52 (7%)
Query 127 WDEFFEKCEDIRPLES---SDTFRVYSSGSSGPLLFLLHGAGHTSLSWACFT 175
W ++FE ED+ +ES D+FRVY SGS GP+L LLHG GH++LSWA FT
Sbjct 44 WSQYFESMEDV-VVESETGKDSFRVYRSGSEGPVLLLLHGGGHSALSWAVFT 94
> xla:446717 ppme1, MGC84506; protein phosphatase methylesterase
1; K13617 protein phosphatase methylesterase 1 [EC:3.1.1.-]
Length=386
Score = 64.3 bits (155), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 31/56 (55%), Positives = 40/56 (71%), Gaps = 5/56 (8%)
Query 123 PLPTWDEFFEKCEDIRPLESSD---TFRVYSSGSSGPLLFLLHGAGHTSLSWACFT 175
P+P W ++FE ED+ +ES +FRVY SGS GP+L LLHG GH++LSWA FT
Sbjct 41 PVP-WSQYFESMEDV-VVESETGKASFRVYRSGSEGPVLLLLHGGGHSALSWAVFT 94
> cel:B0464.9 hypothetical protein; K13617 protein phosphatase
methylesterase 1 [EC:3.1.1.-]
Length=364
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 33/104 (31%), Positives = 51/104 (49%), Gaps = 11/104 (10%)
Query 71 DFETEAATEHRPHSRQPRSASSARQRTGRHRDAQTPHSGSSSRRSSCNASCGPLPTWDEF 130
D ++E + PH R RQ R P + +S ++ + LP W +F
Sbjct 11 DLQSETSHVTTPH----RQNDLLRQAVTHGRPPPVPSTSTSGKKREMSE----LP-WSDF 61
Query 131 FEKCEDIRPLESSDTFRVYSSGSSGPLLFLLHGAGHTSLSWACF 174
F++ +D D F VY G+ GP+ +LLHG G++ L+WACF
Sbjct 62 FDEKKDANI--DGDVFNVYIKGNEGPIFYLLHGGGYSGLTWACF 103
> ath:AT4G10050 hydrolase, alpha/beta fold family protein; K13617
protein phosphatase methylesterase 1 [EC:3.1.1.-]
Length=350
Score = 52.8 bits (125), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 31/48 (64%), Gaps = 0/48 (0%)
Query 127 WDEFFEKCEDIRPLESSDTFRVYSSGSSGPLLFLLHGAGHTSLSWACF 174
W +F+K +DI S D F VY +G+ GP++F LHG G++ LS++
Sbjct 50 WKSYFDKEDDISITGSDDVFHVYMAGNEGPVVFCLHGGGYSGLSFSIV 97
> sce:YHR075C PPE1; Ppe1p (EC:3.1.1.-); K13617 protein phosphatase
methylesterase 1 [EC:3.1.1.-]
Length=400
Score = 34.3 bits (77), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 24/76 (31%), Positives = 39/76 (51%), Gaps = 9/76 (11%)
Query 105 TPHSGSSSRRSSCNASCGPLPTWDEFFEKCEDI----RPLESSDTFR----VYSSGSSGP 156
T +GS++ R S LPTW +FF+ E + R L+ + + + S+ +S P
Sbjct 56 TGKTGSTTDRISSKEKSS-LPTWSDFFDNKELVSLPDRDLDVNTYYTLPTSLLSNTTSIP 114
Query 157 LLFLLHGAGHTSLSWA 172
+ HGAG + LS+A
Sbjct 115 IFIFHHGAGSSGLSFA 130
> xla:100037169 hypothetical protein LOC100037169
Length=959
Score = 32.7 bits (73), Expect = 0.87, Method: Composition-based stats.
Identities = 24/89 (26%), Positives = 39/89 (43%), Gaps = 9/89 (10%)
Query 32 SPHQSRRKSAGCSMRPVKEERQQAEPEHAPEGSASHANLDFETEAATEHRPHSRQPR--- 88
S + RRKS P K + + +P +P + +T++ E R SR +
Sbjct 207 SKSKERRKSKS----PAKRSKSEDQPRRSPALHRQSPEKNIKTKSPQEERVKSRDRKKSP 262
Query 89 --SASSARQRTGRHRDAQTPHSGSSSRRS 115
S S +R+RT + + + S S RRS
Sbjct 263 VASESKSRERTKKSKSPVSRRSKSKERRS 291
> ath:AT4G24410 hypothetical protein
Length=169
Score = 31.6 bits (70), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 24/38 (63%), Gaps = 2/38 (5%)
Query 27 GSPSESPHQSRRKSAGCS--MRPVKEERQQAEPEHAPE 62
GSPS P + RRK+ G S +RP++E+R+ E A E
Sbjct 121 GSPSRDPIEIRRKTVGKSRDVRPIQEKREARARERASE 158
> hsa:89795 NAV3, KIAA0938, POMFIL1, STEERIN3, unc53H3; neuron
navigator 3
Length=2363
Score = 31.2 bits (69), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 33/62 (53%), Gaps = 1/62 (1%)
Query 29 PSESPHQSRRKSAGCSMRPVKEERQQAEPEHAPEGSASHANLDFETEAATEHRPHSRQPR 88
P E ++R K+ C+ +PVKEE+ Q E AP+ ++ A+L + T + S P
Sbjct 465 PKEKEEKNRDKNKVCTEKPVKEEKDQVT-EMAPKKTSKIASLIPKGSKTTAAKKESLIPS 523
Query 89 SA 90
S+
Sbjct 524 SS 525
> mmu:260315 Nav3, 4732483H20Rik, 9630020C08Rik, POMFIL1, Pomfil1p,
mKIAA0938, unc53H3; neuron navigator 3
Length=2366
Score = 30.8 bits (68), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 34/64 (53%), Gaps = 4/64 (6%)
Query 9 KELPPITRGRSSNFPDR--VGSPSESPHQSRRKSAGCSMRPVKEERQQAEPEHAPEGSAS 66
K PP + S NF ++ + P E ++R K+ C+ + KEE+ Q E AP+ ++
Sbjct 445 KLTPP--KAGSKNFSNKKSLLQPKEKEEKTRDKNKACAEKSGKEEKDQVTTEAAPKKTSK 502
Query 67 HANL 70
A+L
Sbjct 503 IASL 506
> dre:565592 pds5b; PDS5, regulator of cohesion maintenance, homolog
B (S. cerevisiae); K11267 sister chromatid cohesion protein
PDS5
Length=1408
Score = 30.4 bits (67), Expect = 3.6, Method: Composition-based stats.
Identities = 26/117 (22%), Positives = 52/117 (44%), Gaps = 2/117 (1%)
Query 2 ARMPPDIKELPPITRGRSSNFPDRVGSPSESPHQSR--RKSAGCSMRPVKEERQQAEPEH 59
A + + ++ P RGR P + E+P + R R+ + E+ QQ E +
Sbjct 1244 ANLETEDEQNSPPKRGRRGRPPKSAAAKEETPSKGRRGRRKTAPPVEEDNEDEQQEEEDE 1303
Query 60 APEGSASHANLDFETEAATEHRPHSRQPRSASSARQRTGRHRDAQTPHSGSSSRRSS 116
E A N++ + + +P +AS + + T + R + P S +++R++S
Sbjct 1304 DEEEDAGSENMEVKPKGRRGRKPKVDVSETASDSVETTPQKRRGRPPKSQTAARKAS 1360
> hsa:10913 EDAR, DL, ED1R, ED3, ED5, EDA-A1R, EDA1R, EDA3, FLJ94390,
HRM1; ectodysplasin A receptor; K05162 ectodysplasin
1, anhidrotic receptor
Length=448
Score = 30.0 bits (66), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 5 PPDIKELPPITRGRSSNFPDRVGSPSESPHQSRRK 39
PP+ KE T G S+NFP GS + SP Q K
Sbjct 142 PPNTKECVGATSGASANFPGTSGSSTLSPFQHAHK 176
> ath:AT1G09610 hypothetical protein
Length=282
Score = 30.0 bits (66), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 31/79 (39%), Gaps = 8/79 (10%)
Query 104 QTPHSGSSSRRSSCNASCGPLP---TWDEFFEKCEDIRPLESSDTFRVYS--SGSSGPLL 158
QTP S+R C+ +C LP I P ++ V S G P
Sbjct 39 QTPQETRSTR---CSGACNKLPRSLAQALIHYSTSVITPQQTLKEIAVSSRVLGKKSPCN 95
Query 159 FLLHGAGHTSLSWACFTVG 177
FL+ G GH SL W+ G
Sbjct 96 FLVFGLGHDSLMWSSLNYG 114
> tgo:TGME49_016000 inner membrane complex protein IMC3 (EC:1.12.98.2)
Length=538
Score = 29.3 bits (64), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 40/107 (37%), Gaps = 32/107 (29%)
Query 1 VARMPPDIKELPPITRGRSSNFPDRVGSPSESPHQSRRKSAGCSMRPVKEERQQAEPEHA 60
+ PPD LPP+ G P RV + +P Q + P A
Sbjct 274 IPYQPPDAATLPPLMVG---GLPSRVSATMPTPFQ---------LGP------------A 309
Query 61 PEGSASHANLDFETEA--------ATEHRPHSRQPRSASSARQRTGR 99
P G A+ NL FE A H P S P++ ++A + GR
Sbjct 310 PAGHANEVNLPFEARVDVTVSQSPAGPHSPLSASPQTGTAASLKLGR 356
Lambda K H
0.314 0.127 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5493959020
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40