bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1612_orf1
Length=101
Score E
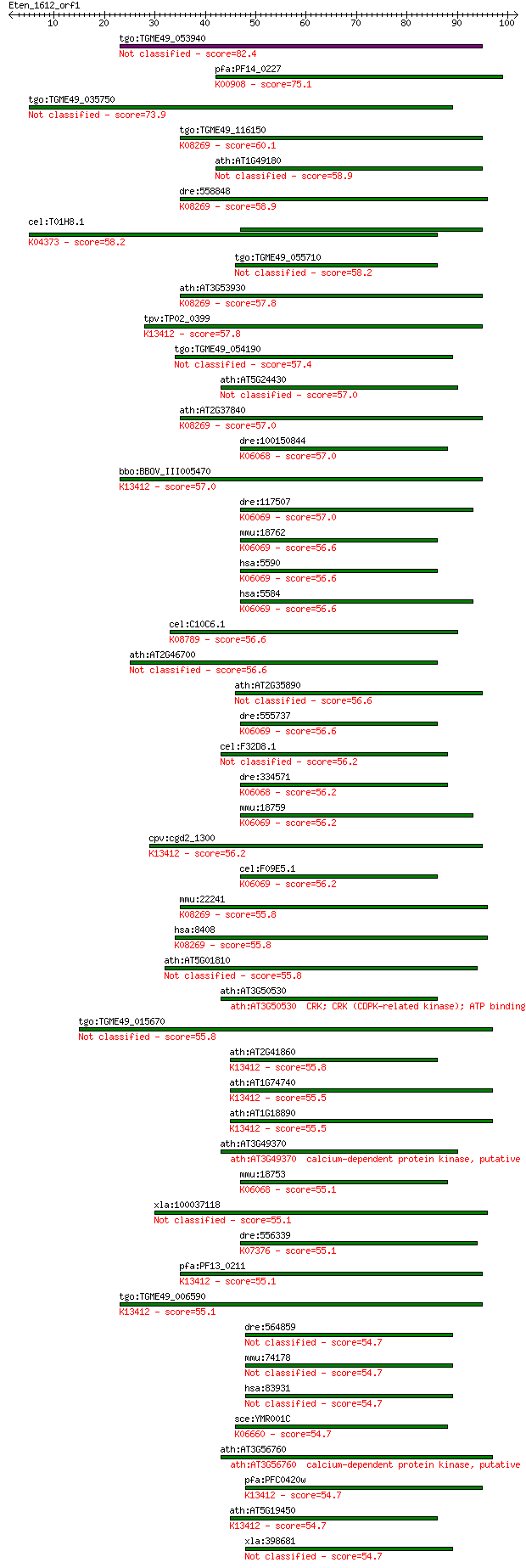
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_053940 calcium/calmodulin-dependent protein kinase,... 82.4 3e-16
pfa:PF14_0227 calcium/calmodulin-dependent protein kinase, put... 75.1 5e-14
tgo:TGME49_035750 calcium/calmodulin-dependent protein kinase ... 73.9 1e-13
tgo:TGME49_116150 protein kinase domain-containing protein (EC... 60.1 2e-09
ath:AT1G49180 protein kinase family protein 58.9 4e-09
dre:558848 ulk1b, ulk1; unc-51-like kinase 1b (C. elegans); K0... 58.9 4e-09
cel:T01H8.1 rskn-1; RSK-pNinety (RSK-p90 kinase) homolog famil... 58.2 6e-09
tgo:TGME49_055710 calcium-dependent protein kinase, putative (... 58.2 6e-09
ath:AT3G53930 protein kinase family protein; K08269 unc51-like... 57.8 9e-09
tpv:TP02_0399 calcium-dependent protein kinase; K13412 calcium... 57.8 9e-09
tgo:TGME49_054190 protein kinase, putative (EC:2.7.11.1) 57.4 1e-08
ath:AT5G24430 calcium-dependent protein kinase, putative / CDP... 57.0 1e-08
ath:AT2G37840 protein kinase family protein; K08269 unc51-like... 57.0 1e-08
dre:100150844 protein kinase C, delta-like; K06068 novel prote... 57.0 1e-08
bbo:BBOV_III005470 17.m07489; protein kinase domain containing... 57.0 1e-08
dre:117507 prkci, aPKC[l], hal, has; protein kinase C, iota (E... 57.0 1e-08
mmu:18762 Prkcz, AI098070, C80388, Pkcz, R74924, aPKCzeta, zet... 56.6 2e-08
hsa:5590 PRKCZ, PKC-ZETA, PKC2; protein kinase C, zeta (EC:2.7... 56.6 2e-08
hsa:5584 PRKCI, DXS1179E, MGC26534, PKCI, nPKC-iota; protein k... 56.6 2e-08
cel:C10C6.1 kin-4; protein KINase family member (kin-4); K0878... 56.6 2e-08
ath:AT2G46700 calcium-dependent protein kinase, putative / CDP... 56.6 2e-08
ath:AT2G35890 CPK25; CPK25; ATP binding / calmodulin-dependent... 56.6 2e-08
dre:555737 prkcz, pkcz, si:ch211-150o23.4; protein kinase C, z... 56.6 2e-08
cel:F32D8.1 hypothetical protein 56.2 2e-08
dre:334571 prkcd, fb09d10, fv43b11, wu:fb09d10, wu:fv43b11, zg... 56.2 2e-08
mmu:18759 Prkci, 2310021H13Rik, AI427505, KIAA4165, PKClambda,... 56.2 2e-08
cpv:cgd2_1300 calcium/calmodulin dependent protein kinase with... 56.2 3e-08
cel:F09E5.1 pkc-3; Protein Kinase C family member (pkc-3); K06... 56.2 3e-08
mmu:22241 Ulk1, AU041434, Unc51.1, mKIAA0722; Unc-51 like kina... 55.8 3e-08
hsa:8408 ULK1, ATG1, ATG1A, FLJ38455, FLJ46475, UNC51, Unc51.1... 55.8 3e-08
ath:AT5G01810 CIPK15; CIPK15 (CBL-INTERACTING PROTEIN KINASE 1... 55.8 3e-08
ath:AT3G50530 CRK; CRK (CDPK-related kinase); ATP binding / ca... 55.8 3e-08
tgo:TGME49_015670 protein kinase, putative (EC:2.7.12.1 2.7.11... 55.8 3e-08
ath:AT2G41860 CPK14; CPK14; ATP binding / calcium ion binding ... 55.8 4e-08
ath:AT1G74740 CPK30; CPK30 (CALCIUM-DEPENDENT PROTEIN KINASE 3... 55.5 4e-08
ath:AT1G18890 ATCDPK1 (CALCIUM-DEPENDENT PROTEIN KINASE 1); ca... 55.5 4e-08
ath:AT3G49370 calcium-dependent protein kinase, putative / CDP... 55.1 5e-08
mmu:18753 Prkcd, AI385711, D14Ertd420e, PKC[d], PKCdelta, Pkcd... 55.1 5e-08
xla:100037118 hypothetical protein LOC100037118 55.1 6e-08
dre:556339 protein kinase, cGMP-dependent, type II-like; K0737... 55.1 6e-08
pfa:PF13_0211 calcium-dependent protein kinase, putative (EC:2... 55.1 6e-08
tgo:TGME49_006590 calcium-dependent protein kinase, putative (... 55.1 6e-08
dre:564859 stk40, cb988, fd99h11, wu:fd99h11; serine/threonine... 54.7 6e-08
mmu:74178 Stk40, 2310004N11Rik, AI428792; serine/threonine kin... 54.7 6e-08
hsa:83931 STK40, MGC4796, RP11-268J15.4, SHIK, SgK495; serine/... 54.7 6e-08
sce:YMR001C CDC5, MSD2, PKX2; Cdc5p (EC:2.7.11.21); K06660 cel... 54.7 7e-08
ath:AT3G56760 calcium-dependent protein kinase, putative / CDP... 54.7 7e-08
pfa:PFC0420w PfCDPK3; calcium-dependent protein kinase-3 (EC:2... 54.7 7e-08
ath:AT5G19450 CDPK19; CDPK19 (CALCIUM-DEPENDENT PROTEIN KINASE... 54.7 8e-08
xla:398681 stk40; serine/threonine kinase 40 (EC:2.7.11.1) 54.7 8e-08
> tgo:TGME49_053940 calcium/calmodulin-dependent protein kinase,
putative (EC:2.7.11.17)
Length=529
Score = 82.4 bits (202), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 38/72 (52%), Positives = 51/72 (70%), Gaps = 8/72 (11%)
Query 23 FATSCRPRTSDTVLEYGFLHLMQICGSPHYLSPEILRRSYSFPADMWSLGVLVFVMLYGR 82
F S R R +T L ++CGSPHYL+PE+LRR Y+ PADMW+LGVLVF+MLYGR
Sbjct 227 FGMSVRVRPDET--------LSEVCGSPHYLAPELLRRQYACPADMWALGVLVFLMLYGR 278
Query 83 YPFQGSSSLDVL 94
YP+ G ++ ++
Sbjct 279 YPYDGHTTSRIV 290
> pfa:PF14_0227 calcium/calmodulin-dependent protein kinase, putative;
K00908 Ca2+/calmodulin-dependent protein kinase [EC:2.7.11.17]
Length=284
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 28/60 (46%), Positives = 47/60 (78%), Gaps = 3/60 (5%)
Query 42 HLMQICGSPHYLSPEILRRSYSFPADMWSLGVLVFVMLYGRYPFQGSSS---LDVLLART 98
+L ++CGSPHY+SPE++R+ Y+ +D+W+LGV+VF ML G+YPF+G ++ +D +L +
Sbjct 178 YLTELCGSPHYISPELIRKKYTMSSDIWALGVMVFFMLTGKYPFEGKNTPKVVDEILNKN 237
> tgo:TGME49_035750 calcium/calmodulin-dependent protein kinase
type I, putative (EC:2.7.11.17)
Length=376
Score = 73.9 bits (180), Expect = 1e-13, Method: Composition-based stats.
Identities = 33/91 (36%), Positives = 55/91 (60%), Gaps = 8/91 (8%)
Query 5 YLYERYGIGNNNTACTPPFATSCRPRTSDTVLEYGFL-------HLMQICGSPHYLSPEI 57
YL+ER GI + + S P + ++++G +L +CGSPHY+SPE+
Sbjct 147 YLHER-GIVHRDVKAENFLFKSKSPDSLIKLIDFGMSAKIPPCGYLTDLCGSPHYISPEL 205
Query 58 LRRSYSFPADMWSLGVLVFVMLYGRYPFQGS 88
+ + Y DMW+ GV++++ML+GRYPF+G+
Sbjct 206 ISKHYGTGVDMWAFGVMIYLMLFGRYPFEGA 236
> tgo:TGME49_116150 protein kinase domain-containing protein (EC:2.7.11.1);
K08269 unc51-like kinase [EC:2.7.11.1]
Length=1463
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 29/68 (42%), Positives = 42/68 (61%), Gaps = 8/68 (11%)
Query 35 VLEYGFLHLMQ-------ICGSPHYLSPEILRRSY-SFPADMWSLGVLVFVMLYGRYPFQ 86
+ ++GF +Q ICGSP Y++PEIL+ Y AD+WS+G + F ML+GR PF
Sbjct 236 IADFGFARSLQPWDLAATICGSPLYMAPEILQHQYYDAKADLWSVGAIFFEMLHGRPPFS 295
Query 87 GSSSLDVL 94
G + L +L
Sbjct 296 GQNPLQLL 303
> ath:AT1G49180 protein kinase family protein
Length=408
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 24/54 (44%), Positives = 42/54 (77%), Gaps = 1/54 (1%)
Query 42 HLMQICGSPHYLSPEILR-RSYSFPADMWSLGVLVFVMLYGRYPFQGSSSLDVL 94
+L +CGSP Y++PE+L+ + Y+ ADMWS+G ++F +L+G PF+G++++ VL
Sbjct 162 YLETVCGSPFYMAPEVLQFQRYNEKADMWSVGAILFELLHGYPPFRGNNNVQVL 215
> dre:558848 ulk1b, ulk1; unc-51-like kinase 1b (C. elegans);
K08269 unc51-like kinase [EC:2.7.11.1]
Length=974
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 28/69 (40%), Positives = 43/69 (62%), Gaps = 8/69 (11%)
Query 35 VLEYGFLHLMQ-------ICGSPHYLSPE-ILRRSYSFPADMWSLGVLVFVMLYGRYPFQ 86
+ ++GF +Q +CGSP Y++PE I+ ++Y AD+WS+G +VF L G+ PFQ
Sbjct 132 IADFGFARYLQNNMMAATLCGSPMYMAPEVIMSQNYDAKADLWSIGTIVFQCLTGKAPFQ 191
Query 87 GSSSLDVLL 95
SS D+ L
Sbjct 192 ASSPQDLRL 200
> cel:T01H8.1 rskn-1; RSK-pNinety (RSK-p90 kinase) homolog family
member (rskn-1); K04373 p90 ribosomal S6 kinase [EC:2.7.11.1]
Length=804
Score = 58.2 bits (139), Expect = 6e-09, Method: Composition-based stats.
Identities = 25/49 (51%), Positives = 33/49 (67%), Gaps = 1/49 (2%)
Query 47 CGSPHYLSPEIL-RRSYSFPADMWSLGVLVFVMLYGRYPFQGSSSLDVL 94
CG+ Y++PE++ RR +S AD WSLGVL+F ML G PFQG D +
Sbjct 259 CGTVEYMAPEVINRRGHSMAADFWSLGVLMFEMLTGHLPFQGRDRNDTM 307
Score = 45.8 bits (107), Expect = 3e-05, Method: Composition-based stats.
Identities = 29/90 (32%), Positives = 45/90 (50%), Gaps = 9/90 (10%)
Query 5 YLYERYGIGNNNTACTPPFATSCRPRTSDTVLEYGFLH--------LMQICGSPHYLSPE 56
YL+ + + TA FA +S ++++GF LM C + +++PE
Sbjct 556 YLHSQQVAHRDLTAANILFALKDGDPSSLRIVDFGFAKQSRAENGMLMTPCYTAQFVAPE 615
Query 57 ILRRS-YSFPADMWSLGVLVFVMLYGRYPF 85
+LR+ Y D+WSLGVL+ ML G PF
Sbjct 616 VLRKQGYDRSCDVWSLGVLLHTMLTGCTPF 645
> tgo:TGME49_055710 calcium-dependent protein kinase, putative
(EC:2.7.11.17)
Length=361
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 21/40 (52%), Positives = 31/40 (77%), Gaps = 0/40 (0%)
Query 46 ICGSPHYLSPEILRRSYSFPADMWSLGVLVFVMLYGRYPF 85
ICGSP Y++PE+ ++Y D+WSLG++V+ MLYG +PF
Sbjct 240 ICGSPRYIAPEVFEQAYDEKCDLWSLGIVVYAMLYGTHPF 279
> ath:AT3G53930 protein kinase family protein; K08269 unc51-like
kinase [EC:2.7.11.1]
Length=712
Score = 57.8 bits (138), Expect = 9e-09, Method: Composition-based stats.
Identities = 25/68 (36%), Positives = 44/68 (64%), Gaps = 8/68 (11%)
Query 35 VLEYGFLHLMQ-------ICGSPHYLSPEILR-RSYSFPADMWSLGVLVFVMLYGRYPFQ 86
+ ++GF +Q +CGSP Y++PEI++ + Y AD+WS+G ++F ++ GR PF
Sbjct 161 IADFGFARSLQPRGLAETLCGSPLYMAPEIMQLQKYDAKADLWSVGAILFQLVTGRTPFT 220
Query 87 GSSSLDVL 94
G+S + +L
Sbjct 221 GNSQIQLL 228
> tpv:TP02_0399 calcium-dependent protein kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=844
Score = 57.8 bits (138), Expect = 9e-09, Method: Composition-based stats.
Identities = 23/74 (31%), Positives = 46/74 (62%), Gaps = 7/74 (9%)
Query 28 RPRTSDTVLEYGFL-------HLMQICGSPHYLSPEILRRSYSFPADMWSLGVLVFVMLY 80
+P ++ ++++GF + G+P+Y++PE+L +Y D+WS GV++F++L
Sbjct 532 KPSSTIKIIDWGFATKCYRTHKFSSLVGTPYYVAPEVLLGNYDKACDIWSAGVILFILLV 591
Query 81 GRYPFQGSSSLDVL 94
G PF GS++ ++L
Sbjct 592 GYPPFHGSNNTEIL 605
> tgo:TGME49_054190 protein kinase, putative (EC:2.7.11.1)
Length=338
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 44/64 (68%), Gaps = 9/64 (14%)
Query 34 TVLEYGFLHLMQI-------CGSPHYLSPEIL-RRSYS-FPADMWSLGVLVFVMLYGRYP 84
V+++GF +++ CG+P Y+ PEIL ++ Y F AD+WSLGV+++ ML+G YP
Sbjct 123 KVIDFGFASIVKNDARCRMRCGTPSYMPPEILLKKEYDGFAADIWSLGVVLYAMLHGGYP 182
Query 85 FQGS 88
F+G+
Sbjct 183 FRGN 186
> ath:AT5G24430 calcium-dependent protein kinase, putative / CDPK,
putative
Length=594
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 23/47 (48%), Positives = 34/47 (72%), Gaps = 0/47 (0%)
Query 43 LMQICGSPHYLSPEILRRSYSFPADMWSLGVLVFVMLYGRYPFQGSS 89
L + GS +Y++PE+L RSYS ADMWS+GV+ +++L G PF G +
Sbjct 305 LNDVVGSAYYVAPEVLHRSYSTEADMWSIGVISYILLCGSRPFYGRT 351
> ath:AT2G37840 protein kinase family protein; K08269 unc51-like
kinase [EC:2.7.11.1]
Length=733
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 25/68 (36%), Positives = 44/68 (64%), Gaps = 8/68 (11%)
Query 35 VLEYGFLHLMQ-------ICGSPHYLSPEILR-RSYSFPADMWSLGVLVFVMLYGRYPFQ 86
+ ++GF +Q +CGSP Y++PEI++ + Y AD+WS+G ++F ++ GR PF
Sbjct 153 IADFGFARSLQPRGLAETLCGSPLYMAPEIMQLQKYDAKADLWSVGAILFQLVTGRTPFT 212
Query 87 GSSSLDVL 94
G+S + +L
Sbjct 213 GNSQIQLL 220
> dre:100150844 protein kinase C, delta-like; K06068 novel protein
kinase C [EC:2.7.11.13]
Length=683
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 24/42 (57%), Positives = 31/42 (73%), Gaps = 1/42 (2%)
Query 47 CGSPHYLSPEILR-RSYSFPADMWSLGVLVFVMLYGRYPFQG 87
CG+P Y++PEIL + YSF D WS GVL++ ML G+ PFQG
Sbjct 516 CGTPDYIAPEILLGQKYSFSVDWWSFGVLLYEMLIGQSPFQG 557
> bbo:BBOV_III005470 17.m07489; protein kinase domain containing
protein; K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=755
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 27/72 (37%), Positives = 39/72 (54%), Gaps = 10/72 (13%)
Query 23 FATSCRPRTSDTVLEYGFLHLMQICGSPHYLSPEILRRSYSFPADMWSLGVLVFVMLYGR 82
FAT C Y + G+P+Y++PE+L SY DMWS GV++F+ML G
Sbjct 444 FATKC----------YKTHKFTSLVGTPYYVAPEVLIGSYDKSCDMWSAGVIMFIMLVGY 493
Query 83 YPFQGSSSLDVL 94
PF G+ + +L
Sbjct 494 PPFHGNDNATIL 505
> dre:117507 prkci, aPKC[l], hal, has; protein kinase C, iota
(EC:2.7.11.13); K06069 atypical protein kinase C [EC:2.7.11.13]
Length=588
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 25/47 (53%), Positives = 32/47 (68%), Gaps = 1/47 (2%)
Query 47 CGSPHYLSPEILR-RSYSFPADMWSLGVLVFVMLYGRYPFQGSSSLD 92
CG+P+Y++PEILR Y F D W+LGVL+F M+ GR PF S D
Sbjct 406 CGTPNYIAPEILRGEDYGFSVDWWALGVLMFEMMAGRSPFDIVGSSD 452
> mmu:18762 Prkcz, AI098070, C80388, Pkcz, R74924, aPKCzeta, zetaPKC;
protein kinase C, zeta (EC:2.7.11.13); K06069 atypical
protein kinase C [EC:2.7.11.13]
Length=409
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 23/40 (57%), Positives = 30/40 (75%), Gaps = 1/40 (2%)
Query 47 CGSPHYLSPEILR-RSYSFPADMWSLGVLVFVMLYGRYPF 85
CG+P+Y++PEILR Y F D W+LGVL+F M+ GR PF
Sbjct 229 CGTPNYIAPEILRGEEYGFSVDWWALGVLMFEMMAGRSPF 268
> hsa:5590 PRKCZ, PKC-ZETA, PKC2; protein kinase C, zeta (EC:2.7.11.13);
K06069 atypical protein kinase C [EC:2.7.11.13]
Length=409
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 23/40 (57%), Positives = 30/40 (75%), Gaps = 1/40 (2%)
Query 47 CGSPHYLSPEILR-RSYSFPADMWSLGVLVFVMLYGRYPF 85
CG+P+Y++PEILR Y F D W+LGVL+F M+ GR PF
Sbjct 229 CGTPNYIAPEILRGEEYGFSVDWWALGVLMFEMMAGRSPF 268
> hsa:5584 PRKCI, DXS1179E, MGC26534, PKCI, nPKC-iota; protein
kinase C, iota (EC:2.7.11.13); K06069 atypical protein kinase
C [EC:2.7.11.13]
Length=596
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 25/47 (53%), Positives = 32/47 (68%), Gaps = 1/47 (2%)
Query 47 CGSPHYLSPEILR-RSYSFPADMWSLGVLVFVMLYGRYPFQGSSSLD 92
CG+P+Y++PEILR Y F D W+LGVL+F M+ GR PF S D
Sbjct 414 CGTPNYIAPEILRGEDYGFSVDWWALGVLMFEMMAGRSPFDIVGSSD 460
> cel:C10C6.1 kin-4; protein KINase family member (kin-4); K08789
microtubule-associated serine/threonine kinase [EC:2.7.11.1]
Length=1565
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 25/58 (43%), Positives = 36/58 (62%), Gaps = 1/58 (1%)
Query 33 DTVLEYGFLHLMQICGSPHYLSPE-ILRRSYSFPADMWSLGVLVFVMLYGRYPFQGSS 89
D V+E Q+CG+P Y++PE ILRR Y P D W+LG++++ L G PF G +
Sbjct 714 DAVVETQQFQDKQLCGTPEYIAPEVILRRGYGKPVDWWALGIILYEFLVGIVPFFGET 771
> ath:AT2G46700 calcium-dependent protein kinase, putative / CDPK,
putative
Length=371
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 25/68 (36%), Positives = 42/68 (61%), Gaps = 7/68 (10%)
Query 25 TSCRPRTSDTVLEYGFLHLMQ-------ICGSPHYLSPEILRRSYSFPADMWSLGVLVFV 77
TS R + ++++G ++ I GS +Y++PE+L RSYS AD+WS+GV+ ++
Sbjct 56 TSSREDSDLKLIDFGLSDFIRPDERLNDIVGSAYYVAPEVLHRSYSLEADIWSIGVITYI 115
Query 78 MLYGRYPF 85
+L G PF
Sbjct 116 LLCGSRPF 123
> ath:AT2G35890 CPK25; CPK25; ATP binding / calmodulin-dependent
protein kinase/ kinase/ protein kinase/ protein serine/threonine
kinase
Length=520
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 20/49 (40%), Positives = 35/49 (71%), Gaps = 0/49 (0%)
Query 46 ICGSPHYLSPEILRRSYSFPADMWSLGVLVFVMLYGRYPFQGSSSLDVL 94
+ GSP+Y++PE+L ++Y AD+WS GV+++V+L G PF G + ++
Sbjct 293 VVGSPYYIAPEVLNKNYGPEADIWSAGVMIYVLLSGSAPFWGETEEEIF 341
> dre:555737 prkcz, pkcz, si:ch211-150o23.4; protein kinase C,
zeta (EC:2.7.11.1); K06069 atypical protein kinase C [EC:2.7.11.13]
Length=599
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 23/40 (57%), Positives = 30/40 (75%), Gaps = 1/40 (2%)
Query 47 CGSPHYLSPEILR-RSYSFPADMWSLGVLVFVMLYGRYPF 85
CG+P+Y++PEILR Y F D W+LGVL+F M+ GR PF
Sbjct 419 CGTPNYIAPEILRGEDYGFSVDWWALGVLMFEMMAGRSPF 458
> cel:F32D8.1 hypothetical protein
Length=333
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 24/46 (52%), Positives = 34/46 (73%), Gaps = 1/46 (2%)
Query 43 LMQICGSPHYLSPEILRRS-YSFPADMWSLGVLVFVMLYGRYPFQG 87
L +ICG+P Y +PE+LR + YS D+WSLGV+++VML G PF+
Sbjct 221 LYRICGTPTYCAPEVLRETGYSTRCDIWSLGVVLYVMLVGYAPFRA 266
> dre:334571 prkcd, fb09d10, fv43b11, wu:fb09d10, wu:fv43b11,
zgc:56558; protein kinase C, delta (EC:2.7.11.13); K06068 novel
protein kinase C [EC:2.7.11.13]
Length=684
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 24/42 (57%), Positives = 30/42 (71%), Gaps = 1/42 (2%)
Query 47 CGSPHYLSPEILR-RSYSFPADMWSLGVLVFVMLYGRYPFQG 87
CG+P Y++PEIL + YSF D WS GVLV+ ML G+ PF G
Sbjct 517 CGTPDYIAPEILLGQQYSFSVDWWSFGVLVYEMLIGQSPFHG 558
> mmu:18759 Prkci, 2310021H13Rik, AI427505, KIAA4165, PKClambda,
Pkci, Pkcl, Prkcl, aPKClambda, mKIAA4165; protein kinase
C, iota (EC:2.7.11.13); K06069 atypical protein kinase C [EC:2.7.11.13]
Length=595
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 25/47 (53%), Positives = 32/47 (68%), Gaps = 1/47 (2%)
Query 47 CGSPHYLSPEILR-RSYSFPADMWSLGVLVFVMLYGRYPFQGSSSLD 92
CG+P+Y++PEILR Y F D W+LGVL+F M+ GR PF S D
Sbjct 413 CGTPNYIAPEILRGEDYGFSVDWWALGVLMFEMMAGRSPFDIVGSSD 459
> cpv:cgd2_1300 calcium/calmodulin dependent protein kinase with
a kinase domain and 4 calmodulin like EF hands ; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=677
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 24/73 (32%), Positives = 44/73 (60%), Gaps = 7/73 (9%)
Query 29 PRTSDTVLEYGFL-------HLMQICGSPHYLSPEILRRSYSFPADMWSLGVLVFVMLYG 81
P + V+++GF + G+P+Y++PE+L SYS D+WS GV+++++L G
Sbjct 318 PNSLLKVIDWGFAAKCPKTHKFTSVVGTPYYVAPEVLYGSYSKLCDLWSAGVILYILLCG 377
Query 82 RYPFQGSSSLDVL 94
PF G ++++L
Sbjct 378 YPPFHGKDNVEIL 390
> cel:F09E5.1 pkc-3; Protein Kinase C family member (pkc-3); K06069
atypical protein kinase C [EC:2.7.11.13]
Length=597
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 23/40 (57%), Positives = 30/40 (75%), Gaps = 1/40 (2%)
Query 47 CGSPHYLSPEILR-RSYSFPADMWSLGVLVFVMLYGRYPF 85
CG+P+Y++PEILR Y F D W+LGVL+F M+ GR PF
Sbjct 413 CGTPNYIAPEILRGDEYGFSVDWWALGVLMFEMMAGRSPF 452
> mmu:22241 Ulk1, AU041434, Unc51.1, mKIAA0722; Unc-51 like kinase
1 (C. elegans) (EC:2.7.11.1); K08269 unc51-like kinase
[EC:2.7.11.1]
Length=1051
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 27/69 (39%), Positives = 42/69 (60%), Gaps = 8/69 (11%)
Query 35 VLEYGFLHLMQ-------ICGSPHYLSPE-ILRRSYSFPADMWSLGVLVFVMLYGRYPFQ 86
+ ++GF +Q +CGSP Y++PE I+ + Y AD+WS+G +V+ L G+ PFQ
Sbjct 163 IADFGFARYLQSNMMAATLCGSPMYMAPEVIMSQHYDGKADLWSIGTIVYQCLTGKAPFQ 222
Query 87 GSSSLDVLL 95
SS D+ L
Sbjct 223 ASSPQDLRL 231
> hsa:8408 ULK1, ATG1, ATG1A, FLJ38455, FLJ46475, UNC51, Unc51.1;
unc-51-like kinase 1 (C. elegans) (EC:2.7.11.1); K08269
unc51-like kinase [EC:2.7.11.1]
Length=1050
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 27/70 (38%), Positives = 42/70 (60%), Gaps = 8/70 (11%)
Query 34 TVLEYGFLHLMQ-------ICGSPHYLSPE-ILRRSYSFPADMWSLGVLVFVMLYGRYPF 85
+ ++GF +Q +CGSP Y++PE I+ + Y AD+WS+G +V+ L G+ PF
Sbjct 162 KIADFGFARYLQSNMMAATLCGSPMYMAPEVIMSQHYDGKADLWSIGTIVYQCLTGKAPF 221
Query 86 QGSSSLDVLL 95
Q SS D+ L
Sbjct 222 QASSPQDLRL 231
> ath:AT5G01810 CIPK15; CIPK15 (CBL-INTERACTING PROTEIN KINASE
15); kinase/ protein kinase
Length=421
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 26/64 (40%), Positives = 43/64 (67%), Gaps = 4/64 (6%)
Query 32 SDTVLEYGFLHLMQICGSPHYLSPEILRRSY--SFPADMWSLGVLVFVMLYGRYPFQGSS 89
SD+ + G LH CG+P Y++PE++ R+ F AD+WS GV++FV+L G PF+ S+
Sbjct 159 SDSRRQDGLLH--TTCGTPAYVAPEVISRNGYDGFKADVWSCGVILFVLLAGYLPFRDSN 216
Query 90 SLDV 93
+++
Sbjct 217 LMEL 220
> ath:AT3G50530 CRK; CRK (CDPK-related kinase); ATP binding /
calcium ion binding / calcium-dependent protein serine/threonine
phosphatase/ kinase/ protein kinase/ protein serine/threonine
kinase; K00924 [EC:2.7.1.-]
Length=601
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 23/43 (53%), Positives = 33/43 (76%), Gaps = 0/43 (0%)
Query 43 LMQICGSPHYLSPEILRRSYSFPADMWSLGVLVFVMLYGRYPF 85
L I GS +Y++PE+L RSYS AD+WS+GV+V+++L G PF
Sbjct 310 LNDIVGSAYYVAPEVLHRSYSTEADIWSVGVIVYILLCGSRPF 352
> tgo:TGME49_015670 protein kinase, putative (EC:2.7.12.1 2.7.11.13
3.2.1.17)
Length=1745
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 34/83 (40%), Positives = 48/83 (57%), Gaps = 4/83 (4%)
Query 15 NNTACTPPFATSCRPRTSDTVLEYGFLHLMQICGSPHYLSPEILR-RSYSFPADMWSLGV 73
N TA +P +T P +S T + L + G+PHY++PE+L YSFPAD WS+GV
Sbjct 1187 NETAPSPAPST-LSPASSPTHISR--LEALSQVGTPHYMAPEVLAGLPYSFPADWWSVGV 1243
Query 74 LVFVMLYGRYPFQGSSSLDVLLA 96
++F +YG PF S LL+
Sbjct 1244 ILFECIYGGVPFNTPSYNPQLLS 1266
> ath:AT2G41860 CPK14; CPK14; ATP binding / calcium ion binding
/ calmodulin-dependent protein kinase/ kinase/ protein kinase/
protein serine/threonine kinase; K13412 calcium-dependent
protein kinase [EC:2.7.11.1]
Length=530
Score = 55.8 bits (133), Expect = 4e-08, Method: Composition-based stats.
Identities = 21/41 (51%), Positives = 32/41 (78%), Gaps = 0/41 (0%)
Query 45 QICGSPHYLSPEILRRSYSFPADMWSLGVLVFVMLYGRYPF 85
+I GSP+Y++PE+LRRSY D+WS GV+++++L G PF
Sbjct 214 EIVGSPYYMAPEVLRRSYGQEIDIWSAGVILYILLCGVPPF 254
> ath:AT1G74740 CPK30; CPK30 (CALCIUM-DEPENDENT PROTEIN KINASE
30); calmodulin-dependent protein kinase/ kinase/ protein kinase;
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=541
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 22/52 (42%), Positives = 36/52 (69%), Gaps = 0/52 (0%)
Query 45 QICGSPHYLSPEILRRSYSFPADMWSLGVLVFVMLYGRYPFQGSSSLDVLLA 96
+I GSP+Y++PE+L+R+Y D+WS GV+++++L G PF + V LA
Sbjct 219 EIVGSPYYMAPEVLKRNYGPEVDVWSAGVILYILLCGVPPFWAETEQGVALA 270
> ath:AT1G18890 ATCDPK1 (CALCIUM-DEPENDENT PROTEIN KINASE 1);
calmodulin-dependent protein kinase/ kinase/ protein kinase;
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=545
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 22/52 (42%), Positives = 35/52 (67%), Gaps = 0/52 (0%)
Query 45 QICGSPHYLSPEILRRSYSFPADMWSLGVLVFVMLYGRYPFQGSSSLDVLLA 96
+I GSP+Y++PE+L+R Y D+WS GV+++++L G PF + V LA
Sbjct 223 EIVGSPYYMAPEVLKRDYGPGVDVWSAGVIIYILLCGVPPFWAETEQGVALA 274
> ath:AT3G49370 calcium-dependent protein kinase, putative / CDPK,
putative; K00924 [EC:2.7.1.-]
Length=594
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 22/47 (46%), Positives = 34/47 (72%), Gaps = 0/47 (0%)
Query 43 LMQICGSPHYLSPEILRRSYSFPADMWSLGVLVFVMLYGRYPFQGSS 89
L + GS +Y++PE+L RSYS AD+WS+GV+ +++L G PF G +
Sbjct 304 LNDVVGSAYYVAPEVLHRSYSTEADIWSIGVISYILLCGSRPFYGRT 350
> mmu:18753 Prkcd, AI385711, D14Ertd420e, PKC[d], PKCdelta, Pkcd;
protein kinase C, delta (EC:2.7.11.13); K06068 novel protein
kinase C [EC:2.7.11.13]
Length=674
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 23/42 (54%), Positives = 30/42 (71%), Gaps = 1/42 (2%)
Query 47 CGSPHYLSPEILR-RSYSFPADMWSLGVLVFVMLYGRYPFQG 87
CG+P Y++PEIL+ YSF D WS GVL++ ML G+ PF G
Sbjct 507 CGTPDYIAPEILQGLKYSFSVDWWSFGVLLYEMLIGQSPFHG 548
> xla:100037118 hypothetical protein LOC100037118
Length=358
Score = 55.1 bits (131), Expect = 6e-08, Method: Composition-based stats.
Identities = 28/67 (41%), Positives = 39/67 (58%), Gaps = 6/67 (8%)
Query 30 RTSDTVLEYGFLHLMQICGSPHYLSPEILR-RSYSFPADMWSLGVLVFVMLYGRYPFQGS 88
R DT+ E + +I G+P Y++PE+L S DMWS+GVL +VML G PFQG
Sbjct 175 RRVDTIKE-----VREILGTPEYVAPEVLNYEPISTATDMWSVGVLAYVMLTGVSPFQGE 229
Query 89 SSLDVLL 95
+ + L
Sbjct 230 TKQETFL 236
> dre:556339 protein kinase, cGMP-dependent, type II-like; K07376
protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=730
Score = 55.1 bits (131), Expect = 6e-08, Method: Composition-based stats.
Identities = 22/48 (45%), Positives = 33/48 (68%), Gaps = 1/48 (2%)
Query 47 CGSPHYLSPE-ILRRSYSFPADMWSLGVLVFVMLYGRYPFQGSSSLDV 93
CG+P Y++PE I+ + + F AD WSLG+L+F +L G PF GS + +
Sbjct 579 CGTPEYVAPEVIMNKGHDFGADCWSLGILIFELLIGSPPFTGSDPIRI 626
> pfa:PF13_0211 calcium-dependent protein kinase, putative (EC:2.7.11.17);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=568
Score = 55.1 bits (131), Expect = 6e-08, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 43/67 (64%), Gaps = 7/67 (10%)
Query 35 VLEYGFL-------HLMQICGSPHYLSPEILRRSYSFPADMWSLGVLVFVMLYGRYPFQG 87
++++GF +L + G+P+Y++PEILR Y D+WS GV+++++L G PF G
Sbjct 264 IIDWGFASKCMNNHNLKSVVGTPYYIAPEILRGKYDKRCDIWSSGVIMYILLCGYPPFNG 323
Query 88 SSSLDVL 94
++ ++L
Sbjct 324 KNNDEIL 330
> tgo:TGME49_006590 calcium-dependent protein kinase, putative
(EC:2.7.11.17 3.4.22.53); K13412 calcium-dependent protein
kinase [EC:2.7.11.1]
Length=761
Score = 55.1 bits (131), Expect = 6e-08, Method: Composition-based stats.
Identities = 25/72 (34%), Positives = 41/72 (56%), Gaps = 8/72 (11%)
Query 23 FATSCRPRTSDTVLEYGFLHLMQICGSPHYLSPEILRRSYSFPADMWSLGVLVFVMLYGR 82
F SCR + + V G+P+Y++P++L+ Y F D WSLGV+++++L G
Sbjct 367 FGLSCRFKKGEFVSTKA--------GTPYYVAPQVLQGKYDFRCDAWSLGVILYILLCGY 418
Query 83 YPFQGSSSLDVL 94
PF G + +VL
Sbjct 419 PPFYGETDAEVL 430
> dre:564859 stk40, cb988, fd99h11, wu:fd99h11; serine/threonine
kinase 40
Length=437
Score = 54.7 bits (130), Expect = 6e-08, Method: Composition-based stats.
Identities = 24/43 (55%), Positives = 33/43 (76%), Gaps = 2/43 (4%)
Query 48 GSPHYLSPEILR-RSY-SFPADMWSLGVLVFVMLYGRYPFQGS 88
GSP Y+SP++L R Y P+DMW+LGV++F MLYG++PF S
Sbjct 236 GSPAYISPDVLSGRPYRGKPSDMWALGVVLFTMLYGQFPFYDS 278
> mmu:74178 Stk40, 2310004N11Rik, AI428792; serine/threonine kinase
40 (EC:2.7.11.1)
Length=449
Score = 54.7 bits (130), Expect = 6e-08, Method: Composition-based stats.
Identities = 24/43 (55%), Positives = 33/43 (76%), Gaps = 2/43 (4%)
Query 48 GSPHYLSPEILR-RSY-SFPADMWSLGVLVFVMLYGRYPFQGS 88
GSP Y+SP++L R Y P+DMW+LGV++F MLYG++PF S
Sbjct 249 GSPAYISPDVLSGRPYRGKPSDMWALGVVLFTMLYGQFPFYDS 291
> hsa:83931 STK40, MGC4796, RP11-268J15.4, SHIK, SgK495; serine/threonine
kinase 40 (EC:2.7.11.1)
Length=435
Score = 54.7 bits (130), Expect = 6e-08, Method: Composition-based stats.
Identities = 24/43 (55%), Positives = 33/43 (76%), Gaps = 2/43 (4%)
Query 48 GSPHYLSPEILR-RSY-SFPADMWSLGVLVFVMLYGRYPFQGS 88
GSP Y+SP++L R Y P+DMW+LGV++F MLYG++PF S
Sbjct 235 GSPAYISPDVLSGRPYRGKPSDMWALGVVLFTMLYGQFPFYDS 277
> sce:YMR001C CDC5, MSD2, PKX2; Cdc5p (EC:2.7.11.21); K06660 cell
cycle serine/threonine-protein kinase CDC5/MSD2 [EC:2.7.11.21]
Length=705
Score = 54.7 bits (130), Expect = 7e-08, Method: Composition-based stats.
Identities = 21/45 (46%), Positives = 33/45 (73%), Gaps = 3/45 (6%)
Query 46 ICGSPHYLSPEIL---RRSYSFPADMWSLGVLVFVMLYGRYPFQG 87
ICG+P+Y++PE+L +SF D+WSLGV+++ +L G+ PFQ
Sbjct 239 ICGTPNYIAPEVLMGKHSGHSFEVDIWSLGVMLYALLIGKPPFQA 283
> ath:AT3G56760 calcium-dependent protein kinase, putative / CDPK,
putative; K00924 [EC:2.7.1.-]
Length=577
Score = 54.7 bits (130), Expect = 7e-08, Method: Composition-based stats.
Identities = 23/54 (42%), Positives = 34/54 (62%), Gaps = 0/54 (0%)
Query 43 LMQICGSPHYLSPEILRRSYSFPADMWSLGVLVFVMLYGRYPFQGSSSLDVLLA 96
L I GS +Y++PE+L R+Y ADMWS+GV+ +++L G PF S + A
Sbjct 286 LNDIVGSAYYVAPEVLHRTYGTEADMWSIGVIAYILLCGSRPFWARSESGIFRA 339
> pfa:PFC0420w PfCDPK3; calcium-dependent protein kinase-3 (EC:2.7.11.17);
K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=562
Score = 54.7 bits (130), Expect = 7e-08, Method: Composition-based stats.
Identities = 21/47 (44%), Positives = 34/47 (72%), Gaps = 0/47 (0%)
Query 48 GSPHYLSPEILRRSYSFPADMWSLGVLVFVMLYGRYPFQGSSSLDVL 94
G+P+Y++P++L SY++ DMWS GVL +++L G PF G S ++L
Sbjct 278 GTPYYVAPQVLTGSYNYKCDMWSSGVLFYILLCGYPPFFGESDHEIL 324
> ath:AT5G19450 CDPK19; CDPK19 (CALCIUM-DEPENDENT PROTEIN KINASE
19); ATP binding / calcium ion binding / calmodulin-dependent
protein kinase/ kinase/ protein kinase/ protein serine/threonine
kinase; K13412 calcium-dependent protein kinase [EC:2.7.11.1]
Length=533
Score = 54.7 bits (130), Expect = 8e-08, Method: Composition-based stats.
Identities = 20/41 (48%), Positives = 32/41 (78%), Gaps = 0/41 (0%)
Query 45 QICGSPHYLSPEILRRSYSFPADMWSLGVLVFVMLYGRYPF 85
+I GSP+Y++PE+LRR+Y D+WS GV+++++L G PF
Sbjct 217 EIVGSPYYMAPEVLRRNYGPEVDIWSAGVILYILLCGVPPF 257
> xla:398681 stk40; serine/threonine kinase 40 (EC:2.7.11.1)
Length=443
Score = 54.7 bits (130), Expect = 8e-08, Method: Composition-based stats.
Identities = 24/43 (55%), Positives = 33/43 (76%), Gaps = 2/43 (4%)
Query 48 GSPHYLSPEILR-RSY-SFPADMWSLGVLVFVMLYGRYPFQGS 88
GSP Y+SP++L R Y P+DMW+LGV++F MLYG++PF S
Sbjct 235 GSPAYISPDVLSGRPYRGKPSDMWALGVVLFTMLYGQFPFYDS 277
Lambda K H
0.326 0.142 0.465
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2036602604
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40