bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1637_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
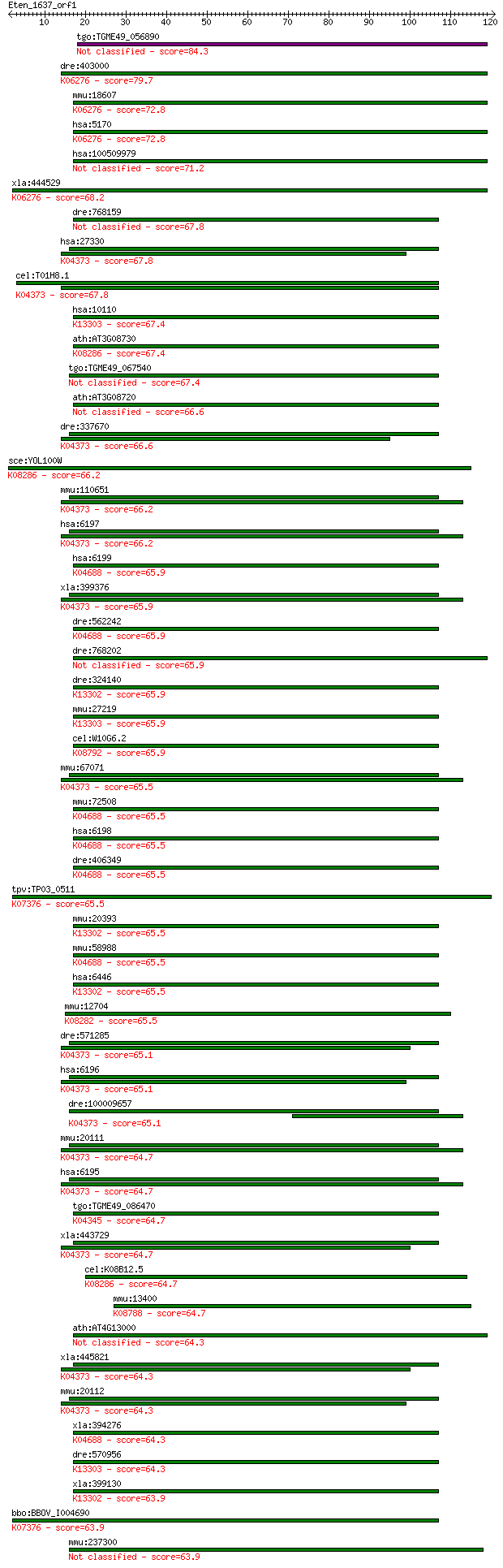
tgo:TGME49_056890 protein kinase domain-containing protein (EC... 84.3 9e-17
dre:403000 pdpk1b, MGC77318, zgc:77318; 3-phosphoinositide dep... 79.7 2e-15
mmu:18607 Pdpk1, Pdk1; 3-phosphoinositide dependent protein ki... 72.8 3e-13
hsa:5170 PDPK1, MGC20087, MGC35290, PDK1, PRO0461; 3-phosphoin... 72.8 3e-13
hsa:100509979 putative 3-phosphoinositide-dependent protein ki... 71.2 7e-13
xla:444529 pdpk1, MGC82080; 3-phosphoinositide dependent prote... 68.2 7e-12
dre:768159 wu:fk56c04, zgc:154030; si:ch211-195b13.1 67.8 8e-12
hsa:27330 RPS6KA6, PP90RSK4, RSK4; ribosomal protein S6 kinase... 67.8 9e-12
cel:T01H8.1 rskn-1; RSK-pNinety (RSK-p90 kinase) homolog famil... 67.8 9e-12
hsa:10110 SGK2, H-SGK2, dJ138B7.2; serum/glucocorticoid regula... 67.4 1e-11
ath:AT3G08730 PK1; PK1 (PROTEIN-SERINE KINASE 1); kinase/ prot... 67.4 1e-11
tgo:TGME49_067540 protein kinase, putative (EC:4.1.1.70 2.7.11... 67.4 1e-11
ath:AT3G08720 S6K2; S6K2 (ARABIDOPSIS THALIANA SERINE/THREONIN... 66.6 2e-11
dre:337670 rps6kal, fj94b02, rps6ka6, wu:fj94b02, zgc:66139; r... 66.6 2e-11
sce:YOL100W PKH2; Pkh2p (EC:2.7.11.1); K08286 protein-serine/t... 66.2 2e-11
mmu:110651 Rps6ka3, MPK-9, Rsk2, S6K-alpha3, pp90RSK2; ribosom... 66.2 2e-11
hsa:6197 RPS6KA3, CLS, HU-3, ISPK-1, MAPKAPK1B, MRX19, RSK, RS... 66.2 3e-11
hsa:6199 RPS6KB2, KLS, P70-beta, P70-beta-1, P70-beta-2, S6K-b... 65.9 3e-11
xla:399376 rps6ka3, MGC82193, S6KII, p90, rsk, rsk2; ribosomal... 65.9 3e-11
dre:562242 ribosomal protein S6 kinase b, polypeptide 1-like; ... 65.9 3e-11
dre:768202 pdpk1a, zgc:153787; 3-phosphoinositide dependent pr... 65.9 3e-11
dre:324140 sgk1, cb1083, sgk, wu:fc20a09; serum/glucocorticoid... 65.9 3e-11
mmu:27219 Sgk2, AI098171, AW146006, Sgkl; serum/glucocorticoid... 65.9 3e-11
cel:W10G6.2 sgk-1; Serum- and Glucocorticoid- inducible kinase... 65.9 3e-11
mmu:67071 Rps6ka6, 2610524K04Rik, RSK4; ribosomal protein S6 k... 65.5 4e-11
mmu:72508 Rps6kb1, 2610318I15Rik, 4732464A07Rik, 70kDa, AA9597... 65.5 4e-11
hsa:6198 RPS6KB1, PS6K, S6K, S6K1, STK14A, p70(S6K)-alpha, p70... 65.5 4e-11
dre:406349 rps6kb1b, fc51h01, rps6kb1, wu:fc51h01, zgc:55713; ... 65.5 4e-11
tpv:TP03_0511 cGMP-dependent protein kinase (EC:2.7.1.37); K07... 65.5 4e-11
mmu:20393 Sgk1, Sgk; serum/glucocorticoid regulated kinase 1 (... 65.5 4e-11
mmu:58988 Rps6kb2, 70kDa, S6K2; ribosomal protein S6 kinase, p... 65.5 4e-11
hsa:6446 SGK1, SGK; serum/glucocorticoid regulated kinase 1 (E... 65.5 4e-11
mmu:12704 Cit, C030025P15Rik, CRIK, CRIK-SK, Cit-k; citron (EC... 65.5 4e-11
dre:571285 ribosomal protein S6 kinase, polypeptide 2-like; K0... 65.1 5e-11
hsa:6196 RPS6KA2, HU-2, MAPKAPK1C, RSK, RSK3, S6K-alpha, S6K-a... 65.1 5e-11
dre:100009657 rps6ka3b, zgc:158386; ribosomal protein S6 kinas... 65.1 5e-11
mmu:20111 Rps6ka1, Rsk1, p90rsk, rsk; ribosomal protein S6 kin... 64.7 6e-11
hsa:6195 RPS6KA1, HU-1, MAPKAPK1A, RSK, RSK1; ribosomal protei... 64.7 6e-11
tgo:TGME49_086470 cAMP-dependent protein kinase catalytic subu... 64.7 6e-11
xla:443729 rps6ka6, MGC79981, rps6ka, rps6ka1; ribosomal prote... 64.7 7e-11
cel:K08B12.5 mrck-1; Myotonic dystrophy-Related, Cdc42-binding... 64.7 7e-11
mmu:13400 Dmpk, DM, DMK, Dm15, MDPK, MT-PK; dystrophia myotoni... 64.7 7e-11
ath:AT4G13000 protein kinase family protein 64.3 8e-11
xla:445821 rps6ka1, MGC81220, hu-1, mapkapk1a, rsk, rsk1; ribo... 64.3 8e-11
mmu:20112 Rps6ka2, 90kDa, D17Wsu134e, Rps6ka-rs1, Rsk3, p90rsk... 64.3 8e-11
xla:394276 rps6kb1, p70-alpha, p70-s6k, p70s6k, ps6k, rps6kb1-... 64.3 9e-11
dre:570956 sgk2a, fm82b06, sgk2, wu:fm82b06, zgc:172111, zgc:1... 64.3 9e-11
xla:399130 sgk1-a, Sgk, sgk-A, sgk1; serum/glucocorticoid regu... 63.9 1e-10
bbo:BBOV_I004690 19.m02237; cGMP dependent protein kinase (EC:... 63.9 1e-10
mmu:237300 Gm4922, 4933423E17, EG237300; predicted gene 4922 (... 63.9 1e-10
> tgo:TGME49_056890 protein kinase domain-containing protein (EC:2.7.11.1
2.7.11.11)
Length=1590
Score = 84.3 bits (207), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 45/111 (40%), Positives = 71/111 (63%), Gaps = 11/111 (9%)
Query 18 PSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAG----------VSLSIGAARLW 67
P + L +Q + L+ V++ +G L+++LRR+S A + LS AA+ W
Sbjct 313 PFIPRLLHAFQDAQRLFLVMDLSGFSNLQEILRRVSPAHADSESSPSCLIQLSELAAQTW 372
Query 68 AAEIVAALATLRQKGVLHRDIRPENIVVSEQGHLTLVDFYSALHLVPGTQQ 118
AE+VAAL LR++GV+HRD++P+N+V+S +GHL +VDF +AL L+P Q
Sbjct 373 TAELVAALEWLRERGVVHRDLKPQNLVISPEGHLCVVDFGAAL-LLPAEDQ 422
> dre:403000 pdpk1b, MGC77318, zgc:77318; 3-phosphoinositide dependent
protein kinase-1b (EC:2.7.11.1); K06276 3-phosphoinositide
dependent protein kinase-1 [EC:2.7.11.1]
Length=537
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 41/105 (39%), Positives = 60/105 (57%), Gaps = 4/105 (3%)
Query 14 HSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVA 73
H HP L +Q EE LYF L+Y L +R++ S R ++AEIV
Sbjct 114 HLDHPFFVKLYFTFQDEEKLYFGLSYAKNGELLKYIRKIG----SFDETCTRFYSAEIVC 169
Query 74 ALATLRQKGVLHRDIRPENIVVSEQGHLTLVDFYSALHLVPGTQQ 118
AL L QKG++HRD++PENI++SE+ H+ + DF +A L ++Q
Sbjct 170 ALEYLHQKGIIHRDLKPENILLSEEMHIQITDFGTAKQLSSDSKQ 214
> mmu:18607 Pdpk1, Pdk1; 3-phosphoinositide dependent protein
kinase 1 (EC:2.7.11.1); K06276 3-phosphoinositide dependent
protein kinase-1 [EC:2.7.11.1]
Length=522
Score = 72.8 bits (177), Expect = 3e-13, Method: Composition-based stats.
Identities = 38/102 (37%), Positives = 58/102 (56%), Gaps = 4/102 (3%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP L +Q +E LYF L+Y L +R++ S R + AEIV+AL
Sbjct 142 HPFFVKLYFTFQDDEKLYFGLSYAKNGELLKYIRKIG----SFDETCTRFYTAEIVSALE 197
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDFYSALHLVPGTQQ 118
L KG++HRD++PENI+++E H+ + DF +A L P ++Q
Sbjct 198 YLHGKGIIHRDLKPENILLNEDMHIQITDFGTAKVLSPESKQ 239
> hsa:5170 PDPK1, MGC20087, MGC35290, PDK1, PRO0461; 3-phosphoinositide
dependent protein kinase-1 (EC:2.7.11.1); K06276 3-phosphoinositide
dependent protein kinase-1 [EC:2.7.11.1]
Length=556
Score = 72.8 bits (177), Expect = 3e-13, Method: Composition-based stats.
Identities = 38/102 (37%), Positives = 58/102 (56%), Gaps = 4/102 (3%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP L +Q +E LYF L+Y L +R++ S R + AEIV+AL
Sbjct 139 HPFFVKLYFTFQDDEKLYFGLSYAKNGELLKYIRKIG----SFDETCTRFYTAEIVSALE 194
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDFYSALHLVPGTQQ 118
L KG++HRD++PENI+++E H+ + DF +A L P ++Q
Sbjct 195 YLHGKGIIHRDLKPENILLNEDMHIQITDFGTAKVLSPESKQ 236
> hsa:100509979 putative 3-phosphoinositide-dependent protein
kinase 2-like
Length=442
Score = 71.2 bits (173), Expect = 7e-13, Method: Composition-based stats.
Identities = 38/102 (37%), Positives = 57/102 (55%), Gaps = 4/102 (3%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP L +QT LYF L+Y L +R++ S R + AEIV+AL
Sbjct 158 HPFFVKLYFTFQTTRRLYFGLSYAKNGELLKYIRKIG----SFDETCTRFYTAEIVSALE 213
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDFYSALHLVPGTQQ 118
L KG++HRD++PENI+++E H+ + DF +A L P ++Q
Sbjct 214 YLHGKGIIHRDLKPENILLNEDMHIQITDFGTAKVLSPESKQ 255
> xla:444529 pdpk1, MGC82080; 3-phosphoinositide dependent protein
kinase-1 (EC:2.7.11.1); K06276 3-phosphoinositide dependent
protein kinase-1 [EC:2.7.11.1]
Length=506
Score = 68.2 bits (165), Expect = 7e-12, Method: Composition-based stats.
Identities = 41/117 (35%), Positives = 60/117 (51%), Gaps = 10/117 (8%)
Query 2 EKETLLSLSLGGHSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSI 61
EKE + L H L +Q +E LYF L+Y L +R++ S
Sbjct 81 EKEVMSCLD------HSFFVKLYFTFQDDEKLYFGLSYAKNGELLKYIRKIG----SFDE 130
Query 62 GAARLWAAEIVAALATLRQKGVLHRDIRPENIVVSEQGHLTLVDFYSALHLVPGTQQ 118
R + AEIV AL L +KG++HRD++PENI++SE H+ + DF +A L +Q
Sbjct 131 TCTRFYTAEIVCALEYLHEKGIIHRDLKPENILLSEDMHIQITDFGTAKVLSSDIKQ 187
> dre:768159 wu:fk56c04, zgc:154030; si:ch211-195b13.1
Length=423
Score = 67.8 bits (164), Expect = 8e-12, Method: Composition-based stats.
Identities = 37/90 (41%), Positives = 53/90 (58%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + L +QT + LYFVL+Y V ++ L R V L AR +AAEI +AL
Sbjct 149 HPFLVGLHYSFQTTDKLYFVLDY---VNGGELFYHLQRERVFLE-PRARFYAAEIASALG 204
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L +++RD++PENI++ QGH+ L DF
Sbjct 205 YLHSLHIVYRDLKPENILLDSQGHIVLTDF 234
> hsa:27330 RPS6KA6, PP90RSK4, RSK4; ribosomal protein S6 kinase,
90kDa, polypeptide 6 (EC:2.7.11.1); K04373 p90 ribosomal
S6 kinase [EC:2.7.11.1]
Length=745
Score = 67.8 bits (164), Expect = 9e-12, Method: Composition-based stats.
Identities = 35/91 (38%), Positives = 55/91 (60%), Gaps = 4/91 (4%)
Query 16 SHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAAL 75
+HP + L +QTE LY +L++ +R DV RLS+ V + + + AE+ AL
Sbjct 131 NHPFIVKLHYAFQTEGKLYLILDF---LRGGDVFTRLSKE-VLFTEEDVKFYLAELALAL 186
Query 76 ATLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L Q G+++RD++PENI++ E GH+ L DF
Sbjct 187 DHLHQLGIVYRDLKPENILLDEIGHIKLTDF 217
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 21/85 (24%), Positives = 40/85 (47%), Gaps = 4/85 (4%)
Query 14 HSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVA 73
+ HP++ L+ ++ +Y V + L D R L + S + L+ I
Sbjct 474 YGQHPNIITLKDVFDDGRYVYLVTDLMKGGELLD--RILKQKCFSEREASDILYV--ISK 529
Query 74 ALATLRQKGVLHRDIRPENIVVSEQ 98
+ L +GV+HRD++P NI+ ++
Sbjct 530 TVDYLHCQGVVHRDLKPSNILYMDE 554
> cel:T01H8.1 rskn-1; RSK-pNinety (RSK-p90 kinase) homolog family
member (rskn-1); K04373 p90 ribosomal S6 kinase [EC:2.7.11.1]
Length=804
Score = 67.8 bits (164), Expect = 9e-12, Method: Composition-based stats.
Identities = 35/104 (33%), Positives = 59/104 (56%), Gaps = 4/104 (3%)
Query 3 KETLLSLSLGGHSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIG 62
+ T L ++ H SHP + L +QTE LY +L++ +R D+ RLS+ V +
Sbjct 143 QRTKLERNILAHISHPFIVKLHYAFQTEGKLYLILDF---LRGGDLFTRLSKE-VMFTED 198
Query 63 AARLWAAEIVAALATLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
+ + AE+ AL L G+++RD++PENI++ GH+ + DF
Sbjct 199 DVKFYLAELTLALEHLHSLGIVYRDLKPENILLDADGHIKVTDF 242
Score = 32.7 bits (73), Expect = 0.32, Method: Composition-based stats.
Identities = 26/97 (26%), Positives = 40/97 (41%), Gaps = 7/97 (7%)
Query 14 HSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVA 73
HS H V L +Y+ E +Y + L D L G + A A ++
Sbjct 496 HSHHQFVVKLFDVYEDETAIYMIEELCEGGELLDKLVNKKSLGSEKEVAAI---MANLLN 552
Query 74 ALATLRQKGVLHRDIRPENIVVS----EQGHLTLVDF 106
A+ L + V HRD+ NI+ + + L +VDF
Sbjct 553 AVQYLHSQQVAHRDLTAANILFALKDGDPSSLRIVDF 589
> hsa:10110 SGK2, H-SGK2, dJ138B7.2; serum/glucocorticoid regulated
kinase 2 (EC:2.7.11.1); K13303 serum/glucocorticoid-regulated
kinase 2 [EC:2.7.11.1]
Length=367
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 36/90 (40%), Positives = 52/90 (57%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + LR +QT E LYFVL+Y L L+R R AR +AAE+ +A+
Sbjct 93 HPFLVGLRYSFQTPEKLYFVLDYVNGGELFFHLQRERR----FLEPRARFYAAEVASAIG 148
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L +++RD++PENI++ QGH+ L DF
Sbjct 149 YLHSLNIIYRDLKPENILLDCQGHVVLTDF 178
> ath:AT3G08730 PK1; PK1 (PROTEIN-SERINE KINASE 1); kinase/ protein
binding / protein kinase/ protein serine/threonine kinase;
K08286 protein-serine/threonine kinase [EC:2.7.11.-]
Length=465
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 34/90 (37%), Positives = 55/90 (61%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + L+ +QT+ LY VL++ L +L G+ AR++ AEIV+A++
Sbjct 191 HPFIVQLKYSFQTKYRLYLVLDFINGGHL---FFQLYHQGL-FREDLARVYTAEIVSAVS 246
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L +KG++HRD++PENI++ GH+ L DF
Sbjct 247 HLHEKGIMHRDLKPENILMDTDGHVMLTDF 276
> tgo:TGME49_067540 protein kinase, putative (EC:4.1.1.70 2.7.11.13)
Length=951
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 37/91 (40%), Positives = 53/91 (58%), Gaps = 4/91 (4%)
Query 16 SHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAAL 75
SHP + + +QT + LYFVL Y L LSRAG AA +AAE++ AL
Sbjct 515 SHPFIVQMHYAFQTRKKLYFVLEYCPGGEL---FFHLSRAG-RFKEYAACFYAAEVLLAL 570
Query 76 ATLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L + V++RD++PEN+++ E GH+ L DF
Sbjct 571 EHLHKYNVVYRDLKPENVLLDEHGHVRLTDF 601
> ath:AT3G08720 S6K2; S6K2 (ARABIDOPSIS THALIANA SERINE/THREONINE
PROTEIN KINASE 2); kinase/ protein kinase
Length=471
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 34/90 (37%), Positives = 55/90 (61%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + L+ +QT+ LY VL++ L +L G+ AR++ AEIV+A++
Sbjct 197 HPFIVQLKYSFQTKYRLYLVLDFINGGHL---FFQLYHQGL-FREDLARVYTAEIVSAVS 252
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L +KG++HRD++PENI++ GH+ L DF
Sbjct 253 HLHEKGIMHRDLKPENILMDVDGHVMLTDF 282
> dre:337670 rps6kal, fj94b02, rps6ka6, wu:fj94b02, zgc:66139;
ribosomal protein S6 kinase, like (EC:2.7.11.1); K04373 p90
ribosomal S6 kinase [EC:2.7.11.1]
Length=740
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 34/91 (37%), Positives = 54/91 (59%), Gaps = 4/91 (4%)
Query 16 SHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAAL 75
+HP + L +QTE LY +L++ +R DV RLS+ V + + + AE+ AL
Sbjct 125 NHPFIVKLHYAFQTEGKLYLILDF---LRGGDVFTRLSKE-VMFTEEDVKFYLAELALAL 180
Query 76 ATLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L G+++RD++PENI++ E GH+ L DF
Sbjct 181 DHLHNLGIVYRDLKPENILLDEAGHIKLTDF 211
Score = 32.7 bits (73), Expect = 0.29, Method: Composition-based stats.
Identities = 22/81 (27%), Positives = 38/81 (46%), Gaps = 4/81 (4%)
Query 14 HSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVA 73
+ HP++ L+ +Y +Y V L D + R + S +A L+ I
Sbjct 468 YGQHPNIITLKDVYDEGRFVYLVTELMKGGELLDKILR--QKFFSEREASAVLYT--ITK 523
Query 74 ALATLRQKGVLHRDIRPENIV 94
+ L +GV+HRD++P NI+
Sbjct 524 TVDYLHCQGVVHRDLKPSNIL 544
> sce:YOL100W PKH2; Pkh2p (EC:2.7.11.1); K08286 protein-serine/threonine
kinase [EC:2.7.11.-]
Length=1081
Score = 66.2 bits (160), Expect = 2e-11, Method: Composition-based stats.
Identities = 42/114 (36%), Positives = 62/114 (54%), Gaps = 9/114 (7%)
Query 1 VEKETLLSLSLGGHSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLS 60
+EK L L+ + PSV L +Q E +LYF+L Y D L + + G SL
Sbjct 226 IEKTALQKLN-----NSPSVVRLFSTFQDESSLYFLLEYAPN---GDFLSLMKKYG-SLD 276
Query 61 IGAARLWAAEIVAALATLRQKGVLHRDIRPENIVVSEQGHLTLVDFYSALHLVP 114
AR +AA+I+ A+ L G++HRDI+PENI++ + + L DF +A L P
Sbjct 277 ETCARYYAAQIIDAIDYLHSNGIIHRDIKPENILLDGEMKIKLTDFGTAKLLNP 330
> mmu:110651 Rps6ka3, MPK-9, Rsk2, S6K-alpha3, pp90RSK2; ribosomal
protein S6 kinase polypeptide 3 (EC:2.7.11.1); K04373 p90
ribosomal S6 kinase [EC:2.7.11.1]
Length=740
Score = 66.2 bits (160), Expect = 2e-11, Method: Composition-based stats.
Identities = 33/91 (36%), Positives = 55/91 (60%), Gaps = 4/91 (4%)
Query 16 SHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAAL 75
+HP + L +QTE LY +L++ +R D+ RLS+ V + + + AE+ AL
Sbjct 126 NHPFIVKLHYAFQTEGKLYLILDF---LRGGDLFTRLSKE-VMFTEEDVKFYLAELALAL 181
Query 76 ATLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L G+++RD++PENI++ E+GH+ L DF
Sbjct 182 DHLHSLGIIYRDLKPENILLDEEGHIKLTDF 212
Score = 34.7 bits (78), Expect = 0.069, Method: Composition-based stats.
Identities = 29/103 (28%), Positives = 49/103 (47%), Gaps = 8/103 (7%)
Query 14 HSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVA 73
+ HP++ L+ +Y + +Y V L D + R + S +A L+ I
Sbjct 470 YGQHPNIITLKDVYDDGKYVYVVTELMKGGELLDKILR--QKFFSEREASAVLFT--ITK 525
Query 74 ALATLRQKGVLHRDIRPENIV-VSEQGH---LTLVDFYSALHL 112
+ L +GV+HRD++P NI+ V E G+ + + DF A L
Sbjct 526 TVEYLHAQGVVHRDLKPSNILYVDESGNPESIRICDFGFAKQL 568
> hsa:6197 RPS6KA3, CLS, HU-3, ISPK-1, MAPKAPK1B, MRX19, RSK,
RSK2, S6K-alpha3, p90-RSK2, pp90RSK2; ribosomal protein S6 kinase,
90kDa, polypeptide 3 (EC:2.7.11.1); K04373 p90 ribosomal
S6 kinase [EC:2.7.11.1]
Length=740
Score = 66.2 bits (160), Expect = 3e-11, Method: Composition-based stats.
Identities = 33/91 (36%), Positives = 55/91 (60%), Gaps = 4/91 (4%)
Query 16 SHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAAL 75
+HP + L +QTE LY +L++ +R D+ RLS+ V + + + AE+ AL
Sbjct 126 NHPFIVKLHYAFQTEGKLYLILDF---LRGGDLFTRLSKE-VMFTEEDVKFYLAELALAL 181
Query 76 ATLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L G+++RD++PENI++ E+GH+ L DF
Sbjct 182 DHLHSLGIIYRDLKPENILLDEEGHIKLTDF 212
Score = 34.7 bits (78), Expect = 0.070, Method: Composition-based stats.
Identities = 29/103 (28%), Positives = 49/103 (47%), Gaps = 8/103 (7%)
Query 14 HSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVA 73
+ HP++ L+ +Y + +Y V L D + R + S +A L+ I
Sbjct 470 YGQHPNIITLKDVYDDGKYVYVVTELMKGGELLDKILR--QKFFSEREASAVLFT--ITK 525
Query 74 ALATLRQKGVLHRDIRPENIV-VSEQGH---LTLVDFYSALHL 112
+ L +GV+HRD++P NI+ V E G+ + + DF A L
Sbjct 526 TVEYLHAQGVVHRDLKPSNILYVDESGNPESIRICDFGFAKQL 568
> hsa:6199 RPS6KB2, KLS, P70-beta, P70-beta-1, P70-beta-2, S6K-beta2,
S6K2, SRK, STK14B, p70(S6K)-beta, p70S6Kb; ribosomal
protein S6 kinase, 70kDa, polypeptide 2 (EC:2.7.11.1); K04688
p70 ribosomal S6 kinase [EC:2.7.11.1]
Length=482
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 34/90 (37%), Positives = 51/90 (56%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + L +QT LY +L C+ ++ L R G+ L A + AEI AL
Sbjct 128 HPFIVELAYAFQTGGKLYLILE---CLSGGELFTHLEREGIFLE-DTACFYLAEITLALG 183
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L +G+++RD++PENI++S QGH+ L DF
Sbjct 184 HLHSQGIIYRDLKPENIMLSSQGHIKLTDF 213
> xla:399376 rps6ka3, MGC82193, S6KII, p90, rsk, rsk2; ribosomal
protein S6 kinase, 90kDa, polypeptide 3 (EC:2.7.11.1); K04373
p90 ribosomal S6 kinase [EC:2.7.11.1]
Length=737
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 33/91 (36%), Positives = 55/91 (60%), Gaps = 4/91 (4%)
Query 16 SHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAAL 75
+HP + L +QTE LY +L++ +R D+ RLS+ V + + + AE+ AL
Sbjct 123 NHPFIVKLHYAFQTEGKLYLILDF---LRGGDLFTRLSKE-VMFTEEDVKYYLAELALAL 178
Query 76 ATLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L G+++RD++PENI++ E+GH+ L DF
Sbjct 179 DHLHSLGIIYRDLKPENILLDEEGHIKLTDF 209
Score = 30.8 bits (68), Expect = 0.99, Method: Composition-based stats.
Identities = 27/103 (26%), Positives = 48/103 (46%), Gaps = 8/103 (7%)
Query 14 HSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVA 73
+ HP++ L+ +Y + +Y V T ++ ++L ++ R S A I
Sbjct 466 YGQHPNIITLKDVYDDGKYVYLV---TELMKGGELLDKILRQKF-FSEREASAVLHTITK 521
Query 74 ALATLRQKGVLHRDIRPENIV-VSEQGH---LTLVDFYSALHL 112
L + V+HRD++P NI+ V E G+ + + DF A L
Sbjct 522 TAEYLHSQWVVHRDLKPSNILYVDESGNPESIRICDFGFAKQL 564
> dre:562242 ribosomal protein S6 kinase b, polypeptide 1-like;
K04688 p70 ribosomal S6 kinase [EC:2.7.11.1]
Length=507
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 36/90 (40%), Positives = 51/90 (56%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + L +QT E LY +L Y L +L R G+ L A + AEI AL
Sbjct 127 HPFIVDLIYAFQTGEKLYLILEYLSGGEL---FMQLEREGIFLE-DTACFYLAEISLALG 182
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L QKG+++RD++PENI+++ GH+ L DF
Sbjct 183 HLHQKGIIYRDLKPENIMLNYNGHVKLTDF 212
> dre:768202 pdpk1a, zgc:153787; 3-phosphoinositide dependent
protein kinase-1a (EC:2.7.11.1)
Length=536
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 36/102 (35%), Positives = 55/102 (53%), Gaps = 4/102 (3%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP L +Q + LYF L+Y + ++LR + + G S R + AEIV AL
Sbjct 134 HPFFVKLFFTFQDSQKLYFALSYA---KNGELLRYIRKIG-SFDETCTRFYTAEIVCALE 189
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDFYSALHLVPGTQQ 118
L G++HRD++PENI++ E H+ + DF +A L + Q
Sbjct 190 YLHTVGIIHRDLKPENILLCEDMHIQITDFGTAKQLSSDSAQ 231
> dre:324140 sgk1, cb1083, sgk, wu:fc20a09; serum/glucocorticoid
regulated kinase 1 (EC:2.7.11.1); K13302 serum/glucocorticoid-regulated
kinase 1 [EC:2.7.11.1]
Length=433
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 35/90 (38%), Positives = 52/90 (57%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + L +QT + LYFVL+Y + ++ L R L AR +AAEI +AL
Sbjct 158 HPFLVGLHYSFQTTDKLYFVLDY---INGGELFYHLQRERCFLE-PRARFYAAEIASALG 213
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L +++RD++PENI++ QGH+ L DF
Sbjct 214 YLHSLNIVYRDLKPENILLDSQGHIILTDF 243
> mmu:27219 Sgk2, AI098171, AW146006, Sgkl; serum/glucocorticoid
regulated kinase 2 (EC:2.7.11.1); K13303 serum/glucocorticoid-regulated
kinase 2 [EC:2.7.11.1]
Length=367
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 35/90 (38%), Positives = 51/90 (56%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + LR +QT E LYFVL+Y L L+R R AR + AE+ +A+
Sbjct 93 HPFLVGLRYSFQTPEKLYFVLDYVNGGELFFHLQRERR----FLEPRARFYTAEVASAIG 148
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L +++RD++PENI++ QGH+ L DF
Sbjct 149 YLHSLNIIYRDLKPENILLDCQGHVVLTDF 178
> cel:W10G6.2 sgk-1; Serum- and Glucocorticoid- inducible kinase
homolog family member (sgk-1); K08792 serum/glucocorticoid-regulated
kinase, invertebrate [EC:2.7.11.1]
Length=463
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 34/90 (37%), Positives = 53/90 (58%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + L +Q +E LYFVL++ L L+R S S R +AAEI AL
Sbjct 193 HPFLVSLHFSFQNKEKLYFVLDHLNGGELFSHLQREKHFSESRS----RFYAAEIACALG 248
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L +K +++RD++PEN+++ ++G+L L DF
Sbjct 249 YLHEKNIIYRDLKPENLLLDDKGYLVLTDF 278
> mmu:67071 Rps6ka6, 2610524K04Rik, RSK4; ribosomal protein S6
kinase polypeptide 6 (EC:2.7.11.1); K04373 p90 ribosomal S6
kinase [EC:2.7.11.1]
Length=860
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 34/91 (37%), Positives = 55/91 (60%), Gaps = 4/91 (4%)
Query 16 SHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAAL 75
+HP + L +QTE LY +L++ +R DV RLS+ V + + + AE+ AL
Sbjct 247 NHPFIVKLHYAFQTEGKLYLILDF---LRGGDVFTRLSKE-VLFTEEDVKFYLAELALAL 302
Query 76 ATLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L + G+++RD++PENI++ E GH+ L DF
Sbjct 303 DHLHRLGIVYRDLKPENILLDEIGHIKLTDF 333
Score = 36.2 bits (82), Expect = 0.023, Method: Composition-based stats.
Identities = 27/103 (26%), Positives = 49/103 (47%), Gaps = 8/103 (7%)
Query 14 HSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVA 73
+ HP++ L++++ + +Y V + L D R L + S + L+ I
Sbjct 590 YGQHPNIISLKEVFDDGKYVYLVTDLMKGGELLD--RILKKKCFSEQEASNVLYV--ITK 645
Query 74 ALATLRQKGVLHRDIRPENIV-VSEQGH---LTLVDFYSALHL 112
+ L +GV+HRD++P NI+ + E H + + DF A L
Sbjct 646 TVECLHSQGVVHRDLKPSNILYMDESAHPDSIKICDFGFAKQL 688
> mmu:72508 Rps6kb1, 2610318I15Rik, 4732464A07Rik, 70kDa, AA959758,
AI256796, AI314060, S6K1, p70/85s6k, p70s6k; ribosomal
protein S6 kinase, polypeptide 1 (EC:2.7.11.1); K04688 p70
ribosomal S6 kinase [EC:2.7.11.1]
Length=525
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 35/90 (38%), Positives = 51/90 (56%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + L +QT LY +L Y L +L R G+ + A + AEI AL
Sbjct 152 HPFIVDLIYAFQTGGKLYLILEYLSGGEL---FMQLEREGIFME-DTACFYLAEISMALG 207
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L QKG+++RD++PENI+++ QGH+ L DF
Sbjct 208 HLHQKGIIYRDLKPENIMLNHQGHVKLTDF 237
> hsa:6198 RPS6KB1, PS6K, S6K, S6K1, STK14A, p70(S6K)-alpha, p70-S6K,
p70-alpha; ribosomal protein S6 kinase, 70kDa, polypeptide
1 (EC:2.7.11.1); K04688 p70 ribosomal S6 kinase [EC:2.7.11.1]
Length=525
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 35/90 (38%), Positives = 51/90 (56%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + L +QT LY +L Y L +L R G+ + A + AEI AL
Sbjct 152 HPFIVDLIYAFQTGGKLYLILEYLSGGEL---FMQLEREGIFME-DTACFYLAEISMALG 207
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L QKG+++RD++PENI+++ QGH+ L DF
Sbjct 208 HLHQKGIIYRDLKPENIMLNHQGHVKLTDF 237
> dre:406349 rps6kb1b, fc51h01, rps6kb1, wu:fc51h01, zgc:55713;
ribosomal protein S6 kinase b, polypeptide 1b (EC:2.7.11.1);
K04688 p70 ribosomal S6 kinase [EC:2.7.11.1]
Length=502
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 35/90 (38%), Positives = 51/90 (56%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + L +QT LY +L Y L +L R G+ + A + AEI AL
Sbjct 126 HPFIVDLIYAFQTGGKLYLILEYLSGGEL---FMQLEREGIFME-DTACFYLAEISMALG 181
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L QKG+++RD++PENI+++ QGH+ L DF
Sbjct 182 HLHQKGIIYRDLKPENIMLNNQGHVKLTDF 211
> tpv:TP03_0511 cGMP-dependent protein kinase (EC:2.7.1.37); K07376
protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=892
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 36/124 (29%), Positives = 65/124 (52%), Gaps = 10/124 (8%)
Query 2 EKETLLSLSLGGHSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSI 61
+K + + + HP + L + ++ EEN+YF+ L D LR+L L
Sbjct 634 QKNIKMEKEIMAQNDHPFIIQLVKTFKDEENIYFLTELVTGGELFDALRKLEL----LKK 689
Query 62 GAARLWAAEIVAALATLRQKGVLHRDIRPENIVVSEQGHLTLVDFYSALHL------VPG 115
A+ + I+ AL L ++ +++RD++PEN+++ QG++ L+DF SA + V G
Sbjct 690 PQAQFYLGSIILALEYLHERQIIYRDLKPENVMLDHQGYIKLIDFGSAKKIEGRTFTVIG 749
Query 116 TQQW 119
T Q+
Sbjct 750 TPQY 753
> mmu:20393 Sgk1, Sgk; serum/glucocorticoid regulated kinase 1
(EC:2.7.11.1); K13302 serum/glucocorticoid-regulated kinase
1 [EC:2.7.11.1]
Length=524
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 35/90 (38%), Positives = 52/90 (57%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + L +QT + LYFVL+Y + ++ L R L AR +AAEI +AL
Sbjct 249 HPFLVGLHFSFQTADKLYFVLDY---INGGELFYHLQRERCFLE-PRARFYAAEIASALG 304
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L +++RD++PENI++ QGH+ L DF
Sbjct 305 YLHSLNIVYRDLKPENILLDSQGHIVLTDF 334
> mmu:58988 Rps6kb2, 70kDa, S6K2; ribosomal protein S6 kinase,
polypeptide 2 (EC:2.7.11.1); K04688 p70 ribosomal S6 kinase
[EC:2.7.11.1]
Length=485
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 34/90 (37%), Positives = 50/90 (55%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + L +QT LY +L C+ ++ L R G+ L A + AEI AL
Sbjct 128 HPFIVELAYAFQTGGKLYLILE---CLSGGELFTHLEREGIFLE-DTACFYLAEITLALG 183
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L G+++RD++PENI++S QGH+ L DF
Sbjct 184 HLHSHGIIYRDLKPENIMLSSQGHIKLTDF 213
> hsa:6446 SGK1, SGK; serum/glucocorticoid regulated kinase 1
(EC:2.7.11.1); K13302 serum/glucocorticoid-regulated kinase
1 [EC:2.7.11.1]
Length=526
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 35/90 (38%), Positives = 52/90 (57%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + L +QT + LYFVL+Y + ++ L R L AR +AAEI +AL
Sbjct 251 HPFLVGLHFSFQTADKLYFVLDY---INGGELFYHLQRERCFLE-PRARFYAAEIASALG 306
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L +++RD++PENI++ QGH+ L DF
Sbjct 307 YLHSLNIVYRDLKPENILLDSQGHIVLTDF 336
> mmu:12704 Cit, C030025P15Rik, CRIK, CRIK-SK, Cit-k; citron (EC:2.7.11.1);
K08282 non-specific serine/threonine protein kinase
[EC:2.7.11.1]
Length=2055
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 35/95 (36%), Positives = 55/95 (57%), Gaps = 3/95 (3%)
Query 15 SSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAA 74
S+ P + L+ +Q + NLY V+ Y D+L L+R L + + AE++ A
Sbjct 152 STSPWIPQLQYAFQDKNNLYLVMEYQPG---GDLLSLLNRYEDQLDESMIQFYLAELILA 208
Query 75 LATLRQKGVLHRDIRPENIVVSEQGHLTLVDFYSA 109
+ ++ Q G +HRDI+PENI++ GH+ LVDF SA
Sbjct 209 VHSVHQMGYVHRDIKPENILIDRTGHIKLVDFGSA 243
> dre:571285 ribosomal protein S6 kinase, polypeptide 2-like;
K04373 p90 ribosomal S6 kinase [EC:2.7.11.1]
Length=733
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 32/91 (35%), Positives = 55/91 (60%), Gaps = 4/91 (4%)
Query 16 SHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAAL 75
+HP + L +QTE LY +L++ +R D+ RLS+ V + + + AE+ AL
Sbjct 117 NHPFIVKLHYAFQTEGKLYLILDF---LRGGDLFTRLSKE-VMFTEEDVKFYLAELALAL 172
Query 76 ATLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L G+++RD++PENI++ E+GH+ + DF
Sbjct 173 DHLHSLGIIYRDLKPENILLDEEGHIKITDF 203
Score = 37.0 bits (84), Expect = 0.013, Method: Composition-based stats.
Identities = 25/87 (28%), Positives = 43/87 (49%), Gaps = 5/87 (5%)
Query 14 HSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVA 73
+ HP++ L+ +Y + +Y V+ +R ++L R+ R S A I
Sbjct 463 YGQHPNIITLKDVYDDGKFVYLVMEL---MRGGELLDRILRQKC-FSEREASAVLCTITK 518
Query 74 ALATLRQKGVLHRDIRPENIV-VSEQG 99
+ L +GV+HRD++P NI+ V E G
Sbjct 519 TVEYLHSQGVVHRDLKPSNILYVDETG 545
> hsa:6196 RPS6KA2, HU-2, MAPKAPK1C, RSK, RSK3, S6K-alpha, S6K-alpha2,
p90-RSK3, pp90RSK3; ribosomal protein S6 kinase, 90kDa,
polypeptide 2 (EC:2.7.11.1); K04373 p90 ribosomal S6 kinase
[EC:2.7.11.1]
Length=741
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 32/91 (35%), Positives = 55/91 (60%), Gaps = 4/91 (4%)
Query 16 SHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAAL 75
+HP + L +QTE LY +L++ +R D+ RLS+ V + + + AE+ AL
Sbjct 125 NHPFIVKLHYAFQTEGKLYLILDF---LRGGDLFTRLSKE-VMFTEEDVKFYLAELALAL 180
Query 76 ATLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L G+++RD++PENI++ E+GH+ + DF
Sbjct 181 DHLHSLGIIYRDLKPENILLDEEGHIKITDF 211
Score = 35.4 bits (80), Expect = 0.041, Method: Composition-based stats.
Identities = 21/85 (24%), Positives = 43/85 (50%), Gaps = 4/85 (4%)
Query 14 HSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVA 73
+ HP++ L+ +Y + +Y V+ +R ++L R+ R A+ + I
Sbjct 471 YGQHPNIITLKDVYDDGKFVYLVMEL---MRGGELLDRILRQRYFSEREASDVLCT-ITK 526
Query 74 ALATLRQKGVLHRDIRPENIVVSEQ 98
+ L +GV+HRD++P NI+ ++
Sbjct 527 TMDYLHSQGVVHRDLKPSNILYRDE 551
> dre:100009657 rps6ka3b, zgc:158386; ribosomal protein S6 kinase,
polypeptide 3b (EC:2.7.11.1); K04373 p90 ribosomal S6 kinase
[EC:2.7.11.1]
Length=720
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 33/91 (36%), Positives = 54/91 (59%), Gaps = 4/91 (4%)
Query 16 SHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAAL 75
+HP + L +QTE LY +L++ +R D+ RLS+ V + + + AE+ AL
Sbjct 125 NHPFIVKLHYAFQTEGKLYLILDF---LRGGDLFTRLSKE-VMFTEEDVKFYLAELALAL 180
Query 76 ATLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L G+++RD++PENI++ E GH+ L DF
Sbjct 181 DHLHGLGIIYRDLKPENILLDEDGHIKLTDF 211
Score = 31.6 bits (70), Expect = 0.63, Method: Composition-based stats.
Identities = 18/46 (39%), Positives = 27/46 (58%), Gaps = 4/46 (8%)
Query 71 IVAALATLRQKGVLHRDIRPENIV-VSEQGH---LTLVDFYSALHL 112
I +A L +GV+HRD++P NI+ V E G+ + + DF A L
Sbjct 492 ITKTVAYLHAQGVVHRDLKPSNILYVDESGNPESIRICDFGFAKQL 537
> mmu:20111 Rps6ka1, Rsk1, p90rsk, rsk; ribosomal protein S6 kinase
polypeptide 1 (EC:2.7.11.1); K04373 p90 ribosomal S6 kinase
[EC:2.7.11.1]
Length=735
Score = 64.7 bits (156), Expect = 6e-11, Method: Composition-based stats.
Identities = 33/91 (36%), Positives = 54/91 (59%), Gaps = 4/91 (4%)
Query 16 SHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAAL 75
+HP V L +QTE LY +L++ +R D+ RLS+ V + + + AE+ L
Sbjct 120 NHPFVVKLHYAFQTEGKLYLILDF---LRGGDLFTRLSKE-VMFTEEDVKFYLAELALGL 175
Query 76 ATLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L G+++RD++PENI++ E+GH+ L DF
Sbjct 176 DHLHSLGIIYRDLKPENILLDEEGHIKLTDF 206
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 30/103 (29%), Positives = 51/103 (49%), Gaps = 8/103 (7%)
Query 14 HSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVA 73
+ HP++ L+ +Y +++Y V T +R ++L ++ R S A I
Sbjct 466 YGQHPNIITLKDVYDDGKHVYLV---TELMRGGELLDKILRQKF-FSEREASFVLHTISK 521
Query 74 ALATLRQKGVLHRDIRPENIV-VSEQGH---LTLVDFYSALHL 112
+ L +GV+HRD++P NI+ V E G+ L + DF A L
Sbjct 522 TVEYLHSQGVVHRDLKPSNILYVDESGNPECLRICDFGFAKQL 564
> hsa:6195 RPS6KA1, HU-1, MAPKAPK1A, RSK, RSK1; ribosomal protein
S6 kinase, 90kDa, polypeptide 1 (EC:2.7.11.1); K04373 p90
ribosomal S6 kinase [EC:2.7.11.1]
Length=744
Score = 64.7 bits (156), Expect = 6e-11, Method: Composition-based stats.
Identities = 33/91 (36%), Positives = 54/91 (59%), Gaps = 4/91 (4%)
Query 16 SHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAAL 75
+HP V L +QTE LY +L++ +R D+ RLS+ V + + + AE+ L
Sbjct 129 NHPFVVKLHYAFQTEGKLYLILDF---LRGGDLFTRLSKE-VMFTEEDVKFYLAELALGL 184
Query 76 ATLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L G+++RD++PENI++ E+GH+ L DF
Sbjct 185 DHLHSLGIIYRDLKPENILLDEEGHIKLTDF 215
Score = 37.7 bits (86), Expect = 0.008, Method: Composition-based stats.
Identities = 30/103 (29%), Positives = 51/103 (49%), Gaps = 8/103 (7%)
Query 14 HSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVA 73
+ HP++ L+ +Y +++Y V T +R ++L ++ R S A I
Sbjct 475 YGQHPNIITLKDVYDDGKHVYLV---TELMRGGELLDKILRQKF-FSEREASFVLHTIGK 530
Query 74 ALATLRQKGVLHRDIRPENIV-VSEQGH---LTLVDFYSALHL 112
+ L +GV+HRD++P NI+ V E G+ L + DF A L
Sbjct 531 TVEYLHSQGVVHRDLKPSNILYVDESGNPECLRICDFGFAKQL 573
> tgo:TGME49_086470 cAMP-dependent protein kinase catalytic subunit
(EC:2.7.11.11); K04345 protein kinase A [EC:2.7.11.11]
Length=514
Score = 64.7 bits (156), Expect = 6e-11, Method: Composition-based stats.
Identities = 31/90 (34%), Positives = 51/90 (56%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + L +Q E+ L+ ++ Y L LRR + + ARL+AAEI A
Sbjct 261 HPFIVNLLAAFQDEKRLFILMEYVNGGELFSHLRRRN----CIPTDQARLYAAEITLAFQ 316
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L Q+ +++RD++PEN+++ QGH+ + DF
Sbjct 317 YLHQRHIVYRDLKPENLLIDSQGHIKITDF 346
> xla:443729 rps6ka6, MGC79981, rps6ka, rps6ka1; ribosomal protein
S6 kinase, 90kDa, polypeptide 6 (EC:2.7.11.1); K04373 p90
ribosomal S6 kinase [EC:2.7.11.1]
Length=733
Score = 64.7 bits (156), Expect = 7e-11, Method: Composition-based stats.
Identities = 33/90 (36%), Positives = 53/90 (58%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP V L +QTE LY +L++ +R D+ RLS+ V + + + AE+ L
Sbjct 121 HPFVVRLHYAFQTEGKLYLILDF---LRGGDLFTRLSKE-VMFTEEDVKFYLAELALGLD 176
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L G+++RD++PENI++ E+GH+ L DF
Sbjct 177 HLHSLGIIYRDLKPENILLDEEGHIKLTDF 206
Score = 37.4 bits (85), Expect = 0.010, Method: Composition-based stats.
Identities = 25/87 (28%), Positives = 44/87 (50%), Gaps = 5/87 (5%)
Query 14 HSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVA 73
+ HP++ L+ +Y+ ++Y V T +R ++L R+ R S A +
Sbjct 464 YGQHPNIIALKDVYKEGNSIYVV---TELMRGGELLDRILRQKF-FSEREASSVLFTVCK 519
Query 74 ALATLRQKGVLHRDIRPENIV-VSEQG 99
+ L +GV+HRD++P NI+ V E G
Sbjct 520 TVENLHSQGVVHRDLKPSNILYVDESG 546
> cel:K08B12.5 mrck-1; Myotonic dystrophy-Related, Cdc42-binding
Kinase homolog family member (mrck-1); K08286 protein-serine/threonine
kinase [EC:2.7.11.-]
Length=1592
Score = 64.7 bits (156), Expect = 7e-11, Method: Composition-based stats.
Identities = 34/95 (35%), Positives = 58/95 (61%), Gaps = 5/95 (5%)
Query 20 VAYLRQIYQTEENLYFVLNY-TGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALATL 78
+ L +Q E+NLYFV++Y G D+L LS+ + A+ + AE+V A+ +L
Sbjct 143 ITNLHYAFQDEKNLYFVMDYYIG----GDMLTLLSKFVDHIPESMAKFYIAEMVLAIDSL 198
Query 79 RQKGVLHRDIRPENIVVSEQGHLTLVDFYSALHLV 113
+ G +HRD++P+N+++ QGH+ L DF S L ++
Sbjct 199 HRLGYVHRDVKPDNVLLDMQGHIRLADFGSCLRIL 233
> mmu:13400 Dmpk, DM, DMK, Dm15, MDPK, MT-PK; dystrophia myotonica-protein
kinase (EC:2.7.11.1); K08788 dystrophia myotonica-protein
kinase [EC:2.7.11.1]
Length=605
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 36/89 (40%), Positives = 52/89 (58%), Gaps = 5/89 (5%)
Query 27 YQTEENLYFVLNY-TGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALATLRQKGVLH 85
+Q E LY V+ Y G D+L LS+ G + AR + AEIV A+ ++ + G +H
Sbjct 138 FQDENYLYLVMEYYVG----GDLLTLLSKFGERIPAEMARFYLAEIVMAIDSVHRLGYVH 193
Query 86 RDIRPENIVVSEQGHLTLVDFYSALHLVP 114
RDI+P+NI++ GH+ L DF S L L P
Sbjct 194 RDIKPDNILLDRCGHIRLADFGSCLKLQP 222
> ath:AT4G13000 protein kinase family protein
Length=372
Score = 64.3 bits (155), Expect = 8e-11, Method: Composition-based stats.
Identities = 34/102 (33%), Positives = 58/102 (56%), Gaps = 2/102 (1%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP L + T++ + + ++Y L + ++ S S I R +AAE+V AL
Sbjct 78 HPLFPRLHGVISTDKVIGYAIDYCPGRDLNSLRKKQSEEMFSDEI--IRFYAAELVIALE 135
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDFYSALHLVPGTQQ 118
L +G+++RD++P+N+++ E GHL LVDF + +L P T Q
Sbjct 136 YLHNQGIVYRDLKPDNVMIQENGHLMLVDFDLSTNLPPRTPQ 177
> xla:445821 rps6ka1, MGC81220, hu-1, mapkapk1a, rsk, rsk1; ribosomal
protein S6 kinase, 90kDa, polypeptide 1 (EC:2.7.11.1);
K04373 p90 ribosomal S6 kinase [EC:2.7.11.1]
Length=733
Score = 64.3 bits (155), Expect = 8e-11, Method: Composition-based stats.
Identities = 33/90 (36%), Positives = 53/90 (58%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP V L +QTE LY +L++ +R D+ RLS+ V + + + AE+ L
Sbjct 121 HPFVVRLHYAFQTEGKLYLILDF---LRGGDLFTRLSKE-VMFTEEDVKFYLAELALGLD 176
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L G+++RD++PENI++ E+GH+ L DF
Sbjct 177 HLHSLGIIYRDLKPENILLDEEGHIKLTDF 206
Score = 36.2 bits (82), Expect = 0.026, Method: Composition-based stats.
Identities = 24/87 (27%), Positives = 44/87 (50%), Gaps = 5/87 (5%)
Query 14 HSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVA 73
+ HP++ L+ +Y+ ++Y V T +R ++L R+ R A + +
Sbjct 464 YGQHPNIITLKDVYEECNSIYLV---TELMRGGELLDRILRQKFFSEREACSVLFT-VCK 519
Query 74 ALATLRQKGVLHRDIRPENIV-VSEQG 99
+ L +GV+HRD++P NI+ V E G
Sbjct 520 TVEYLHSQGVVHRDLKPSNILYVDESG 546
> mmu:20112 Rps6ka2, 90kDa, D17Wsu134e, Rps6ka-rs1, Rsk3, p90rsk,
pp90rsk; ribosomal protein S6 kinase, polypeptide 2 (EC:2.7.11.1);
K04373 p90 ribosomal S6 kinase [EC:2.7.11.1]
Length=733
Score = 64.3 bits (155), Expect = 8e-11, Method: Composition-based stats.
Identities = 32/91 (35%), Positives = 55/91 (60%), Gaps = 4/91 (4%)
Query 16 SHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAAL 75
+HP + L +QTE LY +L++ +R D+ RLS+ V + + + AE+ AL
Sbjct 117 NHPFIVKLHYAFQTEGKLYLILDF---LRGGDLFTRLSKE-VMFTEEDVKFYLAELALAL 172
Query 76 ATLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L G+++RD++PENI++ E+GH+ + DF
Sbjct 173 DHLHGLGIIYRDLKPENILLDEEGHIKITDF 203
Score = 35.0 bits (79), Expect = 0.064, Method: Composition-based stats.
Identities = 21/85 (24%), Positives = 43/85 (50%), Gaps = 4/85 (4%)
Query 14 HSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVA 73
+ HP++ L+ +Y + +Y V+ +R ++L R+ R A+ + I
Sbjct 463 YGQHPNIITLKDVYDDGKYVYLVMEL---MRGGELLDRILRQRCFSEREASDVLYT-IAR 518
Query 74 ALATLRQKGVLHRDIRPENIVVSEQ 98
+ L +GV+HRD++P NI+ ++
Sbjct 519 TMDYLHSQGVVHRDLKPSNILYMDE 543
> xla:394276 rps6kb1, p70-alpha, p70-s6k, p70s6k, ps6k, rps6kb1-A,
s6K1, s6k, stk14a; ribosomal protein S6 kinase, 70kDa,
polypeptide 1 (EC:2.7.11.1); K04688 p70 ribosomal S6 kinase
[EC:2.7.11.1]
Length=501
Score = 64.3 bits (155), Expect = 9e-11, Method: Composition-based stats.
Identities = 35/90 (38%), Positives = 51/90 (56%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + L +QT LY +L Y L +L R G+ + A + AEI AL
Sbjct 128 HPFIVDLIYAFQTGGKLYLILEYLSGGEL---FMQLEREGIFME-DTACFYLAEISMALG 183
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L QKG+++RD++PENI+++ QGH+ L DF
Sbjct 184 HLHQKGIIYRDLKPENIMLNLQGHVKLTDF 213
> dre:570956 sgk2a, fm82b06, sgk2, wu:fm82b06, zgc:172111, zgc:198213;
serum/glucocorticoid regulated kinase 2a (EC:2.7.11.1);
K13303 serum/glucocorticoid-regulated kinase 2 [EC:2.7.11.1]
Length=361
Score = 64.3 bits (155), Expect = 9e-11, Method: Composition-based stats.
Identities = 34/90 (37%), Positives = 50/90 (55%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + L +QT E LYFVL+Y L L+R S AR + AE+ +A+
Sbjct 88 HPFLVGLHYSFQTSEKLYFVLDYVNGGELFYHLQRER----CFSEPRARFYTAEVASAIG 143
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L +++RD++PENI++ QGH+ L DF
Sbjct 144 YLHSLNIVYRDLKPENILLDYQGHVVLTDF 173
> xla:399130 sgk1-a, Sgk, sgk-A, sgk1; serum/glucocorticoid regulated
kinase 1 (EC:2.7.11.1); K13302 serum/glucocorticoid-regulated
kinase 1 [EC:2.7.11.1]
Length=434
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 34/90 (37%), Positives = 51/90 (56%), Gaps = 4/90 (4%)
Query 17 HPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAALA 76
HP + L +QT LYF+L+Y + ++ L R L AR +AAEI +AL
Sbjct 159 HPFLVGLHFSFQTTSRLYFILDY---INGGELFYHLQRERCFLE-PRARFYAAEIASALG 214
Query 77 TLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
L +++RD++PENI++ QGH+ L DF
Sbjct 215 YLHSLNIVYRDLKPENILLDSQGHIVLTDF 244
> bbo:BBOV_I004690 19.m02237; cGMP dependent protein kinase (EC:2.7.11.1);
K07376 protein kinase, cGMP-dependent [EC:2.7.11.12]
Length=887
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 32/105 (30%), Positives = 57/105 (54%), Gaps = 4/105 (3%)
Query 2 EKETLLSLSLGGHSSHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSI 61
+K L + + HP + L + ++ E N+YF+ L D +R++ LS
Sbjct 619 QKHIKLEREIMAQNDHPFIIQLVKTFKDESNVYFLTELITGGELYDAIRKIGL----LSR 674
Query 62 GAARLWAAEIVAALATLRQKGVLHRDIRPENIVVSEQGHLTLVDF 106
A+ + A IV A L ++ + +RD++PENI++ EQG++ L+DF
Sbjct 675 PQAQFYIASIVLAFEYLHERQIAYRDLKPENILLDEQGYIKLIDF 719
> mmu:237300 Gm4922, 4933423E17, EG237300; predicted gene 4922
(EC:2.7.11.1)
Length=497
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 31/102 (30%), Positives = 59/102 (57%), Gaps = 4/102 (3%)
Query 16 SHPSVAYLRQIYQTEENLYFVLNYTGCVRLRDVLRRLSRAGVSLSIGAARLWAAEIVAAL 75
SHP++ L Q+ +TE+N+Y ++ +L + R+ A L AR ++++A+
Sbjct 79 SHPNIVSLLQVIETEQNIYLIMEVAQGTQLHN---RVQEARC-LKEDEARSIFVQLLSAI 134
Query 76 ATLRQKGVLHRDIRPENIVVSEQGHLTLVDFYSALHLVPGTQ 117
+GV+HRD++P+N++V E G++ +VDF +PG +
Sbjct 135 GYCHGEGVVHRDLKPDNVIVDEHGNVKIVDFGLGARFMPGQK 176
Lambda K H
0.322 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2018002440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40