bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1639_orf2
Length=471
Score E
Sequences producing significant alignments: (Bits) Value
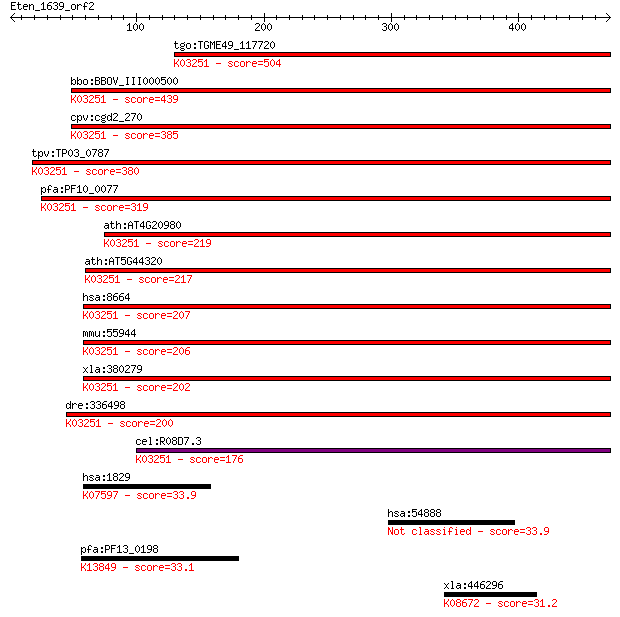
tgo:TGME49_117720 eukaryotic translation initiation factor 3 s... 504 2e-142
bbo:BBOV_III000500 17.m07069; eukaryotic translation initiatio... 439 1e-122
cpv:cgd2_270 translation initiation factor eIF-3 subunit 7 ; K... 385 2e-106
tpv:TP03_0787 eukaryotic translation initiation factor 3 subun... 380 4e-105
pfa:PF10_0077 eukaryotic translation initiation factor 3 subun... 319 1e-86
ath:AT4G20980 eukaryotic translation initiation factor 3 subun... 219 2e-56
ath:AT5G44320 eukaryotic translation initiation factor 3 subun... 217 9e-56
hsa:8664 EIF3D, EIF3S7, MGC126526, MGC17258, eIF3-p66, eIF3-ze... 207 1e-52
mmu:55944 Eif3d, 66/67kDa, AA407891, Eif3s7, MGC115774, eIF3p6... 206 1e-52
xla:380279 eif3d, MGC53375, eif3s7; eukaryotic translation ini... 202 2e-51
dre:336498 eif3d, eif3s7, wu:fb12a03, zgc:64097; eukaryotic tr... 200 8e-51
cel:R08D7.3 eif-3.D; Eukaryotic Initiation Factor family membe... 176 2e-43
hsa:1829 DSG2, ARVC10, ARVD10, CDHF5, CMD1BB, HDGC, MGC117034,... 33.9 1.5
hsa:54888 NSUN2, FLJ20303, MISU, SAKI, TRM4; NOP2/Sun domain f... 33.9 1.6
pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog A... 33.1 2.7
xla:446296 pcsk6, PACE4, PACE4AIIa, Xpace4, spc4; proprotein c... 31.2 9.9
> tgo:TGME49_117720 eukaryotic translation initiation factor 3
subunit 7, putative ; K03251 translation initiation factor
3 subunit D
Length=546
Score = 504 bits (1299), Expect = 2e-142, Method: Compositional matrix adjust.
Identities = 243/360 (67%), Positives = 292/360 (81%), Gaps = 18/360 (5%)
Query 130 VEDLEWRGTLGVYSRQADKISSKLPVPMQNLSSNFYYYWVTTQDDEVIRDYMLNGSKQVD 189
V+DL WRG L Y++QAD+I +KLPV +QNLSSNF YYWVTTQDD++I++ MLN S+ +D
Sbjct 143 VKDLAWRGELRTYNKQADQIRTKLPVALQNLSSNFDYYWVTTQDDDLIKEQMLNSSEPID 202
Query 190 VAATDQVLACLMAAPQSKYSWHLYLTKIDGKIIIDKANGSIVDLLTVNETSIEPPMQDAE 249
VAATDQVLACLMAAPQSKYSWHL+LTK++GK++IDKANGSIVDLLTVNET+ EPPMQDAE
Sbjct 203 VAATDQVLACLMAAPQSKYSWHLHLTKVEGKLLIDKANGSIVDLLTVNETASEPPMQDAE 262
Query 250 NRLNRPAALGFEAVRINQNLRQQLLVPGEVAAEYPPAPFVESGDTPATIAYRYRLFTIPP 309
NRLNRPAALGFEAV+INQNLRQQLLV GE ++ P PFVE+GD PA+IAYRYR+FTIP
Sbjct 263 NRLNRPAALGFEAVKINQNLRQQLLVKGEEPVKFSPPPFVEAGDNPASIAYRYRMFTIPG 322
Query 310 RAGPNA---SRRPINILTRAEVYAKSPM---------------ARADLKEAREGDNFIVD 351
R G SR+PINI+TR EV AK P +E + D F D
Sbjct 323 RTGGTGGGKSRQPINIITRTEVNAKLPQPADGSRKPSPGDEMTGGHSKREKEDSDPFGKD 382
Query 352 GGGYIYTCALNEIEPKGQKNWRTLLESQRGALLATEVRNNACKLRKFVTSAMIAGCDELR 411
GGGY+Y CALNE + KGQ+NWR +E+QRGALLATEVRNNACKL+KFV SAM+AGCD+L+
Sbjct 383 GGGYVYICALNEYDIKGQRNWRVQMETQRGALLATEVRNNACKLKKFVASAMMAGCDDLK 442
Query 412 LGFVARKTPNDNEQHLILTVHNYQTRDLGRQIGLRPENAWGIVRAVLDLLEDKPDGRYVL 471
LGFV+RK NDNE H++L+V +YQTRDLG QIGL+P+NAWGIVRA++D++ +KPDGRYVL
Sbjct 443 LGFVSRKANNDNEHHVVLSVQSYQTRDLGMQIGLKPDNAWGIVRAIVDMVMEKPDGRYVL 502
> bbo:BBOV_III000500 17.m07069; eukaryotic translation initiation
factor 3 subunit 7; K03251 translation initiation factor
3 subunit D
Length=547
Score = 439 bits (1128), Expect = 1e-122, Method: Compositional matrix adjust.
Identities = 212/433 (48%), Positives = 294/433 (67%), Gaps = 27/433 (6%)
Query 49 KQTTAAFNEKTKDEEELKLGKKAKSDQKKQQQQQQARYARIHARHRAFSEWSIQPTNDWQ 108
KQTT AFN+K +EE + + K K + ++Q+ +R R ARH F+EWSI+P++ W
Sbjct 106 KQTTQAFNQKQMEEEAMIMSAKQKHPEMRKQRMMHSRGNRPTARHHTFNEWSIEPSHSWH 165
Query 109 VLAELPLQQLHKQQIETP-RVTVEDLEWRGTLGVYSRQADKISSKLPVPMQNLSSNFYYY 167
+L+++P QL K ++ P +V D+ WRG LGVY ++ D IS+K VP+QNL+ F YY
Sbjct 166 LLSDIPFSQLSK--VDLPGKVVGTDVTWRGKLGVYDKRMDHISAKSEVPLQNLNDQFDYY 223
Query 168 WVTTQDDEVIRDYMLNGSKQ--------VDVAATDQVLACLMAAPQSKYSWHLYLTKIDG 219
W +T DD+ I DY+L+ +Q + + ATDQ+LA LM A +SKYSWHL +TKID
Sbjct 224 WTSTNDDDCIMDYLLSTQRQESADAGDHIILGATDQILAVLMTAARSKYSWHLNVTKIDN 283
Query 220 KIIIDKANGSIVDLLTVNETSIEPPMQDAENRLNRPAALGFEAVRINQNLRQQLLVPGEV 279
KIIIDKANGSI+D+LTVNETS EPP+ DAE ++NRP ALG+EAV++NQ LRQQ+LV G+
Sbjct 284 KIIIDKANGSIIDMLTVNETSPEPPLPDAEIKINRPPALGYEAVKVNQCLRQQVLVSGQF 343
Query 280 AAEYPPAPFVESGDTPATIAYRYRLFTIP-PRAGPNASRRPINILTRAEVYAKSPMARAD 338
+ E+ PFVE GD P+T+AYRYR F +P G SR PI+++TR EV+A+ P
Sbjct 344 SDEFELPPFVEEGDNPSTMAYRYRTFMVPGDNHGHGWSRVPIHVITRGEVHARIPG---- 399
Query 339 LKEAREGDNFIVDGGGYIYTCALNEIEPKGQKNWRTLLESQRGALLATEVRNNACKLRKF 398
V GGY Y CALNE+ K QK+WR +E+Q+GALLA E+RNN K+++F
Sbjct 400 -----------VSNGGYTYVCALNELPHKTQKSWRIQIETQKGALLANEMRNNTAKMQRF 448
Query 399 VTSAMIAGCDELRLGFVARKTPNDNEQHLILTVHNYQTRDLGRQIGLRPENAWGIVRAVL 458
A+I+GCD L+L +V+R++P D E H I+ +H Y T +L Q+GL NAW +VR +
Sbjct 449 AACAIISGCDTLKLAYVSRRSPGDAEHHSIIGIHTYTTENLAVQMGLNMRNAWAVVRHIC 508
Query 459 DLLEDKPDGRYVL 471
L+ D+PDG+Y+L
Sbjct 509 ALIMDRPDGQYIL 521
> cpv:cgd2_270 translation initiation factor eIF-3 subunit 7 ;
K03251 translation initiation factor 3 subunit D
Length=545
Score = 385 bits (988), Expect = 2e-106, Method: Compositional matrix adjust.
Identities = 196/435 (45%), Positives = 294/435 (67%), Gaps = 27/435 (6%)
Query 49 KQTTAAFNEKTKDEEE----LKLGKK-AKSDQKKQQQQQQARYARIHARHRAFSEWSIQP 103
K+T A+++K + +E+ L+ GK+ AK Q QQ Q +R+ R +R R +EWSI+P
Sbjct 94 KETAQAYHQKQQQQEKAEMLLRQGKRSAKMRQAGTQQYQYSRH-RFVSRMRTLTEWSIEP 152
Query 104 TNDWQVLAELPLQQLHKQQIETPRVTVEDLEWRGTLGVYSRQADKISSKLPVPMQNLSSN 163
+ W +AE+PL L KQ ++ + T+ED+ W GTL Y++ D+IS KLP+ +++ N
Sbjct 153 LSSWISVAEIPLSSLPKQFLDVSKSTIEDIGWFGTLKSYNKSMDRISPKLPMVLKDFG-N 211
Query 164 FYYYWVTTQDDEVIRDYMLNGSKQVDVAATDQVLACLMAAPQSKYSWHLYLTKIDGKIII 223
+ TT DDE++RD +L + +DVA TDQ+LACL+AA QSKYSWH+ +T +DGK+I
Sbjct 212 MEFLQPTTSDDEILRDALLQPERNIDVAITDQILACLVAAAQSKYSWHILVTNVDGKLIF 271
Query 224 DKANGSIVDLLTVNETSIEPPMQDAENRLNRPAALGFEAVRINQNLRQQLLVPGE--VAA 281
DK +GSI+DL+TVNETS EPP+ D N+ NRP LG EA++ NQN Q + + G+ +
Sbjct 272 DKQDGSIIDLITVNETSAEPPLPDNVNKNNRPHLLGLEALKSNQNYTQNIQIFGDEGIIK 331
Query 282 EYPPAPFVESG-DTP-ATIAYRYRLFTIPPRAGPNAS---RRPINILTRAEVYAKSPMAR 336
++ P PF E DT + YRYR+F++PPRAG ++S ++ + + R E+ A+ P A
Sbjct 332 QFEPIPFSEENQDTNIESKCYRYRIFSLPPRAGASSSENAQKTVRVAIRTEIDAQIPNAE 391
Query 337 ADLKEAREGDNFIVDGGGYIYTCALNEIEPKGQKNWRTLLESQRGALLATEVRNNACKLR 396
++ G+I+ ALNE +PK K W T LE+Q+GALLATE+RNNAC+L
Sbjct 392 SE-------------NSGFIFARALNEYDPKTVKTWNTQLETQKGALLATEIRNNACQLT 438
Query 397 KFVTSAMIAGCDELRLGFVARKTPNDNEQHLILTVHNYQTRDLGRQIGLRPENAWGIVRA 456
K+ T A+++ CD L+LG++ R+TPND E H I++V +Y+T++L Q+GL+ ENAWGIVRA
Sbjct 439 KWATQALLSSCDILKLGYIVRRTPNDRESHQIISVQSYKTKELSAQMGLKEENAWGIVRA 498
Query 457 VLDLLEDKPDGRYVL 471
++DLL ++P+G+YV+
Sbjct 499 LVDLLSEQPEGQYVI 513
> tpv:TP03_0787 eukaryotic translation initiation factor 3 subunit
7; K03251 translation initiation factor 3 subunit D
Length=596
Score = 380 bits (977), Expect = 4e-105, Method: Compositional matrix adjust.
Identities = 205/515 (39%), Positives = 300/515 (58%), Gaps = 80/515 (15%)
Query 18 SSCTSSRLKLHVLQLRIWNGAAPWVSTRGK--LKQTTAAFNEKTKDEEELKLGK-KAKSD 74
S+ L+ + R P + K +KQTT AFN+K +EE+L+ K K D
Sbjct 72 STSMDEELQFQTVDSRTIVKIKPTTHYKKKTPIKQTTQAFNQKVMEEEQLQFSKQKQYPD 131
Query 75 QKKQQQQQQARYARIHARHRAFSEWSIQPTNDWQVLAELPLQQLHKQQIETPRVTVEDLE 134
++Q+ + R R+ R+ +EWSI+PT W V+AE+P L KQ++++ +VTVEDL
Sbjct 132 LRRQKYIAKNRSLRLPTRYHPLNEWSIEPTPAWNVVAEIPFNILLKQEMDSSQVTVEDLF 191
Query 135 WRGTLGVYSRQADKISSKLPVPMQNLSSNFYYYWVTTQDDEVIRDYM------------- 181
WRG LG+Y R+ D+I+ K V + +LS+++ YYW +T DD+ I D +
Sbjct 192 WRGKLGIYDRKMDQITPKNEVYLLHLSNSYDYYWTSTHDDDCIADVLLETYKTSPSKPLE 251
Query 182 ------LNGSKQVD------------------------------------VAATDQVLAC 199
+ G + D +AATDQ+L+
Sbjct 252 SEYQNGVEGEDEFDSGDKKQAGNMPLQGDSASYDQAMKVERDLLRYDKIVLAATDQILSV 311
Query 200 LMAAPQSKYSWHLYLTKIDGKIIIDKANGSIVDLLTVNETSIEPPMQDAENRLNRPAALG 259
LM A +SK+SWHL +TKI+ +IIIDKANGSI+D+LTVNET+ +PP D E ++NRP++L
Sbjct 312 LMTAGRSKHSWHLNVTKIENQIIIDKANGSIIDMLTVNETAPDPPQPDCEIKINRPSSLR 371
Query 260 FEAVRINQNLRQQLLVPGEVAAEYPPAPFVESGDTPATIAYRYRLFTIPPRAGPNAS--- 316
+EAV++NQNLRQQ+L+ E + + PFVE D PATIAYRYR FT+P PNA+
Sbjct 372 YEAVKVNQNLRQQVLLQDEYSETFLDPPFVEESDNPATIAYRYRKFTLP--GTPNATNFE 429
Query 317 RRPINILTRAEVYAKSPMARADLKEAREGDNFIVDGGGYIYTCALNEIEPKGQKNWRTLL 376
+ PI ++TR EV+AK + Y Y CALNE K K+WR+ +
Sbjct 430 KLPILVITRCEVHAK-----------------LQGTDTYTYVCALNECPNKNNKSWRSQI 472
Query 377 ESQRGALLATEVRNNACKLRKFVTSAMIAGCDELRLGFVARKTPNDNEQHLILTVHNYQT 436
E+Q+GALLA E+RNN KL++FV A +AGC +L +V R++PND E H I+ +H + T
Sbjct 473 ETQKGALLANEIRNNTTKLQRFVACANMAGCGVFKLAYVTRRSPNDAENHSIIGIHTHTT 532
Query 437 RDLGRQIGLRPENAWGIVRAVLDLLEDKPDGRYVL 471
++L Q+G +NAWG ++A++ L+ +PDG+YVL
Sbjct 533 QNLALQMGFSLDNAWGSIKAIVHLVMRRPDGQYVL 567
> pfa:PF10_0077 eukaryotic translation initiation factor 3 subunit
7, putative; K03251 translation initiation factor 3 subunit
D
Length=543
Score = 319 bits (818), Expect = 1e-86, Method: Compositional matrix adjust.
Identities = 181/464 (39%), Positives = 280/464 (60%), Gaps = 34/464 (7%)
Query 25 LKLHVLQLRIWNGAAPWVSTRGKL---KQTTAAFNEKTKDEEELKLGKKAKSDQKKQQQQ 81
L+ + LR + + K+ + TT AF +K ++E+ L + ++QKKQ+
Sbjct 78 LQFQTVDLRTGQKQKGSIFNKKKIHNKQNTTQAFTQKQQEEDNLYSSRIKTAEQKKQKLL 137
Query 82 QQARYARIHARHRAFSEWSIQPTNDWQVLAELPLQQLHKQQIETPRVTVEDLEWRGTLGV 141
QQA+ ARI+ARHR F+EWSI+PT W V E+ +L K+ I+ + VEDL ++G
Sbjct 138 QQAKTARINARHRIFTEWSIEPTPLWTVECEIMFNELPKKIIKIDNLKVEDLFFKGKSLF 197
Query 142 YSRQADKISSKLPVPMQNLSSNFYYYWVTTQDDEVIRDYMLNGSKQVDVA---------- 191
Y ++ + I+ K P + + + T++D + + + N +K ++
Sbjct 198 YDKKIENINVKNPPLLFHHTKEGINLIPKTKEDHNLINILNNENKNKIISNEEDPNNINN 257
Query 192 ----ATDQVLACLMAAPQSKYSWHLYLTKIDGKIIIDKANGSIVDLLTVNETSIEPPMQD 247
+TDQ+L+CLM+ QSKYSWHL + K +IIIDK + SI+DLLTVNE SI+ P QD
Sbjct 258 IIVISTDQILSCLMSCVQSKYSWHLLIKKKGNQIIIDKDDDSIIDLLTVNENSIDAPTQD 317
Query 248 AENRLNRPAALGFEAVRINQNLRQQLLVPGEVAAEYPPAPFVESGDTPATIAYRYRLFTI 307
EN++N ALG EAV+INQ R+Q+++ ++A E+ P F I YRYR F I
Sbjct 318 NENKINSLQALGIEAVKINQRFRKQVILKNDIAEEFEPGNFNIKNIKINDILYRYRKFQI 377
Query 308 PPRAGPNASRRPINILTRAEVYAKSPMARADLKEAREGDNFIVDGGGYIYTCALNEIEPK 367
PP N +++ ++TR E++++ +G N Y+Y CALNE + K
Sbjct 378 PPLVQSN-NKKTYTLITRGEIHSQI-----------KGLN-----KSYVYVCALNEYDIK 420
Query 368 GQKNWRTLLESQRGALLATEVRNNACKLRKFVTSAMIAGCDELRLGFVARKTPNDNEQHL 427
KNWR+ +E+Q+GALLA E+RNN KL+KF++ A+++GCD+++LGF++RK ND + H
Sbjct 421 SHKNWRSQIENQKGALLANEIRNNTSKLQKFISQALLSGCDDVKLGFISRKNANDPDNHH 480
Query 428 ILTVHNYQTRDLGRQIGLRPENAWGIVRAVLDLLEDKPDGRYVL 471
IL++ +++T+DL QIGL+ +N WGI R V+D L DKP+G YV+
Sbjct 481 ILSIQSHKTKDLSTQIGLKYDNMWGIFRFVIDNLADKPNGNYVI 524
> ath:AT4G20980 eukaryotic translation initiation factor 3 subunit
7, putative / eIF-3 zeta, putative / eIF3d, putative; K03251
translation initiation factor 3 subunit D
Length=591
Score = 219 bits (557), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 145/413 (35%), Positives = 225/413 (54%), Gaps = 50/413 (12%)
Query 75 QKKQQQQQQARYARIHARHR-----------AF-SEWSIQPTNDWQVLAELPLQQLHK-- 120
+K+ ++++AR R++ +R AF S IQP +W +L ++P K
Sbjct 149 KKRDAEKERARRDRLYNNNRNNIHHQRREAAAFKSSVDIQP--EWNMLEQIPFSTFSKLS 206
Query 121 QQIETPRVTVEDLEWRGTLGVYSRQADKISSKLPVPMQNLSSNFYYYWVTTQDDEVIRDY 180
++ P EDL G L Y+R D+I+ K ++ N ++ VTT DD VIR
Sbjct 207 YTVQEP----EDLLLCGGLEYYNRLFDRITPKNERRLERFK-NRNFFKVTTSDDPVIR-- 259
Query 181 MLNGSKQVDVAATDQVLACLMAAPQSKYSWHLYLTKIDGKIIIDKANGSIVDLLTVNETS 240
L + V ATD +LA LM AP+S YSW + + ++ K+ DK +GS +DLL+V+ETS
Sbjct 260 RLAKEDKATVFATDAILAALMCAPRSVYSWDIVIQRVGNKLFFDKRDGSQLDLLSVHETS 319
Query 241 IEPPMQDAENRLNRPAALGFEAVRINQNLRQQLLVPGEVAAEYPPA-PFVESGDTPATIA 299
EP + ++++ +N +LG EA INQN QQ+LV + A PF G+ A++A
Sbjct 320 QEP-LPESKDDINSAHSLGVEAAYINQNFSQQVLVRDGKKETFDEANPFANEGEEIASVA 378
Query 300 YRYRLFTIPPRAGPNASRRPINILTRAEVYAKSPMARADLKEAREGDNFIVDGGGYIYTC 359
YRYR + + ++++ R E+ + ADL R ++
Sbjct 379 YRYRRWKLDDN---------MHLVARCEL-----QSVADLNNQR----------SFLTLN 414
Query 360 ALNEIEPK-GQKNWRTLLESQRGALLATEVRNNACKLRKFVTSAMIAGCDELRLGFVARK 418
ALNE +PK +WR LE+QRGA+LATE++NN KL K+ A++A D +++GFV+R
Sbjct 415 ALNEFDPKYSGVDWRQKLETQRGAVLATELKNNGNKLAKWTAQALLANADMMKIGFVSRV 474
Query 419 TPNDNEQHLILTVHNYQTRDLGRQIGLRPENAWGIVRAVLDLLEDKPDGRYVL 471
P D+ H+IL+V Y+ +D QI L N WGIV++++DL +G+YVL
Sbjct 475 HPRDHFNHVILSVLGYKPKDFAGQINLNTSNMWGIVKSIVDLCMKLSEGKYVL 527
> ath:AT5G44320 eukaryotic translation initiation factor 3 subunit
7, putative / eIF-3 zeta, putative / eIF3d, putative; K03251
translation initiation factor 3 subunit D
Length=588
Score = 217 bits (552), Expect = 9e-56, Method: Compositional matrix adjust.
Identities = 147/418 (35%), Positives = 220/418 (52%), Gaps = 39/418 (9%)
Query 60 KDEEELKLGKKAKSDQKKQQQQQQARYARIHARHR---AF-SEWSIQPTNDWQVLAELPL 115
+DEE ++A+ D+ ++ + IH + R AF S IQP +W +L ++P
Sbjct 138 RDEEVEAKKREAEKDRARRDRLYNNNRNNIHQQRREAAAFKSSVDIQP--EWNMLEQIPF 195
Query 116 QQLHKQQIETPRVTVEDLEWRGTLGVYSRQADKISSKLPVPMQNLSSNFYYYWVTTQDDE 175
K EDL G L Y R D+I+ K ++ + + VTT DD
Sbjct 196 STFSKLSFTVSEP--EDLLLCGGLESYDRSFDRITPKADRRLERFKNRSFK--VTTSDDL 251
Query 176 VIRDYMLNGSKQVDVAATDQVLACLMAAPQSKYSWHLYLTKIDGKIIIDKANGSIVDLLT 235
VIR L + V ATD +LA LM AP+S YSW L + ++ K+ DK +GS +DLL+
Sbjct 252 VIR--RLAKEDKATVFATDAILAALMCAPRSVYSWDLVIQRVGNKLFFDKRDGSPLDLLS 309
Query 236 VNETSIEPPMQDAENRLNRPAALGFEAVRINQNLRQQLLVP-GEVAAEYPPAPFVESGDT 294
V+ETS EP + + ++ +N +LG EA INQN QQ+LV G+ P P V G+
Sbjct 310 VHETSQEP-LPEGKDDINSAHSLGLEAAYINQNFAQQVLVKNGKRETFDEPIPNVNEGEE 368
Query 295 PATIAYRYRLFTIPPRAGPNASRRPINILTRAEVYAKSPMARADLKEAREGDNFIVDGGG 354
A+IAYRYR + + + ++ R E+ + DL R
Sbjct 369 NASIAYRYRRWKLDDS---------MYLVARCEL-----QSTVDLNNQR----------S 404
Query 355 YIYTCALNEIEPK-GQKNWRTLLESQRGALLATEVRNNACKLRKFVTSAMIAGCDELRLG 413
++ ALNE +PK +WR LE+QRGA+LA E++NN KL K+ A++A D +++G
Sbjct 405 FLTLNALNEFDPKYSGVDWRQKLETQRGAVLANELKNNGNKLAKWTAQALLANADMMKIG 464
Query 414 FVARKTPNDNEQHLILTVHNYQTRDLGRQIGLRPENAWGIVRAVLDLLEDKPDGRYVL 471
FV+R P D+ H+IL+V Y+ +D QI L N WGIV++++DL +G+YVL
Sbjct 465 FVSRVHPRDHFNHVILSVLGYKPKDFAGQINLNTNNMWGIVKSIVDLCMKLSEGKYVL 522
> hsa:8664 EIF3D, EIF3S7, MGC126526, MGC17258, eIF3-p66, eIF3-zeta;
eukaryotic translation initiation factor 3, subunit D;
K03251 translation initiation factor 3 subunit D
Length=548
Score = 207 bits (526), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 140/427 (32%), Positives = 220/427 (51%), Gaps = 58/427 (13%)
Query 58 KTKDEEELKLGKKAKSD----QKKQQQQQQARYARIHARHRAFSEWSIQPTNDWQVLAEL 113
K K+ E ++L KK + QK Q+ Q+ R + + R +DW+V E+
Sbjct 127 KQKERERIRLQKKFQKQFGVRQKWDQKSQKPRDSSVEVR------------SDWEVKEEM 174
Query 114 PLQQLHKQ---QIETPRVTVEDLEWRGTLGVYSRQADKISSKLPVPMQNLSSNFYYYWVT 170
QL K ++ P +D+E G L Y + D+I+++ P++++ F+ VT
Sbjct 175 DFPQLMKMRYLEVSEP----QDIECCGALEYYDKAFDRITTRSEKPLRSIKRIFHT--VT 228
Query 171 TQDDEVIRDYMLNGSKQVDVAATDQVLACLMAAPQSKYSWHLYLTKIDGKIIIDKANGSI 230
T DD VIR Q +V ATD +LA LM+ +S YSW + + ++ K+ DK + S
Sbjct 229 TTDDPVIRKL---AKTQGNVFATDAILATLMSCTRSVYSWDIVVQRVGSKLFFDKRDNSD 285
Query 231 VDLLTVNETSIEPPMQDAENRLNRPAALGFEAVRINQNLRQQLLVPGEVAAEYP-PAPFV 289
DLLTV+ET+ EPP QD N N P L EA IN N QQ L G+ +P P PFV
Sbjct 286 FDLLTVSETANEPP-QDEGNSFNSPRNLAMEATYINHNFSQQCLRMGKERYNFPNPNPFV 344
Query 290 ESG---DTPATIAYRYRLFTIPPRAGPNASRRPINILTRAEVYAKSPMARADLKEAREGD 346
E + A++AYRYR + + I+++ R E A ++
Sbjct 345 EDDMDKNEIASVAYRYRRWKLGD---------DIDLIVRCEHDGVMTGANGEV------- 388
Query 347 NFIVDGGGYIYTCALNEIEPKGQK--NWRTLLESQRGALLATEVRNNACKLRKFVTSAMI 404
+I LNE + + +WR L+SQRGA++ATE++NN+ KL ++ A++
Sbjct 389 -------SFINIKTLNEWDSRHCNGVDWRQKLDSQRGAVIATELKNNSYKLARWTCCALL 441
Query 405 AGCDELRLGFVARKTPNDNEQHLILTVHNYQTRDLGRQIGLRPENAWGIVRAVLDLLEDK 464
AG + L+LG+V+R D+ +H+IL ++ + QI L ENAWGI+R V+D+
Sbjct 442 AGSEYLKLGYVSRYHVKDSSRHVILGTQQFKPNEFASQINLSVENAWGILRCVIDICMKL 501
Query 465 PDGRYVL 471
+G+Y++
Sbjct 502 EEGKYLI 508
> mmu:55944 Eif3d, 66/67kDa, AA407891, Eif3s7, MGC115774, eIF3p66;
eukaryotic translation initiation factor 3, subunit D;
K03251 translation initiation factor 3 subunit D
Length=548
Score = 206 bits (525), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 140/427 (32%), Positives = 219/427 (51%), Gaps = 58/427 (13%)
Query 58 KTKDEEELKLGKKAKSD----QKKQQQQQQARYARIHARHRAFSEWSIQPTNDWQVLAEL 113
K K+ E ++L KK + QK Q+ Q+ R + + R +DW+V E+
Sbjct 127 KQKERERIRLQKKFQKQFGVRQKWDQKSQKPRDSSVEVR------------SDWEVKEEM 174
Query 114 PLQQLHKQ---QIETPRVTVEDLEWRGTLGVYSRQADKISSKLPVPMQNLSSNFYYYWVT 170
QL K ++ P +D+E G L Y + D+I+++ P++++ F+ VT
Sbjct 175 DFPQLMKMRYLEVSEP----QDIECCGALEYYDKAFDRITTRSEKPLRSIKRIFHT--VT 228
Query 171 TQDDEVIRDYMLNGSKQVDVAATDQVLACLMAAPQSKYSWHLYLTKIDGKIIIDKANGSI 230
T DD VIR Q +V ATD +LA LM+ +S YSW + + ++ K+ DK + S
Sbjct 229 TTDDPVIRKL---AKTQGNVFATDAILATLMSCTRSVYSWDIVVQRVGSKLFFDKRDNSD 285
Query 231 VDLLTVNETSIEPPMQDAENRLNRPAALGFEAVRINQNLRQQLLVPGEVAAEYP-PAPFV 289
DLLTV+ET+ EPP QD N N P L EA IN N QQ L G +P P PFV
Sbjct 286 FDLLTVSETANEPP-QDEGNSFNSPRNLAMEATYINHNFSQQCLRMGRERYNFPNPNPFV 344
Query 290 ESG---DTPATIAYRYRLFTIPPRAGPNASRRPINILTRAEVYAKSPMARADLKEAREGD 346
E + A++AYRYR + + I+++ R E A ++
Sbjct 345 EDDMDKNEIASVAYRYRRWKLGD---------DIDLIVRCEHDGVMTGANGEV------- 388
Query 347 NFIVDGGGYIYTCALNEIEPKGQK--NWRTLLESQRGALLATEVRNNACKLRKFVTSAMI 404
+I LNE + + +WR L+SQRGA++ATE++NN+ KL ++ A++
Sbjct 389 -------SFINIKTLNEWDSRHCNGVDWRQKLDSQRGAVIATELKNNSYKLARWTCCALL 441
Query 405 AGCDELRLGFVARKTPNDNEQHLILTVHNYQTRDLGRQIGLRPENAWGIVRAVLDLLEDK 464
AG + L+LG+V+R D+ +H+IL ++ + QI L ENAWGI+R V+D+
Sbjct 442 AGSEYLKLGYVSRYHVKDSSRHVILGTQQFKPNEFASQINLSVENAWGILRCVIDICMKL 501
Query 465 PDGRYVL 471
+G+Y++
Sbjct 502 EEGKYLI 508
> xla:380279 eif3d, MGC53375, eif3s7; eukaryotic translation initiation
factor 3, subunit D; K03251 translation initiation
factor 3 subunit D
Length=550
Score = 202 bits (515), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 138/421 (32%), Positives = 221/421 (52%), Gaps = 43/421 (10%)
Query 58 KTKDEEELKLGKKAKSDQKKQQQQQQARYARIHARHRAFSEWSIQPTNDWQVLAELPLQQ 117
K K+ + L+L KK + +Q+ Q A++ R + S++ +DW+V E+ +
Sbjct 127 KQKERDRLRLQKKFQKQFGVRQKWDQRSQAQLKPR-----DSSVEVRSDWEVKEEMDFPR 181
Query 118 LHKQQ-IETPRVTVEDLEWRGTLGVYSRQADKISSKLPVPMQNLSSNFYYYWVTTQDDEV 176
L K + +E T D+E G + Y + D+I+++ P++++ F+ VTT DD V
Sbjct 182 LMKMRYMEVADPT--DIECCGAVEYYDKAFDRITTRNERPLRSIKRIFHT--VTTTDDPV 237
Query 177 IRDYMLNGSKQVDVAATDQVLACLMAAPQSKYSWHLYLTKIDGKIIIDKANGSIVDLLTV 236
IR Q +V ATD +LA LM +S SW + + ++ KI DK + S DLLTV
Sbjct 238 IRKL---AKTQGNVFATDAILATLMCCTRSVNSWDIVVQRVGSKIFFDKRDNSDFDLLTV 294
Query 237 NETSIEPPMQDAENRLNRPAALGFEAVRINQNLRQQLLVPGEVAAEYP-PAPFVESG--- 292
+ET+ EPP QD N LN P L EA IN N QQ L G+ +P P PF+E
Sbjct 295 SETANEPP-QDEVNSLNSPRNLAMEATYINHNFSQQCLRMGKEKHTFPNPNPFIEDDVDK 353
Query 293 DTPATIAYRYRLFTIPPRAGPNASRRPINILTRAEVYAKSPMARADLKEAREGDNFIVDG 352
+ A++AYRYR + + I+++ R E A ++
Sbjct 354 NEVASVAYRYRRWKLGD---------DIDLVVRCEHDGVMTGANGEV------------- 391
Query 353 GGYIYTCALNEIEPK--GQKNWRTLLESQRGALLATEVRNNACKLRKFVTSAMIAGCDEL 410
+I LNE + K +WR L+SQRGA++ATE++NN+ KL ++ A++AG + L
Sbjct 392 -SFINIKTLNEWDSKYCNGVDWRQKLDSQRGAVIATELKNNSYKLARWTCCALLAGSEYL 450
Query 411 RLGFVARKTPNDNEQHLILTVHNYQTRDLGRQIGLRPENAWGIVRAVLDLLEDKPDGRYV 470
+LG+V+R D+ +H++L ++ + QI L ENAWGI+R V+D+ +G+Y+
Sbjct 451 KLGYVSRYNVKDSTRHVVLGTQQFKPNEFANQINLSMENAWGILRCVVDICMKLDEGKYL 510
Query 471 L 471
+
Sbjct 511 I 511
> dre:336498 eif3d, eif3s7, wu:fb12a03, zgc:64097; eukaryotic
translation initiation factor 3, subunit D; K03251 translation
initiation factor 3 subunit D
Length=552
Score = 200 bits (509), Expect = 8e-51, Method: Compositional matrix adjust.
Identities = 143/444 (32%), Positives = 229/444 (51%), Gaps = 56/444 (12%)
Query 45 RGKLKQTTAAFNEKT-------KDEEELKLGKKAKSDQKKQQQQQQARYARIHARHRAFS 97
R K ++T FN +T K+ + ++L KK + +Q+ Q A++ R
Sbjct 107 RDKDRRTLTQFNMQTLPKSAKQKERDRMRLQKKFQKQFGVRQKWDQKSQAQLKPR----- 161
Query 98 EWSIQPTNDWQVLAELPLQQLHKQQ---IETPRVTVEDLEWRGTLGVYSRQADKISSKLP 154
+ S++ +DW+V E+ +L K + + P D+E G L Y + D+I+++
Sbjct 162 DSSVEVRSDWEVKEEMDFPRLMKMRYMDVADPL----DIECCGALEHYDKAFDRITTRNE 217
Query 155 VPMQNLSSNFYYYWVTTQDDEVIRDYMLNGSKQVDVAATDQVLACLMAAPQSKYSWHLYL 214
P++++ F+ VTT DD VIR Q +V ATD +LA LM +S SW + +
Sbjct 218 KPLKSIKRIFHT--VTTTDDPVIRKL---AKTQGNVFATDAILATLMCCTRSVNSWDIIV 272
Query 215 TKIDGKIIIDKANGSIVDLLTVNETSIEPPMQDAENRLNRPAALGFEAVRINQNLRQQLL 274
++ K+ DK + S DLLTV+ET+ EPP QD + LN P L EA IN N QQ L
Sbjct 273 QRVGNKLFFDKRDNSDFDLLTVSETANEPP-QDEGSSLNSPRNLAMEATYINHNFSQQCL 331
Query 275 VPGEVAAEYP-PAPFVESGDTP----ATIAYRYRLFTIPPRAGPNASRRPINILTRAEVY 329
G ++P P PFVE DT A++AYRYR + + I+++ R E
Sbjct 332 RMGGERYKFPNPNPFVEE-DTEKSEVASVAYRYRRWKLG---------EDIDLIVRCEHD 381
Query 330 AKSPMARADLKEAREGDNFIVDGGGYIYTCALNEIEPK--GQKNWRTLLESQRGALLATE 387
A ++ +I LNE + + +WR L+SQRGA+LATE
Sbjct 382 GVMTGANGEV--------------SFINVKTLNEWDSRYCNGVDWRQKLDSQRGAVLATE 427
Query 388 VRNNACKLRKFVTSAMIAGCDELRLGFVARKTPNDNEQHLILTVHNYQTRDLGRQIGLRP 447
++NN+ KL ++ A++AG + L+LG+V+R D+ +H++L ++ + QI L
Sbjct 428 LKNNSYKLARWTCCAILAGSEYLKLGYVSRYHVKDSSRHVVLGTQQFKPNEFASQINLSM 487
Query 448 ENAWGIVRAVLDLLEDKPDGRYVL 471
ENAWGI+R V+D+ +G+Y++
Sbjct 488 ENAWGILRCVIDICRKLDEGKYLI 511
> cel:R08D7.3 eif-3.D; Eukaryotic Initiation Factor family member
(eif-3.D); K03251 translation initiation factor 3 subunit
D
Length=570
Score = 176 bits (446), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 122/387 (31%), Positives = 193/387 (49%), Gaps = 44/387 (11%)
Query 100 SIQPTNDWQVLAELPLQQLHKQQIETPRV----TVEDLEWRGTLGVYSRQADKISSKLPV 155
S+Q +W VL E+ L K + P + + D ++ G+L Y + D++S K +
Sbjct 178 SVQVRPEWVVLEEMNLSAFSK--LALPNIPGGDDIGDHQY-GSLQYYDKTIDRVSVKNSI 234
Query 156 PMQNLSSNFYYYWVTTQDDEVIRDYMLNGSKQVDVAATDQVLACLMAAPQSKYSWHLYLT 215
P+Q + FY VTT +D VI++ G+ +V TD +LA LM AP+S YSW +
Sbjct 235 PLQRCAGVFYN--VTTTEDPVIQELAQGGAG--NVFGTDIILATLMTAPRSVYSWDIVAY 290
Query 216 KIDGKIIIDKANG----SIVDLLTVNETSIEPPMQDAENRLNRPAALGFEAVRINQNLRQ 271
++ K+ DK N + V+ LTV+ETS EPP D N +N L EA INQN R+
Sbjct 291 RVGDKLFFDKRNTRDILNPVETLTVSETSAEPPSFDG-NGINNAKDLATEAFYINQNFRR 349
Query 272 QLLVPGEVAAEYPPA--PFVESGDTPATIAYRYRLFTIPPRAGPNASRRPINILTRAEVY 329
Q++ + + A PF + + AY+YR + + G +P+ ++ R E+
Sbjct 350 QVVKRNDAGFTFKNARAPFEDEETGESGTAYKYRKWNL----GNGVDGKPVELVCRTEL- 404
Query 330 AKSPMARADLKEAREGDNFIVDGGGYIYTC---ALNEIEP--KGQKNWRTLLESQRGALL 384
D I G T A NE + G +WRT L+ Q+GA++
Sbjct 405 ----------------DGVIHGLGNETQTLTIKAFNEWDSTQSGGVDWRTKLDVQKGAVM 448
Query 385 ATEVRNNACKLRKFVTSAMIAGCDELRLGFVARKTPNDNEQHLILTVHNYQTRDLGRQIG 444
ATE++NN+ K+ K+ A++AG D ++LG+V+R + H IL + + I
Sbjct 449 ATEIKNNSAKVAKWTLQALLAGSDTMKLGYVSRNNARSTQNHSILLTQYVKPTEFASNIA 508
Query 445 LRPENAWGIVRAVLDLLEDKPDGRYVL 471
L +N WGI+R V+D + G+Y+L
Sbjct 509 LNMDNCWGILRCVIDSCMKQKPGKYLL 535
> hsa:1829 DSG2, ARVC10, ARVD10, CDHF5, CMD1BB, HDGC, MGC117034,
MGC117036, MGC117037; desmoglein 2; K07597 desmoglein 2
Length=1118
Score = 33.9 bits (76), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 28/101 (27%), Positives = 50/101 (49%), Gaps = 5/101 (4%)
Query 58 KTKDEEELKLGKKAKSDQKKQQQQQQARYARIH-ARHRAFSEWSIQPTNDWQVLAELPLQ 116
K K E+ LG+K +++ +Q+Q+ A ++ A H + + N + + P+
Sbjct 832 KFKTLAEVCLGQKIDINKEIEQRQKPATETSMNTASHSLCEQTMVNSENTYSSGSSFPVP 891
Query 117 QLHKQQIETPRVTVEDLEWRGTLGVYSRQADKISSKLPVPM 157
+ Q+ +VT E + R V SRQA K+++ LP PM
Sbjct 892 K-SLQEANAEKVTQEIVTERS---VSSRQAQKVATPLPDPM 928
> hsa:54888 NSUN2, FLJ20303, MISU, SAKI, TRM4; NOP2/Sun domain
family, member 2 (EC:2.1.1.29)
Length=732
Score = 33.9 bits (76), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 44/100 (44%), Gaps = 3/100 (3%)
Query 298 IAYRYRLFTIPPRAGPNASRRPINILTRAEVYAKSPMARADLKEAREGDNFIVDGGGYIY 357
I + R+ P +G R+ I++ + + L+ A G + +GG +Y
Sbjct 224 ILFYDRILCDVPCSGDGTMRKNIDVWKKWTTLNSLQLHGLQLRIATRGAEQLAEGGRMVY 283
Query 358 -TCALNEIEPKGQKNWRTLLESQRGALLATEVRNNACKLR 396
TC+LN IE + +LLE GAL +V N L+
Sbjct 284 STCSLNPIE--DEAVIASLLEKSEGALELADVSNELPGLK 321
> pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog
A; K13849 reticulocyte-binding protein
Length=3130
Score = 33.1 bits (74), Expect = 2.7, Method: Composition-based stats.
Identities = 25/123 (20%), Positives = 59/123 (47%), Gaps = 16/123 (13%)
Query 57 EKTKDEEELKLGKKAKSDQKKQQQQQQARYARIHARHRAFSEWSIQPTNDWQVLAELPLQ 116
E+ + EEELK ++ + +++KQ+Q Q+ + + R E +++ ++ E L+
Sbjct 2795 ERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELK 2854
Query 117 QLHKQQIETPRVTVEDLEWRGTLGVYSRQADKISSKLPVPMQNLSSNFYYYWVTTQDDEV 176
+ ++++E ++ + + E I SKL M + + +T + DE+
Sbjct 2855 RQEQERLERKKIELAERE------------QHIKSKLESDMVKIIKD----ELTKEKDEI 2898
Query 177 IRD 179
I++
Sbjct 2899 IKN 2901
> xla:446296 pcsk6, PACE4, PACE4AIIa, Xpace4, spc4; proprotein
convertase subtilisin/kexin type 6; K08672 proprotein convertase
subtilisin/kexin type 6 [EC:3.4.21.-]
Length=911
Score = 31.2 bits (69), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 34/73 (46%), Gaps = 4/73 (5%)
Query 342 AREGDNFIVDG-GGYIYTCALNEIEPKGQKNWRTLLESQRGALLATEVRNNACKLRKFVT 400
REGD DG IYT +++ G K W LE + + LAT + A RK VT
Sbjct 304 GREGDYCSCDGYTNSIYTISISSTTENGYKPW--YLE-ECASTLATTYSSGAFYERKIVT 360
Query 401 SAMIAGCDELRLG 413
+ + GC + G
Sbjct 361 TDLRQGCTDDHTG 373
Lambda K H
0.318 0.133 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 22361397460
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40