bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1655_orf1
Length=141
Score E
Sequences producing significant alignments: (Bits) Value
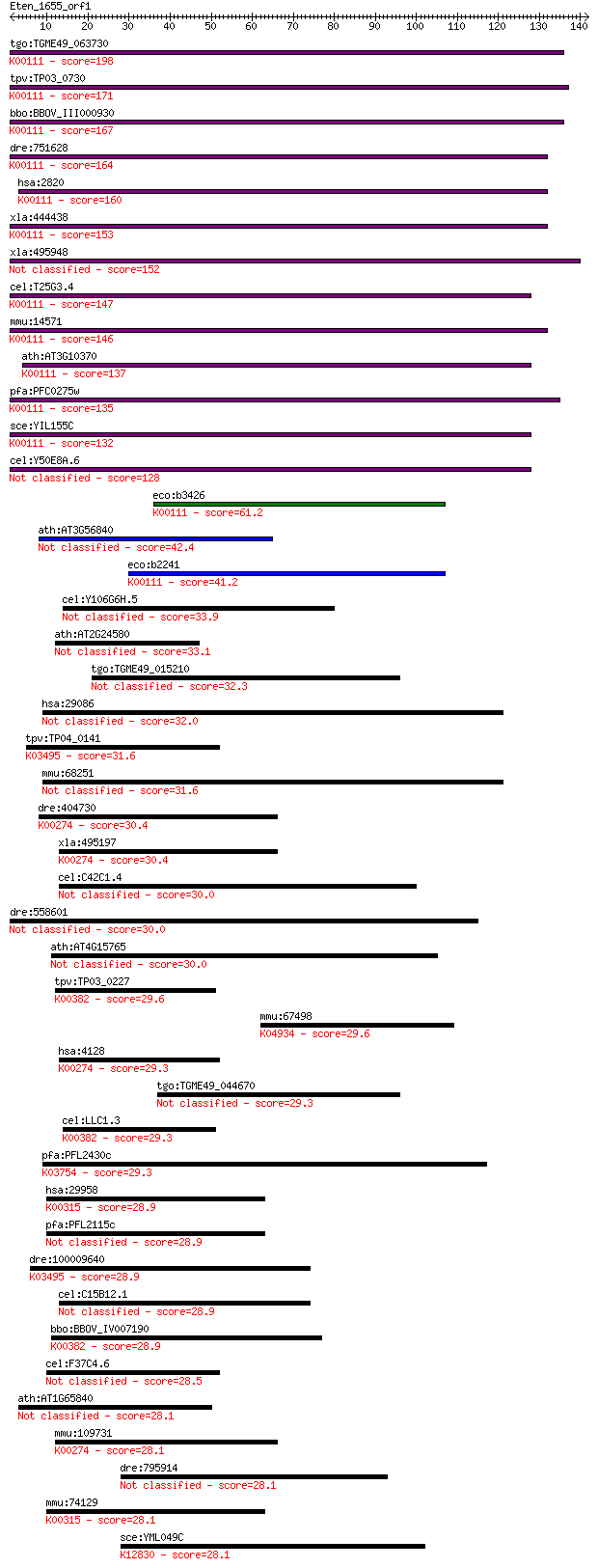
tgo:TGME49_063730 glycerol-3-phosphate dehydrogenase, putative... 198 4e-51
tpv:TP03_0730 glycerol-3-phosphate dehydrogenase (EC:1.1.3.21)... 171 9e-43
bbo:BBOV_III000930 17.m07109; FAD-dependent glycerol-3-phospha... 167 1e-41
dre:751628 gpd2, MGC153729, cb853, fe02f02, im:7147983, wu:fe0... 164 1e-40
hsa:2820 GPD2, GDH2, GPDM, mGPDH; glycerol-3-phosphate dehydro... 160 9e-40
xla:444438 gpd2, MGC83596; glycerol-3-phosphate dehydrogenase ... 153 1e-37
xla:495948 hypothetical LOC495948 152 4e-37
cel:T25G3.4 hypothetical protein; K00111 glycerol-3-phosphate ... 147 8e-36
mmu:14571 Gpd2, AA408484, AI448216, AU021455, AW494132, GPDH, ... 146 2e-35
ath:AT3G10370 SDP6; SDP6 (SUGAR-DEPENDENT 6); glycerol-3-phosp... 137 9e-33
pfa:PFC0275w FAD-dependent glycerol-3-phosphate dehydrogenase,... 135 3e-32
sce:YIL155C GUT2; Gut2p (EC:1.1.5.3); K00111 glycerol-3-phosph... 132 4e-31
cel:Y50E8A.6 hypothetical protein 128 7e-30
eco:b3426 glpD, ECK3412, glyD, JW3389; sn-glycerol-3-phosphate... 61.2 9e-10
ath:AT3G56840 FAD-dependent oxidoreductase family protein 42.4 5e-04
eco:b2241 glpA, ECK2233, JW2235; sn-glycerol-3-phosphate dehyd... 41.2 0.001
cel:Y106G6H.5 hypothetical protein 33.9 0.15
ath:AT2G24580 sarcosine oxidase family protein 33.1 0.28
tgo:TGME49_015210 F-box domain-containing protein 32.3 0.51
hsa:29086 BABAM1, C19orf62, FLJ20571, MERIT40, NBA1; BRISC and... 32.0 0.67
tpv:TP04_0141 glucose inhibited division protein A; K03495 glu... 31.6 0.75
mmu:68251 5430437P03Rik, Babam1, Merit40; RIKEN cDNA 5430437P0... 31.6 0.93
dre:404730 mao, Z-MAO, maob, moa, wu:fb68b05, wu:fo76d11, wu:f... 30.4 1.8
xla:495197 maoa; monoamine oxidase A (EC:1.4.3.4); K00274 mono... 30.4 1.9
cel:C42C1.4 hypothetical protein 30.0 2.2
dre:558601 similar to Peroxisomal sarcosine oxidase (PSO) (L-p... 30.0 2.5
ath:AT4G15765 electron carrier/ monooxygenase/ oxidoreductase 30.0 2.7
tpv:TP03_0227 dihydrolipoamide dehydrogenase (EC:1.8.1.4); K00... 29.6 2.9
mmu:67498 Kcnv1, 2700023A03Rik, vibe; potassium channel, subfa... 29.6 3.6
hsa:4128 MAOA; monoamine oxidase A (EC:1.4.3.4); K00274 monoam... 29.3 4.0
tgo:TGME49_044670 hypothetical protein 29.3 4.3
cel:LLC1.3 hypothetical protein; K00382 dihydrolipoamide dehyd... 29.3 4.4
pfa:PFL2430c eukaryotictranslation initiation factor 2b, subun... 29.3 4.5
hsa:29958 DMGDH, DMGDHD, ME2GLYDH; dimethylglycine dehydrogena... 28.9 5.1
pfa:PFL2115c glucose inhibited division protein A homologue, p... 28.9 5.1
dre:100009640 zgc:158782; K03495 glucose inhibited division pr... 28.9 5.6
cel:C15B12.1 hypothetical protein 28.9 5.6
bbo:BBOV_IV007190 23.m05858; dihydrolipoamide dehydrogenase (E... 28.9 5.6
cel:F37C4.6 hypothetical protein 28.5 7.2
ath:AT1G65840 ATPAO4; ATPAO4 (ARABIDOPSIS THALIANA POLYAMINE O... 28.1 8.5
mmu:109731 Maob, 6330414K01Rik, MAO-B; monoamine oxidase B (EC... 28.1 9.1
dre:795914 si:ch211-278p9.4 28.1 9.6
mmu:74129 Dmgdh, 1200014D15Rik, AI787269, MGC107623; dimethylg... 28.1 9.9
sce:YML049C RSE1; Protein involved in pre-mRNA splicing; compo... 28.1 9.9
> tgo:TGME49_063730 glycerol-3-phosphate dehydrogenase, putative
(EC:1.1.3.21); K00111 glycerol-3-phosphate dehydrogenase
[EC:1.1.5.3]
Length=653
Score = 198 bits (504), Expect = 4e-51, Method: Compositional matrix adjust.
Identities = 90/135 (66%), Positives = 110/135 (81%), Gaps = 0/135 (0%)
Query 1 REEMIAKMKKEKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVHG 60
RE MI +MKKE+FD+LVIGGGA+G GV DA++RGL CLVE DFA GTSS+STKL+HG
Sbjct 60 REAMIERMKKERFDLLVIGGGATGSGVALDAATRGLSCCLVERGDFADGTSSRSTKLIHG 119
Query 61 GIRYLQKACETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAILMPIYKMWQIPYYWFGV 120
G+RYL+KA E D+ QL LV EALEER+H+L AAP++ PLAILMP+YK WQ+PY+W GV
Sbjct 120 GVRYLEKAFEELDFRQLYLVWEALEERSHMLSAAPYMNHPLAILMPVYKYWQVPYFWAGV 179
Query 121 KAYGFLANLVCCGDT 135
K YG L+ LVCC +T
Sbjct 180 KVYGLLSKLVCCFET 194
> tpv:TP03_0730 glycerol-3-phosphate dehydrogenase (EC:1.1.3.21);
K00111 glycerol-3-phosphate dehydrogenase [EC:1.1.5.3]
Length=615
Score = 171 bits (432), Expect = 9e-43, Method: Compositional matrix adjust.
Identities = 76/136 (55%), Positives = 99/136 (72%), Gaps = 0/136 (0%)
Query 1 REEMIAKMKKEKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVHG 60
R EM+ K+K EKFDVL+IGGG +G V D ++RGL LVE+NDFASGTSSKSTKL+HG
Sbjct 48 RNEMLNKLKTEKFDVLIIGGGCTGTSVALDLATRGLNCALVEANDFASGTSSKSTKLLHG 107
Query 61 GIRYLQKACETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAILMPIYKMWQIPYYWFGV 120
GIRYL+ A D+ +L V +ALEERAHL+ + P+ P+ I+MPIY+ WQ+PY+W +
Sbjct 108 GIRYLENAIYKLDFRELRFVWKALEERAHLIGSLPYANYPIPIVMPIYQYWQLPYFWVNI 167
Query 121 KAYGFLANLVCCGDTR 136
K Y LA CC +T+
Sbjct 168 KIYELLARFFCCNETK 183
> bbo:BBOV_III000930 17.m07109; FAD-dependent glycerol-3-phosphate
dehydrogenase (EC:1.1.99.5); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=619
Score = 167 bits (422), Expect = 1e-41, Method: Compositional matrix adjust.
Identities = 77/135 (57%), Positives = 98/135 (72%), Gaps = 0/135 (0%)
Query 1 REEMIAKMKKEKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVHG 60
R M ++ E+FDVLV+GGG +G V D ++RGLK LVE+NDFASGTSSKSTKL+HG
Sbjct 48 RSLMSRALRDEEFDVLVVGGGCTGSAVALDCATRGLKCALVEANDFASGTSSKSTKLLHG 107
Query 61 GIRYLQKACETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAILMPIYKMWQIPYYWFGV 120
GIRYL+ A FD +L V +ALEERAHL+ AAP P+ I++PIY++WQIPY+WF +
Sbjct 108 GIRYLESALLHFDIKELRFVWKALEERAHLIFAAPFANPPIPIVLPIYQLWQIPYFWFNI 167
Query 121 KAYGFLANLVCCGDT 135
K Y LA CC +T
Sbjct 168 KVYELLARFFCCNET 182
> dre:751628 gpd2, MGC153729, cb853, fe02f02, im:7147983, wu:fe02f02,
zgc:153729; glycerol-3-phosphate dehydrogenase 2 (mitochondrial)
(EC:1.1.5.3); K00111 glycerol-3-phosphate dehydrogenase
[EC:1.1.5.3]
Length=536
Score = 164 bits (414), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 74/132 (56%), Positives = 99/132 (75%), Gaps = 1/132 (0%)
Query 1 REEMIAKMKK-EKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVH 59
REE ++ ++ E+FDVLV+GGGA+G G DA +R LK+ LVE +DF+SGTSS+STKL+H
Sbjct 56 REEQLSTLRNTEEFDVLVVGGGATGSGCALDAVTRNLKTALVERSDFSSGTSSRSTKLIH 115
Query 60 GGIRYLQKACETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAILMPIYKMWQIPYYWFG 119
GG+RYLQKA D+ Q +V EAL ERA+LL APH++ PL I++P+YK WQ+PYYW G
Sbjct 116 GGVRYLQKAIMKLDYEQYMMVKEALHERANLLEIAPHLSAPLPIMLPVYKWWQLPYYWAG 175
Query 120 VKAYGFLANLVC 131
+K Y +A C
Sbjct 176 IKMYDLVAGSQC 187
> hsa:2820 GPD2, GDH2, GPDM, mGPDH; glycerol-3-phosphate dehydrogenase
2 (mitochondrial) (EC:1.1.5.3); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=727
Score = 160 bits (406), Expect = 9e-40, Method: Compositional matrix adjust.
Identities = 71/129 (55%), Positives = 94/129 (72%), Gaps = 0/129 (0%)
Query 3 EMIAKMKKEKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVHGGI 62
+++ +FD+LVIGGGA+G G DA +RGLK+ LVE +DF+SGTSS+STKL+HGG+
Sbjct 60 QLLTLQNTSEFDILVIGGGATGSGCALDAVTRGLKTALVERDDFSSGTSSRSTKLIHGGV 119
Query 63 RYLQKACETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAILMPIYKMWQIPYYWFGVKA 122
RYLQKA D Q +V EAL ERA+LL APH++ PL I++P+YK WQ+PYYW G+K
Sbjct 120 RYLQKAIMKLDIEQYRMVKEALHERANLLEIAPHLSAPLPIMLPVYKWWQLPYYWVGIKL 179
Query 123 YGFLANLVC 131
Y +A C
Sbjct 180 YDLVAGSNC 188
> xla:444438 gpd2, MGC83596; glycerol-3-phosphate dehydrogenase
2 (mitochondrial) (EC:1.1.5.3); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=725
Score = 153 bits (387), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 79/132 (59%), Positives = 98/132 (74%), Gaps = 1/132 (0%)
Query 1 REEMIAKMKKEK-FDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVH 59
RE + ++K K FDVLVIGGGA+G G DA SRGLK+ LVE +DF+SGTSS+STKL+H
Sbjct 55 REAQLLTLQKTKEFDVLVIGGGATGCGCALDAVSRGLKTALVEKDDFSSGTSSRSTKLIH 114
Query 60 GGIRYLQKACETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAILMPIYKMWQIPYYWFG 119
GG+RYLQKA D Q LV EAL ERA+LL APH++ PL I++P+YK WQ+PYYW G
Sbjct 115 GGVRYLQKAIMNLDIEQYKLVKEALHERANLLEIAPHLSAPLPIMLPVYKWWQLPYYWIG 174
Query 120 VKAYGFLANLVC 131
+KAY +A C
Sbjct 175 IKAYDLVAGSQC 186
> xla:495948 hypothetical LOC495948
Length=216
Score = 152 bits (383), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 80/140 (57%), Positives = 102/140 (72%), Gaps = 4/140 (2%)
Query 1 REEMIAKMKKEK-FDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVH 59
RE + ++K K FDVLVIGGGA+G G DA SRGLK+ +VE +DF+SGTSS+STKL+H
Sbjct 55 REAQLLTLQKTKEFDVLVIGGGATGCGCALDAVSRGLKTAIVERDDFSSGTSSRSTKLIH 114
Query 60 GGIRYLQKACETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAILMPIYKMWQIPYYWFG 119
GG+RYLQKA D Q LV EAL ERA+LL APH++ PL I++P+YK WQ+PYYW G
Sbjct 115 GGVRYLQKAIMNLDIEQYKLVKEALYERANLLEIAPHLSAPLPIMLPVYKWWQLPYYWIG 174
Query 120 VKAYGFLANLVCCGDTRNSF 139
+KAY +A C R+S+
Sbjct 175 IKAYDLVAGSQCL---RSSY 191
> cel:T25G3.4 hypothetical protein; K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=722
Score = 147 bits (372), Expect = 8e-36, Method: Compositional matrix adjust.
Identities = 74/128 (57%), Positives = 100/128 (78%), Gaps = 1/128 (0%)
Query 1 REEMIAKMKK-EKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVH 59
R++++ + K E+FDVL+IGGGA+G GV DA +RGLK+ LVE +DF+SGTSS+STKL+H
Sbjct 62 RKDILTNLSKGEEFDVLIIGGGATGAGVALDAQTRGLKTALVELDDFSSGTSSRSTKLIH 121
Query 60 GGIRYLQKACETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAILMPIYKMWQIPYYWFG 119
GG+RYLQ A D Q +V EAL ER +LL APH++ PL I++PIYK+WQ+PYYW G
Sbjct 122 GGVRYLQAAIMKLDLEQYRMVKEALFERHNLLEIAPHLSSPLPIMLPIYKLWQVPYYWSG 181
Query 120 VKAYGFLA 127
+KAY F++
Sbjct 182 IKAYDFVS 189
> mmu:14571 Gpd2, AA408484, AI448216, AU021455, AW494132, GPDH,
Gdm1, Gpd-m, Gpdh-m, TISP38; glycerol phosphate dehydrogenase
2, mitochondrial (EC:1.1.5.3); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=727
Score = 146 bits (368), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 74/132 (56%), Positives = 96/132 (72%), Gaps = 1/132 (0%)
Query 1 REEMIAKMKK-EKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVH 59
RE + +K +FD+LVIGGGA+G G DA +RGLK+ LVE +DF+SGTSS+STKL+H
Sbjct 57 REAQLMTLKNTPEFDILVIGGGATGCGCALDAVTRGLKTALVERDDFSSGTSSRSTKLIH 116
Query 60 GGIRYLQKACETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAILMPIYKMWQIPYYWFG 119
GG+RYLQKA D Q +V EAL ERA+LL APH++ PL I++P+YK WQ+PYYW G
Sbjct 117 GGVRYLQKAIMNLDVEQYRMVKEALHERANLLEIAPHLSAPLPIMLPLYKWWQLPYYWVG 176
Query 120 VKAYGFLANLVC 131
+K Y +A C
Sbjct 177 IKMYDLVAGSQC 188
> ath:AT3G10370 SDP6; SDP6 (SUGAR-DEPENDENT 6); glycerol-3-phosphate
dehydrogenase (EC:1.1.5.3); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=629
Score = 137 bits (346), Expect = 9e-33, Method: Compositional matrix adjust.
Identities = 64/124 (51%), Positives = 85/124 (68%), Gaps = 0/124 (0%)
Query 4 MIAKMKKEKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVHGGIR 63
+IA + DVLVIGGGA+G GV DA +RGL+ LVE DF+SGTSS+STKL+HGG+R
Sbjct 65 LIAATASDPLDVLVIGGGATGSGVALDAVTRGLRVGLVEREDFSSGTSSRSTKLIHGGVR 124
Query 64 YLQKACETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAILMPIYKMWQIPYYWFGVKAY 123
YL+KA D+GQL LV ALEER L+ APH+ L + P + +++ Y+W G+K Y
Sbjct 125 YLEKAVFNLDYGQLKLVFHALEERKQLIENAPHLCHALPCMTPCFDWFEVIYFWMGLKMY 184
Query 124 GFLA 127
+A
Sbjct 185 DLVA 188
> pfa:PFC0275w FAD-dependent glycerol-3-phosphate dehydrogenase,
putative; K00111 glycerol-3-phosphate dehydrogenase [EC:1.1.5.3]
Length=653
Score = 135 bits (341), Expect = 3e-32, Method: Composition-based stats.
Identities = 76/134 (56%), Positives = 104/134 (77%), Gaps = 0/134 (0%)
Query 1 REEMIAKMKKEKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVHG 60
R EMI K+++ ++D+L+IGGGA+G G+ DA++RGLK LVE NDF+SGTSSKSTKL+HG
Sbjct 46 RNEMIKKLQENQYDLLIIGGGATGAGLALDAATRGLKCALVEKNDFSSGTSSKSTKLLHG 105
Query 61 GIRYLQKACETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAILMPIYKMWQIPYYWFGV 120
GIRYL+ A D+ +L V EAL ERAH ++ AP+++ P++ILMPIYK WQ+PY+ + +
Sbjct 106 GIRYLENAVNNLDFTELYFVWEALAERAHTMKIAPYLSRPVSILMPIYKYWQVPYFSYNI 165
Query 121 KAYGFLANLVCCGD 134
K Y LA+LVC D
Sbjct 166 KIYDLLADLVCYFD 179
> sce:YIL155C GUT2; Gut2p (EC:1.1.5.3); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=649
Score = 132 bits (332), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 71/128 (55%), Positives = 90/128 (70%), Gaps = 1/128 (0%)
Query 1 REEMIAKM-KKEKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVH 59
R +++ ++ K +FDVL+IGGGA+G G DA++RGL LVE DFASGTSSKSTK++H
Sbjct 55 RRDLLDRLAKTHQFDVLIIGGGATGTGCALDAATRGLNVALVEKGDFASGTSSKSTKMIH 114
Query 60 GGIRYLQKACETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAILMPIYKMWQIPYYWFG 119
GG+RYL+KA F QL LV EAL ER HL+ APH+ L IL+PIY WQ+PY + G
Sbjct 115 GGVRYLEKAFWEFSKAQLDLVIEALNERKHLINTAPHLCTVLPILIPIYSTWQVPYIYMG 174
Query 120 VKAYGFLA 127
K Y F A
Sbjct 175 CKFYDFFA 182
> cel:Y50E8A.6 hypothetical protein
Length=609
Score = 128 bits (321), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 67/128 (52%), Positives = 93/128 (72%), Gaps = 1/128 (0%)
Query 1 REEMIAKMKKEK-FDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVH 59
RE +I +K EK FDVLVIGGG++G GV DA +RGLK+ +VE D+ SGTSSKS+KL+H
Sbjct 50 RESIIESLKTEKRFDVLVIGGGSAGAGVALDAQTRGLKTAMVEYGDYCSGTSSKSSKLLH 109
Query 60 GGIRYLQKACETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAILMPIYKMWQIPYYWFG 119
GG++YL+ A + FD+ +V E L ER +++++AP +++ +L+P YK WQ YYW G
Sbjct 110 GGVKYLETALKEFDYEHYQIVQEGLNERLNIMKSAPFLSQTFPVLVPTYKWWQKIYYWGG 169
Query 120 VKAYGFLA 127
VK Y FLA
Sbjct 170 VKVYDFLA 177
> eco:b3426 glpD, ECK3412, glyD, JW3389; sn-glycerol-3-phosphate
dehydrogenase, aerobic, FAD/NAD(P)-binding (EC:1.1.5.3);
K00111 glycerol-3-phosphate dehydrogenase [EC:1.1.5.3]
Length=501
Score = 61.2 bits (147), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 32/71 (45%), Positives = 43/71 (60%), Gaps = 7/71 (9%)
Query 36 LKSCLVESNDFASGTSSKSTKLVHGGIRYLQKACETFDWGQLSLVCEALEERAHLLRAAP 95
L ++E+ D A TSS S+KL+HGG+RYL+ + LV EAL ER LL+ AP
Sbjct 27 LSVLMLEAQDLACATSSASSKLIHGGLRYLEHY-------EFRLVSEALAEREVLLKMAP 79
Query 96 HIAEPLAILMP 106
HIA P+ +P
Sbjct 80 HIAFPMRFRLP 90
> ath:AT3G56840 FAD-dependent oxidoreductase family protein
Length=483
Score = 42.4 bits (98), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 21/58 (36%), Positives = 38/58 (65%), Gaps = 1/58 (1%)
Query 8 MKKEKFDVLVIGGGASGVGVLFDASSRGLKSCLVE-SNDFASGTSSKSTKLVHGGIRY 64
+ KE+ D +VIG G G+ V + S RG + +++ ++ F + TSS+++++VH GI Y
Sbjct 75 IAKERVDTVVIGAGVVGLAVARELSLRGREVLILDAASSFGTVTSSRNSEVVHAGIYY 132
> eco:b2241 glpA, ECK2233, JW2235; sn-glycerol-3-phosphate dehydrogenase
(anaerobic), large subunit, FAD/NAD(P)-binding (EC:1.1.5.3);
K00111 glycerol-3-phosphate dehydrogenase [EC:1.1.5.3]
Length=542
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 27/80 (33%), Positives = 38/80 (47%), Gaps = 11/80 (13%)
Query 30 DASSRGLKSCLVESNDFASGTSSKSTKLVHGGIRYLQKACETFDWGQLSLVCEALEERAH 89
D + RGL+ LVE +D A+G + ++ L+H G RY E+ E + E
Sbjct 26 DCALRGLRVILVERHDIATGATGRNHGLLHSGARYAVTDAES--------ARECISENQI 77
Query 90 LLRAAPHIAEP---LAILMP 106
L R A H EP L I +P
Sbjct 78 LKRIARHCVEPTNGLFITLP 97
> cel:Y106G6H.5 hypothetical protein
Length=855
Score = 33.9 bits (76), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 31/67 (46%), Gaps = 1/67 (1%)
Query 14 DVLVIGGGASGVGVLFDASSRGLKSCLVESNDFA-SGTSSKSTKLVHGGIRYLQKACETF 72
DV+V GGG SG + + + RG K LVE + SG + S LV I + + +
Sbjct 23 DVVVCGGGISGTSIAYHLAKRGKKVALVEKDSIGCSGATGLSAGLVSSPIFWQDTSLQAI 82
Query 73 DWGQLSL 79
L L
Sbjct 83 AQASLDL 89
> ath:AT2G24580 sarcosine oxidase family protein
Length=416
Score = 33.1 bits (74), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 12 KFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDF 46
+FDV+V+G G G + + RG K+ L+E DF
Sbjct 8 RFDVIVVGAGVMGSSAAYQLAKRGQKTLLLEQFDF 42
> tgo:TGME49_015210 F-box domain-containing protein
Length=683
Score = 32.3 bits (72), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 37/75 (49%), Gaps = 3/75 (4%)
Query 21 GASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVHGGIRYLQKACETFDWGQLSLV 80
GA+G D SRG ++ + F S S + T+ HG + + ++A + G +LV
Sbjct 33 GAAGCTSAQDDVSRGYEAKPAQKRRFFSALSEEETEGAHGTLPH-REAVAREETG--TLV 89
Query 81 CEALEERAHLLRAAP 95
C+ E +HL R P
Sbjct 90 CDDYSEHSHLKRKDP 104
> hsa:29086 BABAM1, C19orf62, FLJ20571, MERIT40, NBA1; BRISC and
BRCA1 A complex member 1
Length=329
Score = 32.0 bits (71), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 27/112 (24%), Positives = 49/112 (43%), Gaps = 6/112 (5%)
Query 9 KKEKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVHGGIRYLQKA 68
K +F ++V+ + + L + R L SCL + T+S ST + G +Q+
Sbjct 140 KSHEFALVVVNDDTAWLSGL-TSDPRELCSCL-----YDLETASCSTFNLEGLFSLIQQK 193
Query 69 CETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAILMPIYKMWQIPYYWFGV 120
E + + R L+ + P ++ P+ KM+Q PY++F V
Sbjct 194 TELPVTENVQTIPPPYVVRTILVYSRPPCQPQFSLTEPMKKMFQCPYFFFDV 245
> tpv:TP04_0141 glucose inhibited division protein A; K03495 glucose
inhibited division protein A
Length=615
Score = 31.6 bits (70), Expect = 0.75, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 5 IAKMKKEKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTS 51
+ K KFDVLV+GGG SG+ ++ G + L+ N + G S
Sbjct 25 VVKPPYPKFDVLVVGGGHSGIEAATASARIGANTLLITPNLSSIGIS 71
> mmu:68251 5430437P03Rik, Babam1, Merit40; RIKEN cDNA 5430437P03
gene
Length=333
Score = 31.6 bits (70), Expect = 0.93, Method: Compositional matrix adjust.
Identities = 26/112 (23%), Positives = 50/112 (44%), Gaps = 6/112 (5%)
Query 9 KKEKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVHGGIRYLQKA 68
K +F ++V+ ++ + L + R L SCL + T+S ST + G +Q+
Sbjct 144 KSHEFALVVVNDDSAWLSGL-TSDPRELCSCL-----YDLETASCSTFNLEGLFSLIQQK 197
Query 69 CETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAILMPIYKMWQIPYYWFGV 120
E + + R L+ + P ++ P+ KM+Q PY++F +
Sbjct 198 TELPVTENVQTIPPPYVVRTILVYSRPPCQPQFSLTEPMKKMFQCPYFFFDI 249
> dre:404730 mao, Z-MAO, maob, moa, wu:fb68b05, wu:fo76d11, wu:fq38g06,
zgc:85761; monoamine oxidase (EC:1.4.3.4); K00274
monoamine oxidase [EC:1.4.3.4]
Length=522
Score = 30.4 bits (67), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 29/62 (46%), Gaps = 4/62 (6%)
Query 8 MKKEKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTS----SKSTKLVHGGIR 63
M +DV+VIGGG SG+ GL ++E+ G + +K TK V G
Sbjct 1 MTANAYDVIVIGGGISGLSAAKLLVDSGLNPVVLEARSRVGGRTYTVQNKETKWVDLGGA 60
Query 64 YL 65
Y+
Sbjct 61 YI 62
> xla:495197 maoa; monoamine oxidase A (EC:1.4.3.4); K00274 monoamine
oxidase [EC:1.4.3.4]
Length=521
Score = 30.4 bits (67), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 29/57 (50%), Gaps = 4/57 (7%)
Query 13 FDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTS----SKSTKLVHGGIRYL 65
+DV+VIGGG SG+ S G+ ++E+ D G + +K K V G Y+
Sbjct 5 YDVIVIGGGISGLSAAKLLSESGISVVVLEARDRVGGRTHTVRNKQVKYVDLGGAYV 61
> cel:C42C1.4 hypothetical protein
Length=1259
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 24/87 (27%), Positives = 43/87 (49%), Gaps = 8/87 (9%)
Query 13 FDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVHGGIRYLQKACETF 72
F ++ G A+G G + +A + +S A+ TS + L H G++YL+K E
Sbjct 387 FSTPILKGSATG-GRVSEAVKAVAEFACYQSVTSATSTSERLIVLAHDGLKYLEKVHE-- 443
Query 73 DWGQLSLVCEALEERAHLLRAAPHIAE 99
W QL E +ER ++ A+ ++ +
Sbjct 444 -WEQL----ERYKERNDVISASLYLLD 465
> dre:558601 similar to Peroxisomal sarcosine oxidase (PSO) (L-pipecolate
oxidase) (L-pipecolic acid oxidase)
Length=508
Score = 30.0 bits (66), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 30/122 (24%), Positives = 53/122 (43%), Gaps = 8/122 (6%)
Query 1 REEMIA-KMKKEKFDVLVIGGGASGVGVLFDASSRGLKSCLVESN--DFASGTSSKSTKL 57
RE+++ +M + FD +VIG G G + + K+ L+E + G+S T++
Sbjct 114 REKVLKLEMSSKVFDCIVIGAGIQGSCTAYQLAKNKQKTLLLEQFVLPHSRGSSHGQTRI 173
Query 58 VHGGIR---YLQKACETFD-WGQLSLVCEA-LEERAHLLRAAPHIAEPLAILMPIYKMWQ 112
+ Y Q E+++ W +L L R LL P +E + L + +
Sbjct 174 IRKAYEEDFYTQMMQESYELWAELEKEAGVELYRRTGLLVMGPEKSEGFSKLKDTMQRHK 233
Query 113 IP 114
IP
Sbjct 234 IP 235
> ath:AT4G15765 electron carrier/ monooxygenase/ oxidoreductase
Length=271
Score = 30.0 bits (66), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 24/97 (24%), Positives = 47/97 (48%), Gaps = 10/97 (10%)
Query 11 EKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSK---STKLVHGGIRYLQK 67
E+ D++++GGG +G+ +G+KS ++E ++ + L+ GI+ ++
Sbjct 2 EELDIVIVGGGIAGLATSLALHRKGIKSVVLERSESVRSEGAAFWIRDVLIEKGIKR-RE 60
Query 68 ACETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAIL 104
+ +G++ V R L+RA H A PL L
Sbjct 61 SVGPASYGEVRGVL-----RNDLVRALAH-ALPLGTL 91
> tpv:TP03_0227 dihydrolipoamide dehydrogenase (EC:1.8.1.4); K00382
dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=499
Score = 29.6 bits (65), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 12 KFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGT 50
K+D+LV+G G G + A+ GLK +VE GT
Sbjct 23 KYDLLVLGAGPGGYTMAIKAAQHGLKVGVVEKRPTLGGT 61
> mmu:67498 Kcnv1, 2700023A03Rik, vibe; potassium channel, subfamily
V, member 1; K04934 potassium channel subfamily V member
1
Length=503
Score = 29.6 bits (65), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 1/47 (2%)
Query 62 IRYLQKACETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAILMPIY 108
+ L+ C ++ G+ L +++R H LR P+I + LAIL P Y
Sbjct 245 LEILEYVCISWFTGEFVLRFLCVKDRCHFLRKVPNIIDLLAIL-PFY 290
> hsa:4128 MAOA; monoamine oxidase A (EC:1.4.3.4); K00274 monoamine
oxidase [EC:1.4.3.4]
Length=527
Score = 29.3 bits (64), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 13 FDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTS 51
FDV+VIGGG SG+ + G+ ++E+ D G +
Sbjct 14 FDVVVIGGGISGLSAAKLLTEYGVSVLVLEARDRVGGRT 52
> tgo:TGME49_044670 hypothetical protein
Length=492
Score = 29.3 bits (64), Expect = 4.3, Method: Composition-based stats.
Identities = 23/59 (38%), Positives = 29/59 (49%), Gaps = 2/59 (3%)
Query 37 KSCLVESNDFASGTSSKSTKLVHGGIRYLQKACETFDWGQLSLVCEALEERAHLLRAAP 95
++C V S FASGT S+S L+ G A ET + L C ER+ LL AP
Sbjct 178 RACNVPSPSFASGTDSRSCLLL-SGTSSSAPAFETPISHECPLACHK-SERSVLLPPAP 234
> cel:LLC1.3 hypothetical protein; K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=495
Score = 29.3 bits (64), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 14 DVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGT 50
D++VIGGG G A+ G+K+ VE N GT
Sbjct 31 DLVVIGGGPGGYVAAIKAAQLGMKTVCVEKNATLGGT 67
> pfa:PFL2430c eukaryotictranslation initiation factor 2b, subunit
2, putative; K03754 translation initiation factor eIF-2B
subunit beta
Length=679
Score = 29.3 bits (64), Expect = 4.5, Method: Composition-based stats.
Identities = 26/110 (23%), Positives = 47/110 (42%), Gaps = 14/110 (12%)
Query 9 KKEKFDVLVIGGGASGVGVLFDA--SSRGLKSCLVESNDFASGTSSKSTKLVHGGIRYLQ 66
KK+ V+V+GG + G S G+ + + S+ K TK+V G +
Sbjct 510 KKDGISVIVVGGDINRNGFRMSQLLSDDGVDTTYI-SDAAVFAVIPKVTKVVLGSVAVSS 568
Query 67 KACETFDWGQLSLVCEALEERAHLLRAAPHIAEPLAILMPIYKMWQIPYY 116
G ++ C A HL ++P+ I++P++K+ +P Y
Sbjct 569 SGGTITKMGGYNIACSA-----HLY------SKPVIIVLPLFKLIYVPLY 607
> hsa:29958 DMGDH, DMGDHD, ME2GLYDH; dimethylglycine dehydrogenase
(EC:1.5.99.2); K00315 dimethylglycine dehydrogenase [EC:1.5.99.2]
Length=866
Score = 28.9 bits (63), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 16/57 (28%), Positives = 32/57 (56%), Gaps = 4/57 (7%)
Query 10 KEKFDVLVIGGGASGVGVLFDASSRGLKS-CLVESNDFASGTSSKS---TKLVHGGI 62
K++ + ++IGGG GV + + + G+K L+E ++ +G++ + T H GI
Sbjct 47 KDRAETVIIGGGCVGVSLAYHLAKAGMKDVVLLEKSELTAGSTWHAAGLTTYFHPGI 103
> pfa:PFL2115c glucose inhibited division protein A homologue,
putative
Length=972
Score = 28.9 bits (63), Expect = 5.1, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 27/53 (50%), Gaps = 2/53 (3%)
Query 10 KEKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVHGGI 62
K+ +DV+VIGGG SG + ++ + LV N G S + + GGI
Sbjct 86 KKNYDVIVIGGGHSGCEASYISAKSNAMTLLVTQNKETIGEMSCNPSI--GGI 136
> dre:100009640 zgc:158782; K03495 glucose inhibited division
protein A
Length=659
Score = 28.9 bits (63), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 32/68 (47%), Gaps = 0/68 (0%)
Query 6 AKMKKEKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTSSKSTKLVHGGIRYL 65
+ + + +DV+V+GGG +G + AS G K+ L+ G S + L G +L
Sbjct 20 SNIHTQHYDVIVVGGGHAGSEAVAAASRIGAKTLLITQKIQTIGALSCNPSLGGVGKGHL 79
Query 66 QKACETFD 73
+ + D
Sbjct 80 VREIDALD 87
> cel:C15B12.1 hypothetical protein
Length=384
Score = 28.9 bits (63), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 16/66 (24%), Positives = 32/66 (48%), Gaps = 5/66 (7%)
Query 13 FDVLVIGGGASGVGVLFDASSRGLKSCLVESNDF-----ASGTSSKSTKLVHGGIRYLQK 67
+DV+V+G G G ++ GLK+ L+E + +S S+ T+ H + Y+
Sbjct 5 YDVVVVGAGIFGSCTAYNCQKIGLKTLLLEQFELGHKNGSSHGKSRITRYAHTEVEYVDL 64
Query 68 ACETFD 73
+ ++
Sbjct 65 VGDAYN 70
> bbo:BBOV_IV007190 23.m05858; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=481
Score = 28.9 bits (63), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 22/80 (27%), Positives = 31/80 (38%), Gaps = 14/80 (17%)
Query 11 EKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGT--------------SSKSTK 56
K+D+ VIGGG G A+ GLK ++ GT +S K
Sbjct 22 HKYDLAVIGGGPGGYTTAIKAAQYGLKVACIDRRTTLGGTCLNVGCIPSKCLLNTSHHYK 81
Query 57 LVHGGIRYLQKACETFDWGQ 76
H GI ++ F+ GQ
Sbjct 82 ASHDGIAGIKFTNVEFNHGQ 101
> cel:F37C4.6 hypothetical protein
Length=544
Score = 28.5 bits (62), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 12/42 (28%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 10 KEKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTS 51
K+ +D ++IGGG +G+ + G K C++E G +
Sbjct 12 KQSYDAIIIGGGHNGLTAAAYLTKAGKKVCVLERRHVVGGAA 53
> ath:AT1G65840 ATPAO4; ATPAO4 (ARABIDOPSIS THALIANA POLYAMINE
OXIDASE 4); amine oxidase/ polyamine oxidase (EC:1.5.3.11)
Length=497
Score = 28.1 bits (61), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 23/47 (48%), Gaps = 0/47 (0%)
Query 3 EMIAKMKKEKFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASG 49
E++ K + V+VIG G SG+ + S K ++ES D G
Sbjct 19 ELMQKQNNVQPSVIVIGSGISGLAAARNLSEASFKVTVLESRDRIGG 65
> mmu:109731 Maob, 6330414K01Rik, MAO-B; monoamine oxidase B (EC:1.4.3.4);
K00274 monoamine oxidase [EC:1.4.3.4]
Length=520
Score = 28.1 bits (61), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 29/58 (50%), Gaps = 4/58 (6%)
Query 12 KFDVLVIGGGASGVGVLFDASSRGLKSCLVESNDFASGTS----SKSTKLVHGGIRYL 65
K DV+V+GGG SG+ GL ++E+ D G + +K+ K V G Y+
Sbjct 4 KSDVIVVGGGISGMAAAKLLHDCGLSVVVLEARDRVGGRTYTIRNKNVKYVDLGGSYV 61
> dre:795914 si:ch211-278p9.4
Length=1008
Score = 28.1 bits (61), Expect = 9.6, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 31/68 (45%), Gaps = 8/68 (11%)
Query 28 LFDASSRGLKSCLVESNDFA---SGTSSKSTKLVHGGIRYLQKACETFDWGQLSLVCEAL 84
LF+ L C +E+N FA SG S + IR L F + L+L+C+ L
Sbjct 885 LFELEKLELDDCSIENNGFADLVSGLKSNPSN-----IRELNLNSNKFIFSGLNLLCDLL 939
Query 85 EERAHLLR 92
++ LR
Sbjct 940 RDQQCKLR 947
> mmu:74129 Dmgdh, 1200014D15Rik, AI787269, MGC107623; dimethylglycine
dehydrogenase precursor (EC:1.5.99.2); K00315 dimethylglycine
dehydrogenase [EC:1.5.99.2]
Length=869
Score = 28.1 bits (61), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 15/57 (26%), Positives = 32/57 (56%), Gaps = 4/57 (7%)
Query 10 KEKFDVLVIGGGASGVGVLFDASSRGLKS-CLVESNDFASGTSSKS---TKLVHGGI 62
K++ + ++IGGG GV + + + G++ L+E ++ +G++ + T H GI
Sbjct 40 KDRAETVIIGGGCVGVSLAYHLAKAGMRDVVLMEKSELTAGSTWHAAGLTTYFHPGI 96
> sce:YML049C RSE1; Protein involved in pre-mRNA splicing; component
of the pre-spliceosome; associates with U2 snRNA; involved
in ER to Golgi transport; K12830 splicing factor 3B subunit
3
Length=1361
Score = 28.1 bits (61), Expect = 9.9, Method: Composition-based stats.
Identities = 27/79 (34%), Positives = 34/79 (43%), Gaps = 7/79 (8%)
Query 28 LFDASSRGLKSCLVESNDFASGTSSKSTKLVHGGIRYLQKACETF-----DWGQLSLVCE 82
L+D + LK N FA+ TS KS L H G R TF D G LS+V
Sbjct 103 LYDTADGELKLIAKFQNLFATITSMKSLDLPHSGSRAKASNWPTFLALTSDSGNLSIV-- 160
Query 83 ALEERAHLLRAAPHIAEPL 101
+ A LR + +PL
Sbjct 161 QIIMHAGALRLKTLVNQPL 179
Lambda K H
0.322 0.137 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2618291680
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40