bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1685_orf1
Length=171
Score E
Sequences producing significant alignments: (Bits) Value
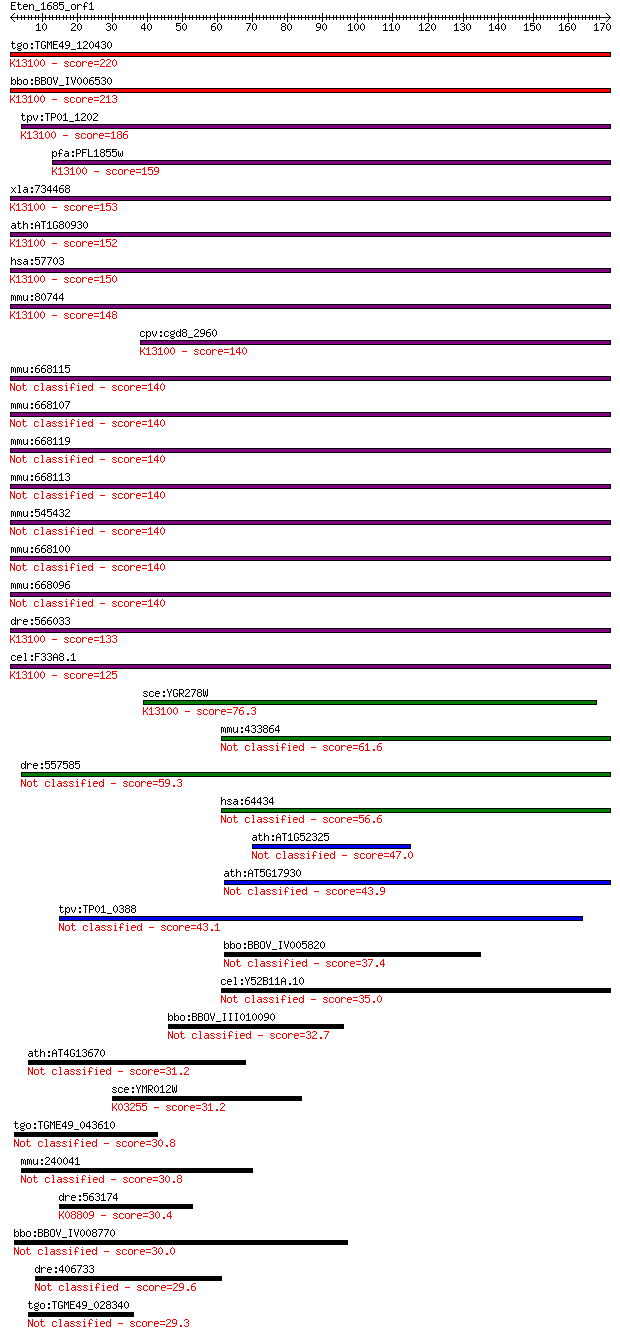
tgo:TGME49_120430 cell cycle control protein, putative ; K1310... 220 2e-57
bbo:BBOV_IV006530 23.m06494; MIF4G/MA3 domains containing prot... 213 2e-55
tpv:TP01_1202 cell cycle control protein; K13100 pre-mRNA-spli... 186 4e-47
pfa:PFL1855w cell cycle control protein, putative; K13100 pre-... 159 4e-39
xla:734468 cwc22, MGC115254; CWC22 spliceosome-associated prot... 153 3e-37
ath:AT1G80930 MIF4G domain-containing protein / MA3 domain-con... 152 6e-37
hsa:57703 CWC22, EIF4GL, KIAA1604, NCM, fSAPb; CWC22 spliceoso... 150 2e-36
mmu:80744 Cwc22, AA684037, AI173004, AL022752, B230213M24, MGC... 148 8e-36
cpv:cgd8_2960 NIC+MI domains containing protein. nucampholin/y... 140 1e-33
mmu:668115 Gm13697, OTTMUSG00000013393; predicted gene 13697 140 1e-33
mmu:668107 Gm13696, OTTMUSG00000013392; predicted gene 13696 140 1e-33
mmu:668119 Gm13691, OTTMUSG00000013386; predicted gene 13691 140 1e-33
mmu:668113 Gm13694, OTTMUSG00000013390; predicted gene 13694 140 1e-33
mmu:545432 Gm13695, Gm5838, OTTMUSG00000013391; predicted gene... 140 1e-33
mmu:668100 Gm13693, OTTMUSG00000013388; predicted gene 13693 140 1e-33
mmu:668096 Gm13698, OTTMUSG00000013395; predicted gene 13698 140 1e-33
dre:566033 MGC153452, cwc22, wu:fb13g08, wu:fi16b02; zgc:15345... 133 3e-31
cel:F33A8.1 let-858; LEThal family member (let-858); K13100 pr... 125 5e-29
sce:YGR278W CWC22; Cwc22p; K13100 pre-mRNA-splicing factor CWC22 76.3 4e-14
mmu:433864 Nom1, D5Kng1, Gm1040; nucleolar protein with MIF4G ... 61.6 1e-09
dre:557585 nom1, si:ch211-266o5.1; nucleolar protein with MIF4... 59.3 5e-09
hsa:64434 NOM1, C7orf3, FLJ16401, SGD1; nucleolar protein with... 56.6 4e-08
ath:AT1G52325 hypothetical protein 47.0 3e-05
ath:AT5G17930 binding / protein binding 43.9 3e-04
tpv:TP01_0388 hypothetical protein 43.1 4e-04
bbo:BBOV_IV005820 23.m05731; hypothetical protein 37.4 0.025
cel:Y52B11A.10 hypothetical protein 35.0 0.13
bbo:BBOV_III010090 17.m07877; hypothetical protein 32.7 0.62
ath:AT4G13670 PTAC5; PTAC5 (PLASTID TRANSCRIPTIONALLY ACTIVE5)... 31.2 1.7
sce:YMR012W CLU1, TIF31; Clu1p; K03255 protein TIF31 31.2 2.0
tgo:TGME49_043610 DNA methyltransferase, putative (EC:3.2.1.11... 30.8 2.0
mmu:240041 Zfp945, A630033E08Rik, C730040L01Rik; zinc finger p... 30.8 2.1
dre:563174 si:ch211-195m20.1; K08809 striated muscle-specific ... 30.4 3.2
bbo:BBOV_IV008770 23.m06331; hypothetical protein 30.0 3.8
dre:406733 hinfp, mizf, wu:fi67c06, zgc:66200; histone H4 tran... 29.6 4.8
tgo:TGME49_028340 elongation factor Tu GTP binding domain-cont... 29.3 7.1
> tgo:TGME49_120430 cell cycle control protein, putative ; K13100
pre-mRNA-splicing factor CWC22
Length=1046
Score = 220 bits (560), Expect = 2e-57, Method: Compositional matrix adjust.
Identities = 125/172 (72%), Positives = 141/172 (81%), Gaps = 5/172 (2%)
Query 1 DEKKWAALSREILGEESTDADASSEDSEAESSSEEEAEETAEPGKKQTV-VKDLTDQEVV 59
DEKKWA LS+EILGEES+D+DASS DSEAE SEEE E K TV + D+TD E++
Sbjct 829 DEKKWARLSKEILGEESSDSDASSSDSEAEDESEEEEGEGE----KATVKILDMTDAEII 884
Query 60 CLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAER 119
+RKTIYLCIMSSLN EECVHKILKMNI+EDL MEV +MLIDCC+MERTFQRFFALQAER
Sbjct 885 HMRKTIYLCIMSSLNFEECVHKILKMNIREDLVMEVVIMLIDCCAMERTFQRFFALQAER 944
Query 120 LAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
LA+L Y EC QEA +RQY HRLET KLRNTAKFFAHLL +DAIPW+V+
Sbjct 945 LARLNPRYSECLQEAMRRQYHTVHRLETTKLRNTAKFFAHLLHTDAIPWTVM 996
> bbo:BBOV_IV006530 23.m06494; MIF4G/MA3 domains containing protein;
K13100 pre-mRNA-splicing factor CWC22
Length=588
Score = 213 bits (542), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 100/175 (57%), Positives = 131/175 (74%), Gaps = 5/175 (2%)
Query 1 DEKKWAALSREILGEESTDADA----SSEDSEAESSSEEEAEETAEPGKKQTVVKDLTDQ 56
+++KW + R+++G+ +D SS DSEAE E+++E+ G TV++D T Q
Sbjct 332 EQEKWTRIRRQLMGDSDDGSDTHDSDSSVDSEAEQHDEDQSEDKPTTGAT-TVIRDSTGQ 390
Query 57 EVVCLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQ 116
++V LRKT+YLCIMSSLN EECVHK+LK+N++E E+E+ MLIDCC+MERTFQ F+ALQ
Sbjct 391 DLVNLRKTVYLCIMSSLNYEECVHKLLKLNVKEGTEIEICTMLIDCCAMERTFQPFYALQ 450
Query 117 AERLAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
AERL+KL Y + FQE F +QY L HRLETAKLRN AKFF HLL +DA+PWSVL
Sbjct 451 AERLSKLSRVYAQNFQECFAKQYQLIHRLETAKLRNVAKFFTHLLATDALPWSVL 505
> tpv:TP01_1202 cell cycle control protein; K13100 pre-mRNA-splicing
factor CWC22
Length=596
Score = 186 bits (471), Expect = 4e-47, Method: Compositional matrix adjust.
Identities = 95/178 (53%), Positives = 122/178 (68%), Gaps = 11/178 (6%)
Query 4 KWAALSREILGEESTDADASSEDSEAESSSEEEAE-------ETAEPGKKQTV---VKDL 53
KW + +E+ G + +SEDSE ++ + + +T + TV +KD
Sbjct 338 KWNKIKQELTGAHTDSESDTSEDSEYDTVESDTVDNLDTVDNDTVDNDTVNTVERGIKDY 397
Query 54 TDQEVVCLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFF 113
T+QE+V LRKTIYLCIMSSLN EECVHKILK+NI ED E+EV +MLIDCC+MERTFQ F+
Sbjct 398 TEQELVNLRKTIYLCIMSSLNYEECVHKILKLNI-EDREIEVCIMLIDCCAMERTFQLFY 456
Query 114 ALQAERLAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
+LQAERL KL+ +Y F++ F RQY L HRLET+KLRN AKFFAHL ++ I W L
Sbjct 457 SLQAERLCKLRQSYRINFEQCFSRQYKLIHRLETSKLRNVAKFFAHLFYTEGISWETL 514
> pfa:PFL1855w cell cycle control protein, putative; K13100 pre-mRNA-splicing
factor CWC22
Length=967
Score = 159 bits (402), Expect = 4e-39, Method: Composition-based stats.
Identities = 87/161 (54%), Positives = 113/161 (70%), Gaps = 2/161 (1%)
Query 13 LGEESTDADA-SSEDSEAESSSEEEAEETAEPGKK-QTVVKDLTDQEVVCLRKTIYLCIM 70
LG + D + SSEDSEAE S + +E + D+T+Q ++ LRK +YL IM
Sbjct 723 LGNDDLDENNNSSEDSEAEKSDSNDDDEDENNDSDNHEEIHDMTEQYLINLRKNVYLSIM 782
Query 71 SSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAERLAKLKTAYCEC 130
SSL+ EECVHK+LK+NI++ E+E+ MLIDCC ME+TFQ+F+ALQAERL KLK Y E
Sbjct 783 SSLSYEECVHKLLKLNIKKGYEIEICNMLIDCCCMEKTFQKFYALQAERLCKLKKIYQEN 842
Query 131 FQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
F++ F + AHRLETAKLRN +KFFAHLL +DAI WS+
Sbjct 843 FEKCFDNSFNTAHRLETAKLRNCSKFFAHLLYTDAISWSIF 883
> xla:734468 cwc22, MGC115254; CWC22 spliceosome-associated protein
homolog; K13100 pre-mRNA-splicing factor CWC22
Length=803
Score = 153 bits (386), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 77/173 (44%), Positives = 111/173 (64%), Gaps = 3/173 (1%)
Query 1 DEKKWAALSREIL--GEESTDADASSEDSEAESSSEEEAEETAEPGKKQTVVKDLTDQEV 58
+E+K+ A+ +EIL G+ ++ DA+ + EE+ +E G+K T+ D T+ +
Sbjct 441 NEEKYKAIKKEILDEGDSDSEGDANEGSEDESEEEEEDGQEAGTEGEKMTI-HDKTEVNL 499
Query 59 VCLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAE 118
V R+TIYL I SSL+ EEC HK++KM+ E E+ M++DCC+ +RT+++FF L A
Sbjct 500 VAFRRTIYLAIQSSLDFEECAHKLIKMDFPESQTKELCNMILDCCAQQRTYEKFFGLLAG 559
Query 119 RLAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
R LK Y E F+ FK Q+ HRLET KLRN AK FAHLL +D++PWSVL
Sbjct 560 RFCLLKKEYLEAFENIFKEQFETIHRLETNKLRNVAKMFAHLLYTDSLPWSVL 612
> ath:AT1G80930 MIF4G domain-containing protein / MA3 domain-containing
protein; K13100 pre-mRNA-splicing factor CWC22
Length=900
Score = 152 bits (383), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 81/172 (47%), Positives = 118/172 (68%), Gaps = 4/172 (2%)
Query 1 DEKKWAALSREILG-EESTDADASSEDSEAESSSEEEAEETAEPGKKQTVVKDLTDQEVV 59
+EKK+ AL +E+LG EES D D S SE E+E++E ++Q ++D T+ +V
Sbjct 592 NEKKYEALKKELLGDEESEDEDGSDASSEDNDEEEDESDEED---EEQMRIRDETETNLV 648
Query 60 CLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAER 119
LR+TIYL IMSS++ EE HK+LK+ ++ EME+ +ML++CCS ERT+ R++ L +R
Sbjct 649 NLRRTIYLTIMSSVDFEEAGHKLLKIKLEPGQEMELCIMLLECCSQERTYLRYYGLLGQR 708
Query 120 LAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
+ + E F++ F +QY++ HRLET KLRN AKFFAHLL +DA+PW VL
Sbjct 709 FCMINKIHQENFEKCFVQQYSMIHRLETNKLRNVAKFFAHLLGTDALPWHVL 760
> hsa:57703 CWC22, EIF4GL, KIAA1604, NCM, fSAPb; CWC22 spliceosome-associated
protein homolog (S. cerevisiae); K13100 pre-mRNA-splicing
factor CWC22
Length=908
Score = 150 bits (379), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 82/172 (47%), Positives = 110/172 (63%), Gaps = 1/172 (0%)
Query 1 DEKKWAALSREILGEESTDADASSEDSEAESSSEEEAEETAEPGKKQTV-VKDLTDQEVV 59
+E+K+ A+ +EIL E TD++ + +E EEE EE E + Q V + D T+ +V
Sbjct 394 NEEKYKAIKKEILDEGDTDSNTDQDAGSSEEDEEEEEEEGEEDEEGQKVTIHDKTEINLV 453
Query 60 CLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAER 119
R+TIYL I SSL+ EEC HK+LKM E E+ M++DCC+ +RT+++FF L A R
Sbjct 454 SFRRTIYLAIQSSLDFEECAHKLLKMEFPESQTKELCNMILDCCAQQRTYEKFFGLLAGR 513
Query 120 LAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
LK Y E F+ FK QY HRLET KLRN AK FAHLL +D++PWSVL
Sbjct 514 FCMLKKEYMESFEGIFKEQYDTIHRLETNKLRNVAKMFAHLLYTDSLPWSVL 565
> mmu:80744 Cwc22, AA684037, AI173004, AL022752, B230213M24, MGC107219,
MGC107450, MGC7531, mKIAA1604; CWC22 spliceosome-associated
protein homolog (S. cerevisiae); K13100 pre-mRNA-splicing
factor CWC22
Length=908
Score = 148 bits (374), Expect = 8e-36, Method: Compositional matrix adjust.
Identities = 80/173 (46%), Positives = 110/173 (63%), Gaps = 2/173 (1%)
Query 1 DEKKWAALSREIL--GEESTDADASSEDSEAESSSEEEAEETAEPGKKQTVVKDLTDQEV 58
+E+K+ A+ +EIL G+ ++ D + SE E +EE E E G ++ + D T+ +
Sbjct 392 NEEKYKAIKKEILDEGDSDSNTDQGAGSSEDEEEEDEEEEGEDEEGGQKVTIHDKTEINL 451
Query 59 VCLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAE 118
V R+TIYL I SSL+ EEC HK+LKM E E+ M++DCC+ +RT+++FF L A
Sbjct 452 VSFRRTIYLAIQSSLDFEECAHKLLKMEFAESQTKELCNMILDCCAQQRTYEKFFGLLAG 511
Query 119 RLAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
R LK Y E F+ FK QY HRLET KLRN AK FAHLL +D++PWSVL
Sbjct 512 RFCMLKKEYMESFESIFKEQYDTIHRLETNKLRNVAKMFAHLLYTDSLPWSVL 564
> cpv:cgd8_2960 NIC+MI domains containing protein. nucampholin/yeast
Cwc22p like protein involved in mRNA splicing ; K13100
pre-mRNA-splicing factor CWC22
Length=619
Score = 140 bits (354), Expect = 1e-33, Method: Composition-based stats.
Identities = 68/134 (50%), Positives = 94/134 (70%), Gaps = 0/134 (0%)
Query 38 EETAEPGKKQTVVKDLTDQEVVCLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAV 97
E+T E K + DL++++ V LRK IYLCIM+SLN EEC H++LK+NI +D E
Sbjct 304 EDTEEQLIKNNPMIDLSEKDFVILRKKIYLCIMNSLNYEECTHRLLKLNIPKDQISETCA 363
Query 98 MLIDCCSMERTFQRFFALQAERLAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFF 157
M++DCCSMERT+Q+FF+L AERL +K Y E F + F + HRLET +LR+ KF+
Sbjct 364 MILDCCSMERTYQKFFSLVAERLCIIKKEYQESFAKLFSESFETVHRLETNRLRHVTKFY 423
Query 158 AHLLMSDAIPWSVL 171
++LL DAIPW++L
Sbjct 424 SYLLSKDAIPWNLL 437
> mmu:668115 Gm13697, OTTMUSG00000013393; predicted gene 13697
Length=830
Score = 140 bits (354), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 81/181 (44%), Positives = 110/181 (60%), Gaps = 13/181 (7%)
Query 1 DEKKWAALSREILGEESTDADASSEDSEAESSSEEEAEETAEPGK----------KQTVV 50
+E+K+ A+ +EIL E +D S+ D A SS +EE EE E + ++ +
Sbjct 393 NEEKYKAIKKEILDEGDSD---SNTDQGAGSSEDEEEEEEEEEEEEEEGEDEEGGQKVTI 449
Query 51 KDLTDQEVVCLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQ 110
D T+ +V R+TIYL I SSL+ EEC HK+LKM E E+ M++DCC+ +RT++
Sbjct 450 HDKTEINLVSFRRTIYLAIQSSLDFEECAHKLLKMEFAESQTKELCNMILDCCAQQRTYE 509
Query 111 RFFALQAERLAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSV 170
FF L A R L+ Y E F+ FK QY HRLET KLRN AK FAHLL +D++PWSV
Sbjct 510 NFFGLLAGRFCMLEKEYMESFESIFKEQYDTIHRLETNKLRNVAKVFAHLLYTDSLPWSV 569
Query 171 L 171
L
Sbjct 570 L 570
> mmu:668107 Gm13696, OTTMUSG00000013392; predicted gene 13696
Length=830
Score = 140 bits (354), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 81/181 (44%), Positives = 110/181 (60%), Gaps = 13/181 (7%)
Query 1 DEKKWAALSREILGEESTDADASSEDSEAESSSEEEAEETAEPGK----------KQTVV 50
+E+K+ A+ +EIL E +D S+ D A SS +EE EE E + ++ +
Sbjct 393 NEEKYKAIKKEILDEGDSD---SNTDQGAGSSEDEEEEEEEEEEEEEEGEDEEGGQKVTI 449
Query 51 KDLTDQEVVCLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQ 110
D T+ +V R+TIYL I SSL+ EEC HK+LKM E E+ M++DCC+ +RT++
Sbjct 450 HDKTEINLVSFRRTIYLAIQSSLDFEECAHKLLKMEFAESQTKELCNMILDCCAQQRTYE 509
Query 111 RFFALQAERLAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSV 170
FF L A R L+ Y E F+ FK QY HRLET KLRN AK FAHLL +D++PWSV
Sbjct 510 NFFGLLAGRFCMLEKEYMESFESIFKEQYDTIHRLETNKLRNVAKVFAHLLYTDSLPWSV 569
Query 171 L 171
L
Sbjct 570 L 570
> mmu:668119 Gm13691, OTTMUSG00000013386; predicted gene 13691
Length=830
Score = 140 bits (354), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 81/181 (44%), Positives = 110/181 (60%), Gaps = 13/181 (7%)
Query 1 DEKKWAALSREILGEESTDADASSEDSEAESSSEEEAEETAEPGK----------KQTVV 50
+E+K+ A+ +EIL E +D S+ D A SS +EE EE E + ++ +
Sbjct 393 NEEKYKAIKKEILDEGDSD---SNTDQGAGSSEDEEEEEEEEEEEEEEGEDEEGGQKVTI 449
Query 51 KDLTDQEVVCLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQ 110
D T+ +V R+TIYL I SSL+ EEC HK+LKM E E+ M++DCC+ +RT++
Sbjct 450 HDKTEINLVSFRRTIYLAIQSSLDFEECAHKLLKMEFAESQTKELCNMILDCCAQQRTYE 509
Query 111 RFFALQAERLAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSV 170
FF L A R L+ Y E F+ FK QY HRLET KLRN AK FAHLL +D++PWSV
Sbjct 510 NFFGLLAGRFCMLEKEYMESFESIFKEQYDTIHRLETNKLRNVAKVFAHLLYTDSLPWSV 569
Query 171 L 171
L
Sbjct 570 L 570
> mmu:668113 Gm13694, OTTMUSG00000013390; predicted gene 13694
Length=830
Score = 140 bits (354), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 81/181 (44%), Positives = 110/181 (60%), Gaps = 13/181 (7%)
Query 1 DEKKWAALSREILGEESTDADASSEDSEAESSSEEEAEETAEPGK----------KQTVV 50
+E+K+ A+ +EIL E +D S+ D A SS +EE EE E + ++ +
Sbjct 393 NEEKYKAIKKEILDEGDSD---SNTDQGAGSSEDEEEEEEEEEEEEEEGEDEEGGQKVTI 449
Query 51 KDLTDQEVVCLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQ 110
D T+ +V R+TIYL I SSL+ EEC HK+LKM E E+ M++DCC+ +RT++
Sbjct 450 HDKTEINLVSFRRTIYLAIQSSLDFEECAHKLLKMEFAESQTKELCNMILDCCAQQRTYE 509
Query 111 RFFALQAERLAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSV 170
FF L A R L+ Y E F+ FK QY HRLET KLRN AK FAHLL +D++PWSV
Sbjct 510 NFFGLLAGRFCMLEKEYMESFESIFKEQYDTIHRLETNKLRNVAKVFAHLLYTDSLPWSV 569
Query 171 L 171
L
Sbjct 570 L 570
> mmu:545432 Gm13695, Gm5838, OTTMUSG00000013391; predicted gene
13695
Length=830
Score = 140 bits (354), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 80/181 (44%), Positives = 109/181 (60%), Gaps = 13/181 (7%)
Query 1 DEKKWAALSREILGEESTDADASSEDSEAESSSEEEAEETAEPGK----------KQTVV 50
+E+K+ A+ +EIL E +D S+ D A SS +EE E E + ++ +
Sbjct 393 NEEKYKAIKKEILDEGDSD---SNTDQGAGSSEDEEEVEEEEEEEEEEGEDEEGGQKVTI 449
Query 51 KDLTDQEVVCLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQ 110
D T+ +V R+TIYL I SSL+ EEC HK+LKM E E+ M++DCC+ +RT++
Sbjct 450 HDKTEINLVSFRRTIYLAIQSSLDFEECAHKLLKMEFAESQTKELCNMILDCCAQQRTYE 509
Query 111 RFFALQAERLAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSV 170
FF L A R L+ Y E F+ FK QY HRLET KLRN AK FAHLL +D++PWSV
Sbjct 510 NFFGLLAGRFCMLEKEYMESFESIFKEQYDTIHRLETNKLRNVAKVFAHLLYTDSLPWSV 569
Query 171 L 171
L
Sbjct 570 L 570
> mmu:668100 Gm13693, OTTMUSG00000013388; predicted gene 13693
Length=830
Score = 140 bits (354), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 81/181 (44%), Positives = 110/181 (60%), Gaps = 13/181 (7%)
Query 1 DEKKWAALSREILGEESTDADASSEDSEAESSSEEEAEETAEPGK----------KQTVV 50
+E+K+ A+ +EIL E +D S+ D A SS +EE EE E + ++ +
Sbjct 393 NEEKYKAIKKEILDEGDSD---SNTDQGAGSSEDEEEEEEEEEEEEEEGEDEEGGQKVTI 449
Query 51 KDLTDQEVVCLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQ 110
D T+ +V R+TIYL I SSL+ EEC HK+LKM E E+ M++DCC+ +RT++
Sbjct 450 HDKTEINLVSFRRTIYLAIQSSLDFEECAHKLLKMEFAESQTKELCNMILDCCAQQRTYE 509
Query 111 RFFALQAERLAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSV 170
FF L A R L+ Y E F+ FK QY HRLET KLRN AK FAHLL +D++PWSV
Sbjct 510 NFFGLLAGRFCMLEKEYMESFESIFKEQYDTIHRLETNKLRNVAKVFAHLLYTDSLPWSV 569
Query 171 L 171
L
Sbjct 570 L 570
> mmu:668096 Gm13698, OTTMUSG00000013395; predicted gene 13698
Length=830
Score = 140 bits (354), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 81/181 (44%), Positives = 110/181 (60%), Gaps = 13/181 (7%)
Query 1 DEKKWAALSREILGEESTDADASSEDSEAESSSEEEAEETAEPGK----------KQTVV 50
+E+K+ A+ +EIL E +D S+ D A SS +EE EE E + ++ +
Sbjct 393 NEEKYKAIKKEILDEGDSD---SNTDQGAGSSEDEEEEEEEEEEEEEEGEDEEGGQKVTI 449
Query 51 KDLTDQEVVCLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQ 110
D T+ +V R+TIYL I SSL+ EEC HK+LKM E E+ M++DCC+ +RT++
Sbjct 450 HDKTEINLVSFRRTIYLAIQSSLDFEECAHKLLKMEFAESQTKELCNMILDCCAQQRTYE 509
Query 111 RFFALQAERLAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSV 170
FF L A R L+ Y E F+ FK QY HRLET KLRN AK FAHLL +D++PWSV
Sbjct 510 NFFGLLAGRFCMLEKEYMESFESIFKEQYDTIHRLETNKLRNVAKVFAHLLYTDSLPWSV 569
Query 171 L 171
L
Sbjct 570 L 570
> dre:566033 MGC153452, cwc22, wu:fb13g08, wu:fi16b02; zgc:153452;
K13100 pre-mRNA-splicing factor CWC22
Length=985
Score = 133 bits (334), Expect = 3e-31, Method: Composition-based stats.
Identities = 77/172 (44%), Positives = 111/172 (64%), Gaps = 1/172 (0%)
Query 1 DEKKWAALSREILGEESTDADASSEDSEAESSSEEEAEETAEPGKKQTV-VKDLTDQEVV 59
+E+K+ + REIL E S+D+ + S + +EE EE A +++ V + D T+ +V
Sbjct 488 NEEKYKTIKREILDEGSSDSGDDAGGSGDDEDDDEEDEEAAAGEEQEKVTIFDQTEVNLV 547
Query 60 CLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAER 119
R+TIYL I SSL+ EEC HK++KM+ E E+ M++DCC+ +RT+++FF L A R
Sbjct 548 AFRRTIYLAIQSSLDFEECAHKLIKMDFPESQTKELCNMILDCCAQQRTYEKFFGLLAGR 607
Query 120 LAKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
LK Y E F+ F+ QY HRLET KLRN A+ FAHLL +D++PWSVL
Sbjct 608 FCLLKKEYMESFEAIFQEQYETIHRLETNKLRNVARIFAHLLYTDSVPWSVL 659
> cel:F33A8.1 let-858; LEThal family member (let-858); K13100
pre-mRNA-splicing factor CWC22
Length=897
Score = 125 bits (315), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 66/171 (38%), Positives = 105/171 (61%), Gaps = 4/171 (2%)
Query 1 DEKKWAALSREILGEESTDADASSEDSEAESSSEEEAEETAEPGKKQTVVKDLTDQEVVC 60
+E+ + + +EI+G +AD S ED E EEE + E KK T + D TDQ +
Sbjct 430 NEEVYEEIRKEIIG----NADISDEDGGDELDDEEEGSDVEEAPKKTTEIIDNTDQNLTA 485
Query 61 LRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAERL 120
R+ +YL + SSL+ +E HK+LKM I + ++ E+ ML+DCC+ +RT++RF+ + ER
Sbjct 486 FRREVYLTMQSSLDYQEAAHKLLKMKIPDSMQNELCAMLVDCCAQQRTYERFYGMLIERF 545
Query 121 AKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
+L+ Y + F++ + Y+ HR++ KLRN A+ AHLL +DAI W +L
Sbjct 546 CRLRLEYQQYFEKLCQDTYSTIHRIDITKLRNLARLIAHLLSTDAIDWKIL 596
> sce:YGR278W CWC22; Cwc22p; K13100 pre-mRNA-splicing factor CWC22
Length=577
Score = 76.3 bits (186), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 40/130 (30%), Positives = 69/130 (53%), Gaps = 1/130 (0%)
Query 39 ETAEPGKKQTVVKDLTDQEVVCLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVM 98
+T E Q + D+T V +K IYL + SSL+ +E HK+LK+ I +L+ V +
Sbjct 269 DTNEGSNSQLQIYDMTSTNDVEFKKKIYLVLKSSLSGDEAAHKLLKLKIANNLKKSVVDI 328
Query 99 LIDCCSMERTFQRFFALQAERLAKLKTAYCECFQEAFKRQYAL-AHRLETAKLRNTAKFF 157
+I E TF +F+++ +ER+ ++ + E F++ Y ET +LR KF+
Sbjct 329 IIKSSLQESTFSKFYSILSERMITFHRSWQTAYNETFEQNYTQDIEDYETDQLRILGKFW 388
Query 158 AHLLMSDAIP 167
HL+ + +P
Sbjct 389 GHLISYEFLP 398
> mmu:433864 Nom1, D5Kng1, Gm1040; nucleolar protein with MIF4G
domain 1
Length=854
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/111 (27%), Positives = 58/111 (52%), Gaps = 0/111 (0%)
Query 61 LRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAERL 120
+R+ I+ +M+S + + K+LK+ +++ E E+ +L+DCC E+T+ F+A A +
Sbjct 649 VRRIIFCTLMTSEDFLDAFEKLLKLGLKDQQEREIVHILMDCCLQEKTYNPFYAFLASKF 708
Query 121 AKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
+ + FQ + ++ L K N AHLL + ++P SVL
Sbjct 709 CDYERRFQMTFQFSIWDKFRDLENLPDTKFSNLVHLLAHLLRTKSLPLSVL 759
> dre:557585 nom1, si:ch211-266o5.1; nucleolar protein with MIF4G
domain 1
Length=487
Score = 59.3 bits (142), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 39/168 (23%), Positives = 77/168 (45%), Gaps = 9/168 (5%)
Query 4 KWAALSREILGEESTDADASSEDSEAESSSEEEAEETAEPGKKQTVVKDLTDQEVVCLRK 63
+W + G D D ++ + + E+ + + E +KQ + D+ R+
Sbjct 233 RWWIMGSSWSGAPMID-DQGNKTTTPSTKGEQYSAKMLELARKQRMNTDV--------RR 283
Query 64 TIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAERLAKL 123
I+ +MSS + + K+L++ +++ E E+ +L+DCC E+ F RF+A+ AE+L
Sbjct 284 NIFCVLMSSEDFLDAYEKLLRLGLKDQQEREIVHVLMDCCLQEKMFNRFYAVLAEKLCSH 343
Query 124 KTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
+ FQ + ++ L N + HLL ++ S+L
Sbjct 344 DRRFQMTFQFSLWDKFRDLANLSNRSFSNLVQLVTHLLHRKSLSLSIL 391
> hsa:64434 NOM1, C7orf3, FLJ16401, SGD1; nucleolar protein with
MIF4G domain 1
Length=860
Score = 56.6 bits (135), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 57/111 (51%), Gaps = 0/111 (0%)
Query 61 LRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAERL 120
+R+ I+ IM+S + + K+LK+ +++ E E+ +L+DCC E+T+ F+A A +
Sbjct 655 IRRNIFCTIMTSEDFLDAFEKLLKLGLKDQQEREIIHVLMDCCLQEKTYNPFYAFLASKF 714
Query 121 AKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
+ + + FQ + ++ L N AHLL + ++ S+L
Sbjct 715 CEYERRFQMTFQFSIWDKFRDLENLPATNFSNLVHLVAHLLKTKSLSLSIL 765
> ath:AT1G52325 hypothetical protein
Length=145
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 19/45 (42%), Positives = 34/45 (75%), Gaps = 0/45 (0%)
Query 70 MSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFA 114
MSSL+ EE HK+LK+ +++ ME+ VM+++CC+ E+T++ F+
Sbjct 1 MSSLDFEEAGHKLLKIRLEQGQGMELCVMVLECCTEEKTYRSFYV 45
> ath:AT5G17930 binding / protein binding
Length=784
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 32/119 (26%), Positives = 60/119 (50%), Gaps = 18/119 (15%)
Query 62 RKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAERL- 120
RK I+ IMSS + + K+L++++ + E+ +L++CC E+ F +F+ + A +L
Sbjct 587 RKAIFCVIMSSEDYIDAFEKLLRLDLPGKQDREIMRVLVECCLQEKAFNKFYTVLASKLC 646
Query 121 -----AKLKTAYCECFQEAFKRQYALAHRLETAKLRNT---AKFFAHLLMSDAIPWSVL 171
K Y C + FK LE+ L+ + AKF A ++++ + +VL
Sbjct 647 EHDKNHKFTLQY--CIWDHFK-------ELESMSLQRSMHLAKFVAEIIVTFNLSLAVL 696
> tpv:TP01_0388 hypothetical protein
Length=509
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 36/158 (22%), Positives = 73/158 (46%), Gaps = 9/158 (5%)
Query 15 EESTDADASSEDSEAESSSEEEAEETAEPGK---KQTVVKDLTDQEVVC-LRKTIYLCIM 70
E + + +S+ ++ +S E E P K K ++K T + ++KTI++CIM
Sbjct 212 ESIINCNNNSDGADGNNSGVTEFETKIAPKKNSRKDELIKKATKIGLNKDIQKTIFICIM 271
Query 71 SSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFA-LQAERLAKLKTAYCE 129
S++ E + ILK+ ++ EV ++I C +E+ F ++ L +KL C+
Sbjct 272 DSISPENALDNILKIKLKTQQFNEVINIVIYCYLVEKEFNYYYIKLLLILTSKLNLNVCK 331
Query 130 CFQEAFKRQYALAHR----LETAKLRNTAKFFAHLLMS 163
++ L + L + +K F H++++
Sbjct 332 KYKRHINNNIVLYFKNFESLTQNQFSLLSKLFYHMILN 369
> bbo:BBOV_IV005820 23.m05731; hypothetical protein
Length=771
Score = 37.4 bits (85), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 15/73 (20%), Positives = 38/73 (52%), Gaps = 0/73 (0%)
Query 62 RKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAERLA 121
+K+ ++ IM +LN + V +I+++ I+ EV ++ C E + +++ + L+
Sbjct 569 QKSTFIAIMGALNPQHAVERIMQIGIKSAQYTEVVYTIVHCLMSENAYNPYYSEVGKLLS 628
Query 122 KLKTAYCECFQEA 134
++ + + F A
Sbjct 629 RMPSTTGKRFTRA 641
> cel:Y52B11A.10 hypothetical protein
Length=819
Score = 35.0 bits (79), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 22/111 (19%), Positives = 55/111 (49%), Gaps = 0/111 (0%)
Query 61 LRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEVAVMLIDCCSMERTFQRFFALQAERL 120
+R+ I+ + S+ + ++ ++LK+ ++ + E E+ +LI E+T+ F+A +R
Sbjct 628 VRRHIFCSVASADDEDDAFERLLKLQLKGEKERELVHVLIAMMMKEKTYNAFYATLLQRF 687
Query 121 AKLKTAYCECFQEAFKRQYALAHRLETAKLRNTAKFFAHLLMSDAIPWSVL 171
+ + Q A + +L+ + + A+ HL+ ++ + +VL
Sbjct 688 CEFNKRFVITLQFALWDRLRECDQLKPFQRGSLAQLLQHLISNEVMSITVL 738
> bbo:BBOV_III010090 17.m07877; hypothetical protein
Length=342
Score = 32.7 bits (73), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 25/50 (50%), Gaps = 0/50 (0%)
Query 46 KQTVVKDLTDQEVVCLRKTIYLCIMSSLNAEECVHKILKMNIQEDLEMEV 95
KQ V D TD ++ T+ LC S + EEC+ I K N+ L +V
Sbjct 120 KQVAVPDDTDVYCGSMKHTLVLCTTSRKHEEECLEAIRKSNVPHTLVYDV 169
> ath:AT4G13670 PTAC5; PTAC5 (PLASTID TRANSCRIPTIONALLY ACTIVE5);
heat shock protein binding / unfolded protein binding
Length=387
Score = 31.2 bits (69), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 35/69 (50%), Gaps = 7/69 (10%)
Query 6 AALSREILGEESTDADASSEDSEAESSSEEEAEE-------TAEPGKKQTVVKDLTDQEV 58
++ E+L D D ++ EA + +EEA T P KKQ++VKD +D+EV
Sbjct 232 GVMTAELLQRLFMDEDVETDKDEASTMKKEEAGNGAVFTSVTQVPEKKQSIVKDQSDREV 291
Query 59 VCLRKTIYL 67
+ ++L
Sbjct 292 DVTQNRVFL 300
> sce:YMR012W CLU1, TIF31; Clu1p; K03255 protein TIF31
Length=1277
Score = 31.2 bits (69), Expect = 2.0, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 32/54 (59%), Gaps = 2/54 (3%)
Query 30 ESSSEEEAEETAEPGKKQTVVKDLTDQEVVCLRKTIYLCIMSSLNAEECVHKIL 83
E S +EE +E ++P +K+ KD+TD+E + + ++ S N+ ++K+L
Sbjct 155 ERSKQEEKDEKSDPEEKKNTFKDVTDEEKLKFNEMVHEVFSSFKNSS--INKLL 206
> tgo:TGME49_043610 DNA methyltransferase, putative (EC:3.2.1.11
2.3.1.97)
Length=1172
Score = 30.8 bits (68), Expect = 2.0, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 2 EKKWAALSREILGEESTDADASSEDSEAESSSEEEAEETAE 42
E++WAA +R++ ++ AD + E E + EEAE E
Sbjct 891 EQRWAAFARKLRAKQGGIADGEGDVGEGERTQFEEAEAKDE 931
> mmu:240041 Zfp945, A630033E08Rik, C730040L01Rik; zinc finger
protein 945
Length=794
Score = 30.8 bits (68), Expect = 2.1, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 31/67 (46%), Gaps = 1/67 (1%)
Query 4 KWAALSREIL-GEESTDADASSEDSEAESSSEEEAEETAEPGKKQTVVKDLTDQEVVCLR 62
KW + I +ST ++ D+ ESS+ + E EP K + V L + C
Sbjct 107 KWKECGKMIHESSKSTPYQTNNRDASIESSNRHKTENAREPCKYKDCVNGLNLYPITCHN 166
Query 63 KTIYLCI 69
+T+YL I
Sbjct 167 QTVYLEI 173
> dre:563174 si:ch211-195m20.1; K08809 striated muscle-specific
serine/threonine protein kinase [EC:2.7.11.1]
Length=3712
Score = 30.4 bits (67), Expect = 3.2, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 15 EESTDADASSEDSEAESSSEEEAEETAEPGKKQTVVKD 52
++S D+DA +D + EE+E+ AE KK ++V +
Sbjct 2453 QKSWDSDAQKKDQSKSEETREESEKVAEIEKKTSIVDE 2490
> bbo:BBOV_IV008770 23.m06331; hypothetical protein
Length=1281
Score = 30.0 bits (66), Expect = 3.8, Method: Composition-based stats.
Identities = 25/106 (23%), Positives = 50/106 (47%), Gaps = 11/106 (10%)
Query 2 EKKWAALSREILGEESTDADASSEDSEAESSSEEEAEETAEPGKKQTV-----VKDLTDQ 56
+ K ++ E+ E DAS++ + E SE++ +P QTV + +L+++
Sbjct 362 DNKRININEELADENPLTPDASNDTPKHELHSEKDGNNIVDPSITQTVGAVSGMMNLSNR 421
Query 57 EVVCLRKTIYLC------IMSSLNAEECVHKILKMNIQEDLEMEVA 96
E+ LR L ++S+LN + +H I ++ ++VA
Sbjct 422 ELSRLRHMKVLANKEKAKLISNLNHRKNIHAIFFPTKDDESTLKVA 467
> dre:406733 hinfp, mizf, wu:fi67c06, zgc:66200; histone H4 transcription
factor
Length=487
Score = 29.6 bits (65), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 25/53 (47%), Gaps = 1/53 (1%)
Query 8 LSREILGEESTDADASSEDSEAESSSEEEAEETAEPGKKQTVVKDLTDQEVVC 60
L+ +++ E D D S E S+ EE EET EP T K D+ VC
Sbjct 391 LTEQLIRERQGDGDGS-ESSQQNVHPGEELEETPEPFSAITEAKSRKDETEVC 442
> tgo:TGME49_028340 elongation factor Tu GTP binding domain-containing
protein (EC:3.2.1.3)
Length=1771
Score = 29.3 bits (64), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 17/31 (54%), Positives = 22/31 (70%), Gaps = 1/31 (3%)
Query 6 AALSREILGEES-TDADASSEDSEAESSSEE 35
A+L+REILGEES DADA +D E+ E+
Sbjct 98 ASLAREILGEESVPDADALEDDQREETGEEK 128
Lambda K H
0.317 0.127 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4276754328
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40