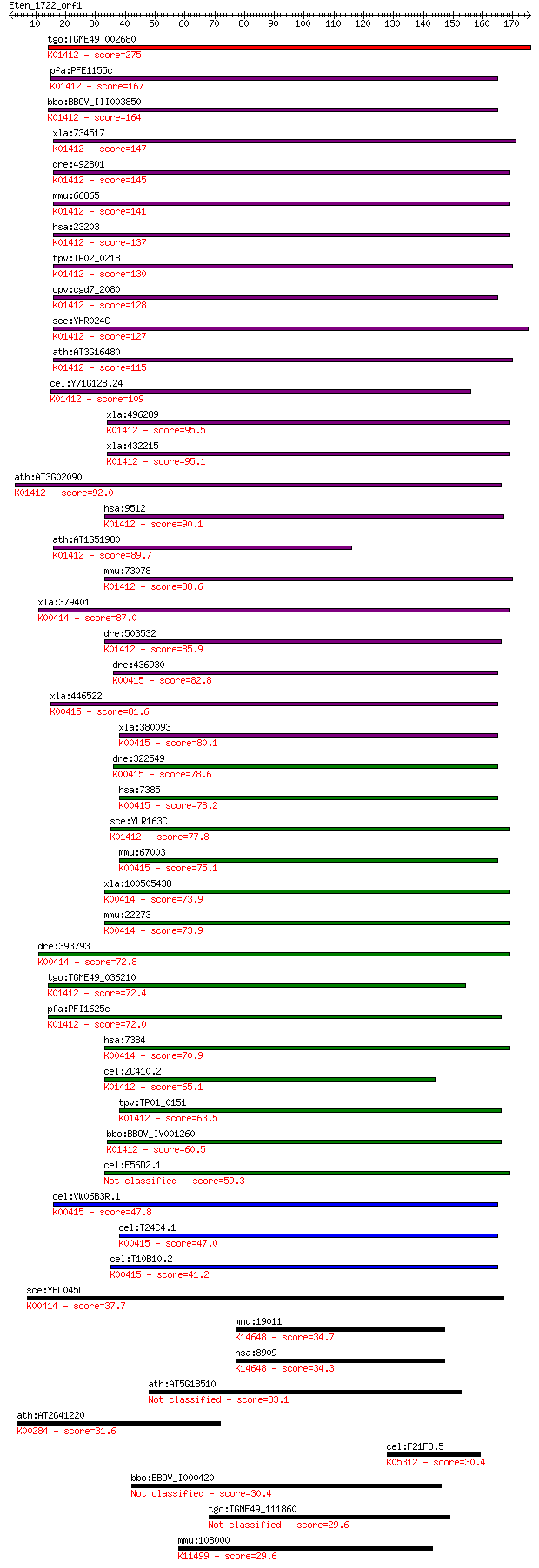
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1722_orf1
Length=175
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_002680 mitochondrial-processing peptidase alpha sub... 275 7e-74
pfa:PFE1155c mitochondrial processing peptidase alpha subunit,... 167 1e-41
bbo:BBOV_III003850 17.m07356; mitochondrial processing peptida... 164 2e-40
xla:734517 pmpca, MGC114896; peptidase (mitochondrial processi... 147 2e-35
dre:492801 pmpca, wu:fi19e06, wu:fj83d11, zgc:101647; peptidas... 145 5e-35
mmu:66865 Pmpca, 1200002L24Rik, 4933435E07Rik, Alpha-MPP, P-55... 141 1e-33
hsa:23203 PMPCA, Alpha-MPP, FLJ26258, INPP5E, KIAA0123, MGC104... 137 2e-32
tpv:TP02_0218 ubiquinol-cytochrome C reductase complex core pr... 130 3e-30
cpv:cgd7_2080 mitochondrial processing peptidase, insulinase l... 128 1e-29
sce:YHR024C MAS2, MIF2; Mas2p (EC:3.4.24.64); K01412 mitochond... 127 2e-29
ath:AT3G16480 MPPalpha (mitochondrial processing peptidase alp... 115 6e-26
cel:Y71G12B.24 mppa-1; Mitochondrial Processing Peptidase Alph... 109 6e-24
xla:496289 hypothetical LOC496289; K01412 mitochondrial proces... 95.5 9e-20
xla:432215 pmpcb, MGC78954; peptidase (mitochondrial processin... 95.1 1e-19
ath:AT3G02090 MPPBETA; mitochondrial processing peptidase beta... 92.0 8e-19
hsa:9512 PMPCB, Beta-MPP, MPP11, MPPB, MPPP52, P-52; peptidase... 90.1 3e-18
ath:AT1G51980 mitochondrial processing peptidase alpha subunit... 89.7 5e-18
mmu:73078 Pmpcb, 3110004O18Rik, MPP11, MPPB, MPPP52; peptidase... 88.6 1e-17
xla:379401 uqcrc1, MGC53748; ubiquinol-cytochrome c reductase ... 87.0 3e-17
dre:503532 pmpcb, zgc:110738; peptidase (mitochondrial process... 85.9 6e-17
dre:436930 uqcrc2a, fb33d01, wu:fb33d01, wu:fc43c05, zgc:92453... 82.8 5e-16
xla:446522 uqcrc2, MGC130698, MGC80228; ubiquinol-cytochrome c... 81.6 1e-15
xla:380093 uqcrc2, MGC53943; Ubiquinol-cytochrome C reductase ... 80.1 4e-15
dre:322549 uqcrc2b, fi06a11, uqcrc2, wu:fb64g04, wu:fi06a11; u... 78.6 1e-14
hsa:7385 UQCRC2, QCR2, UQCR2; ubiquinol-cytochrome c reductase... 78.2 1e-14
sce:YLR163C MAS1, MIF1; Mas1p (EC:3.4.24.64); K01412 mitochond... 77.8 2e-14
mmu:67003 Uqcrc2, 1500004O06Rik; ubiquinol cytochrome c reduct... 75.1 1e-13
xla:100505438 hypothetical protein LOC100505438; K00414 ubiqui... 73.9 3e-13
mmu:22273 Uqcrc1, 1110032G10Rik, MGC97899; ubiquinol-cytochrom... 73.9 3e-13
dre:393793 uqcrc1, MGC73404, zgc:73404, zgc:85750; ubiquinol-c... 72.8 6e-13
tgo:TGME49_036210 mitochondrial-processing peptidase beta subu... 72.4 7e-13
pfa:PFI1625c organelle processing peptidase, putative; K01412 ... 72.0 9e-13
hsa:7384 UQCRC1, D3S3191, QCR1, UQCR1; ubiquinol-cytochrome c ... 70.9 2e-12
cel:ZC410.2 mppb-1; Mitochondrial Processing Peptidase Beta fa... 65.1 1e-10
tpv:TP01_0151 biquinol-cytochrome C reductase complex core pro... 63.5 3e-10
bbo:BBOV_IV001260 21.m02910; mitochondrial processing peptidas... 60.5 3e-09
cel:F56D2.1 ucr-1; Ubiquinol-Cytochrome c oxidoReductase compl... 59.3 7e-09
cel:VW06B3R.1 ucr-2.1; Ubiquinol-Cytochrome c oxidoReductase c... 47.8 2e-05
cel:T24C4.1 ucr-2.3; Ubiquinol-Cytochrome c oxidoReductase com... 47.0 3e-05
cel:T10B10.2 ucr-2.2; Ubiquinol-Cytochrome c oxidoReductase co... 41.2 0.002
sce:YBL045C COR1, QCR1; Cor1p; K00414 ubiquinol-cytochrome c r... 37.7 0.021
mmu:19011 Endou, Pp11r, Tcl-30; endonuclease, polyU-specific; ... 34.7 0.18
hsa:8909 ENDOU, MGC133268, P11, PP11, PRSS26; endonuclease, po... 34.3 0.24
ath:AT5G18510 hypothetical protein 33.1 0.53
ath:AT2G41220 GLU2; GLU2 (GLUTAMATE SYNTHASE 2); glutamate syn... 31.6 1.5
cel:F21F3.5 unc-38; UNCoordinated family member (unc-38); K053... 30.4 3.3
bbo:BBOV_I000420 16.m00758; hypothetical protein 30.4 3.4
tgo:TGME49_111860 hypothetical protein 29.6 5.3
mmu:108000 Cenpf, 6530404A22Rik, AI325968, CENF, Lek1, PRO1779... 29.6 6.0
> tgo:TGME49_002680 mitochondrial-processing peptidase alpha subunit,
putative (EC:3.4.24.64); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=563
Score = 275 bits (702), Expect = 7e-74, Method: Compositional matrix adjust.
Identities = 128/162 (79%), Positives = 145/162 (89%), Gaps = 0/162 (0%)
Query 14 PGKGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAG 73
PGKGMYTRLYLNVLN+ +WVESAMAFNTQYTD+G+FGLYMLA+P K+++AVKVMAEQF
Sbjct 402 PGKGMYTRLYLNVLNQNEWVESAMAFNTQYTDSGIFGLYMLADPTKSANAVKVMAEQFGK 461
Query 74 MTRVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEA 133
M VT EELQRAKNS+KSSI+MNLE RGIVMED+GRQLLMS RVISP EFC AID V EA
Sbjct 462 MVSVTKEELQRAKNSLKSSIFMNLECRGIVMEDVGRQLLMSNRVISPQEFCTAIDAVTEA 521
Query 134 DIRRVAEKMFSKPPTVVAYGDICSVPHYEEIRAALRAAGVGK 175
DI+RV + M+ KPPTVVAYGD+ +VPHYEE+RAALRAAGVGK
Sbjct 522 DIKRVVDAMYKKPPTVVAYGDVSTVPHYEEVRAALRAAGVGK 563
> pfa:PFE1155c mitochondrial processing peptidase alpha subunit,
putative; K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=534
Score = 167 bits (424), Expect = 1e-41, Method: Composition-based stats.
Identities = 76/150 (50%), Positives = 107/150 (71%), Gaps = 0/150 (0%)
Query 15 GKGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGM 74
GKGMY+RL+LNVLN Y+++ES MAF+TQ++DTGLFGLY P+ S +K MA +F M
Sbjct 378 GKGMYSRLFLNVLNSYNFIESCMAFSTQHSDTGLFGLYFTGEPSNTSDIIKAMALEFQKM 437
Query 75 TRVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEAD 134
RVT EEL RAK S+KS ++M+LE + I+MED+ RQ+++ R+++ + AID++ + D
Sbjct 438 NRVTDEELNRAKKSLKSFMWMSLEYKSILMEDLARQMMILNRILTGKQLSDAIDSITKED 497
Query 135 IRRVAEKMFSKPPTVVAYGDICSVPHYEEI 164
I+RV PTVV YG+I PHY+EI
Sbjct 498 IQRVVHNFLKTKPTVVVYGNINYSPHYDEI 527
> bbo:BBOV_III003850 17.m07356; mitochondrial processing peptidase
alpha subunit; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=496
Score = 164 bits (414), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 76/152 (50%), Positives = 111/152 (73%), Gaps = 1/152 (0%)
Query 14 PGKGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAG 73
PGKGM++RL+LNVLNK+++VES MAF+T Y+D G+FG+YM+ P + A+ VM+ +F
Sbjct 339 PGKGMHSRLFLNVLNKHEFVESCMAFSTVYSDAGMFGMYMVVAPQASRGAIDVMSNEFRN 398
Query 74 MTRVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEA 133
M VT +EL+RAKNS+KS ++M+LE++ + MEDI RQLL+ RV++ E RAID+V
Sbjct 399 MLSVTPKELERAKNSLKSFLHMSLEHKAVQMEDIARQLLLCDRVLTVPELERAIDSVTAL 458
Query 134 DIRRVAEKMFS-KPPTVVAYGDICSVPHYEEI 164
DI+R + M P+VVA G++ +PH EE+
Sbjct 459 DIQRCVQSMLKGSKPSVVALGNLAFMPHPEEL 490
> xla:734517 pmpca, MGC114896; peptidase (mitochondrial processing)
alpha (EC:3.4.24.64); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=518
Score = 147 bits (371), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 69/156 (44%), Positives = 101/156 (64%), Gaps = 1/156 (0%)
Query 16 KGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMT 75
KGM+TRLYLNVLN++ W+ +A +++ Y DTGL ++ A+P + V+++ +F M
Sbjct 350 KGMFTRLYLNVLNRHHWMYNATSYHHSYEDTGLLCIHASADPRQVRDMVEIITREFTLMA 409
Query 76 RVTAE-ELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEAD 134
E EL RA+ +KS + MNLE+R ++ ED+GRQ+L +G P E C I+NVK +D
Sbjct 410 GSVGEVELNRARTQLKSMLMMNLESRPVIFEDVGRQVLATGTRKLPHELCNLINNVKASD 469
Query 135 IRRVAEKMFSKPPTVVAYGDICSVPHYEEIRAALRA 170
I+RVA KM P V A GD+ +P YE I+AAL +
Sbjct 470 IKRVATKMLRNKPAVAALGDLTELPDYEHIQAALSS 505
> dre:492801 pmpca, wu:fi19e06, wu:fj83d11, zgc:101647; peptidase
(mitochondrial processing) alpha (EC:3.4.24.64); K01412
mitochondrial processing peptidase [EC:3.4.24.64]
Length=517
Score = 145 bits (367), Expect = 5e-35, Method: Compositional matrix adjust.
Identities = 67/154 (43%), Positives = 100/154 (64%), Gaps = 1/154 (0%)
Query 16 KGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMT 75
KGM+TRLYLNVLN++ W+ +A +++ Y D+GL ++ A+P + V+++ +F MT
Sbjct 349 KGMFTRLYLNVLNRHHWMYNATSYHHSYEDSGLLCIHASADPRQVREMVEIITREFIQMT 408
Query 76 RVTAE-ELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEAD 134
E EL+RAK +KS + MNLE+R ++ ED+GRQ+L +G+ P E C I V +D
Sbjct 409 GTAGEMELERAKTQLKSMLMMNLESRPVIFEDVGRQVLATGKRKLPHELCELISTVTASD 468
Query 135 IRRVAEKMFSKPPTVVAYGDICSVPHYEEIRAAL 168
I+RV KM P V A GD+ +P YE+I+AAL
Sbjct 469 IKRVTMKMLRSKPAVAALGDLTELPSYEDIQAAL 502
> mmu:66865 Pmpca, 1200002L24Rik, 4933435E07Rik, Alpha-MPP, P-55;
peptidase (mitochondrial processing) alpha (EC:3.4.24.64);
K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=524
Score = 141 bits (356), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 68/154 (44%), Positives = 99/154 (64%), Gaps = 1/154 (0%)
Query 16 KGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMT 75
KGM++RLYLNVLN++ W+ +A +++ Y DTGL ++ A+P + V+++ ++F M
Sbjct 356 KGMFSRLYLNVLNRHHWMYNATSYHHSYEDTGLLCIHASADPRQVREMVEIITKEFILMG 415
Query 76 R-VTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEAD 134
R V EL+RAK + S + MNLE+R ++ ED+GRQ+L + P E C I NVK D
Sbjct 416 RTVDLVELERAKTQLMSMLMMNLESRPVIFEDVGRQVLATHSRKLPHELCTLIRNVKPED 475
Query 135 IRRVAEKMFSKPPTVVAYGDICSVPHYEEIRAAL 168
I+RVA KM P V A GD+ +P YE I+AAL
Sbjct 476 IKRVASKMLRGKPAVAALGDLTDLPTYEHIQAAL 509
> hsa:23203 PMPCA, Alpha-MPP, FLJ26258, INPP5E, KIAA0123, MGC104197;
peptidase (mitochondrial processing) alpha (EC:3.4.24.64);
K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=525
Score = 137 bits (344), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 65/154 (42%), Positives = 97/154 (62%), Gaps = 1/154 (0%)
Query 16 KGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMT 75
KGM++RLYLNVLN++ W+ +A +++ Y DTGL ++ A+P + V+++ ++F M
Sbjct 357 KGMFSRLYLNVLNRHHWMYNATSYHHSYEDTGLLCIHASADPRQVREMVEIITKEFILMG 416
Query 76 -RVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEAD 134
V EL+RAK + S + MNLE+R ++ ED+GRQ+L + P E C I NVK D
Sbjct 417 GTVDTVELERAKTQLTSMLMMNLESRPVIFEDVGRQVLATRSRKLPHELCTLIRNVKPED 476
Query 135 IRRVAEKMFSKPPTVVAYGDICSVPHYEEIRAAL 168
++RVA KM P V A GD+ +P YE I+ AL
Sbjct 477 VKRVASKMLRGKPAVAALGDLTDLPTYEHIQTAL 510
> tpv:TP02_0218 ubiquinol-cytochrome C reductase complex core
protein II, mitochondrial precursor (EC:1.10.2.2); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=525
Score = 130 bits (326), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 65/159 (40%), Positives = 101/159 (63%), Gaps = 5/159 (3%)
Query 16 KGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKAS----SAVKVMAEQF 71
KG+ T LY NVLN+Y++VES MAFNT ++ +GLFG+Y++ N AS ++ ++F
Sbjct 367 KGLTTSLYNNVLNRYEFVESCMAFNTVHSTSGLFGIYLVVNGAYASGNMDQVFTLVRDEF 426
Query 72 AGMTRVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVK 131
M ++T EL KNS+KS ++M+LE++ +V ED+GRQLL RV+ P++ ID V
Sbjct 427 ERMKKITNHELSGGKNSLKSFLHMSLEHKAVVCEDVGRQLLFCNRVLDPSDLENLIDEVT 486
Query 132 EADIRRVAEKM-FSKPPTVVAYGDICSVPHYEEIRAALR 169
DI+ V ++ ++ P+VV YG + VPH + + L
Sbjct 487 LDDIKAVVNELRVNQTPSVVVYGKLSRVPHPDTLLQLLH 525
> cpv:cgd7_2080 mitochondrial processing peptidase, insulinase
like metalloprotease ; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=497
Score = 128 bits (321), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 65/152 (42%), Positives = 98/152 (64%), Gaps = 3/152 (1%)
Query 16 KGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMT 75
KG++++L+L+VLNK+DWVES F QY+DTGLFG+++ + P + ++KV+A+Q M
Sbjct 335 KGIHSKLFLDVLNKFDWVESCNCFVNQYSDTGLFGIHITSYPGYSLESIKVIAKQLGKMK 394
Query 76 RVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADI 135
++ EL+RAKN + S+I ENR ME+I +Q+L I E I ++ DI
Sbjct 395 NISERELERAKNLVLSTICTAYENRSHYMEEISKQILSYSEFIELDEIINCIKSIGIEDI 454
Query 136 RRVAEKMFSKP--PTVVAYG-DICSVPHYEEI 164
++VA+ + SK PTVVA G D+ VP+Y EI
Sbjct 455 KKVADLILSKADRPTVVAVGTDMNQVPNYNEI 486
> sce:YHR024C MAS2, MIF2; Mas2p (EC:3.4.24.64); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=482
Score = 127 bits (318), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 74/174 (42%), Positives = 109/174 (62%), Gaps = 18/174 (10%)
Query 16 KGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQ----F 71
KGMY+RLY +VLN+Y +VE+ +AFN Y+D+G+FG+ + P A AV+V+A+Q F
Sbjct 296 KGMYSRLYTHVLNQYYFVENCVAFNHSYSDSGIFGISLSCIPQAAPQAVEVIAQQMYNTF 355
Query 72 AGMT-RVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNV 130
A R+T +E+ RAKN +KSS+ MNLE++ + +ED+GRQ+LM GR I E I+++
Sbjct 356 ANKDLRLTEDEVSRAKNQLKSSLLMNLESKLVELEDMGRQVLMHGRKIPVNEMISKIEDL 415
Query 131 KEADIRRVAEKMFSKP----------PTVVAYGDICSVPHYEEIRAALRAAGVG 174
K DI RVAE +F+ TVV GD S + ++ L+A G+G
Sbjct 416 KPDDISRVAEMIFTGNVNNAGNGKGRATVVMQGDRGS---FGDVENVLKAYGLG 466
> ath:AT3G16480 MPPalpha (mitochondrial processing peptidase alpha
subunit); catalytic/ metal ion binding / metalloendopeptidase/
zinc ion binding; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=499
Score = 115 bits (289), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 57/156 (36%), Positives = 92/156 (58%), Gaps = 2/156 (1%)
Query 16 KGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMT 75
KGM++ LYL +LN++ +S AF + + +TGLFG+Y +P AS ++++A + +
Sbjct 344 KGMHSWLYLRLLNQHQQFQSCTAFTSVFNNTGLFGIYGCTSPEFASQGIELVASEMNAVA 403
Query 76 --RVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEA 133
+V + L RAK + KS+I MNLE+R I EDIGRQ+L G +F + +D +
Sbjct 404 DGKVNQKHLDRAKAATKSAILMNLESRMIAAEDIGRQILTYGERKPVDQFLKTVDQLTLK 463
Query 134 DIRRVAEKMFSKPPTVVAYGDICSVPHYEEIRAALR 169
DI K+ +KP T+ +GD+ +VP Y+ + R
Sbjct 464 DIADFTSKVITKPLTMATFGDVLNVPSYDSVSKRFR 499
> cel:Y71G12B.24 mppa-1; Mitochondrial Processing Peptidase Alpha
family member (mppa-1); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=477
Score = 109 bits (272), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 52/142 (36%), Positives = 86/142 (60%), Gaps = 1/142 (0%)
Query 15 GKGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGM 74
GKGMY R+Y ++N++ W+ SA+A N Y+D+G+F + + P + A+ ++ Q +
Sbjct 313 GKGMYARMYTELMNRHHWIYSAIAHNHSYSDSGVFTVTASSPPENINDALILLVHQILQL 372
Query 75 TR-VTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEA 133
+ V EL RA+ ++S + MNLE R ++ ED+ RQ+L G P E+ I+ V +
Sbjct 373 QQGVEPTELARARTQLRSHLMMNLEVRPVLFEDMVRQVLGHGDRKQPEEYAEKIEKVTNS 432
Query 134 DIRRVAEKMFSKPPTVVAYGDI 155
DI RV E++ + P++V YGDI
Sbjct 433 DIIRVTERLLASKPSLVGYGDI 454
> xla:496289 hypothetical LOC496289; K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=479
Score = 95.5 bits (236), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 52/137 (37%), Positives = 78/137 (56%), Gaps = 2/137 (1%)
Query 34 ESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGM-TRVTAEELQRAKNSIKSS 92
S +FNT YTDTGL+GLYM+ PN + + ++ + T VT E+ RAKN +K++
Sbjct 338 HSFQSFNTCYTDTGLWGLYMVCEPNTVEDMMHFVQREWIRLCTSVTENEVARAKNLLKTN 397
Query 93 IYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEK-MFSKPPTVVA 151
+ + L+ + EDIGRQ+L R I E ID + IR V K +++K P V A
Sbjct 398 MLLQLDGSTPICEDIGRQMLCYNRRIPLPELEARIDLISAETIREVCTKYIYNKSPAVAA 457
Query 152 YGDICSVPHYEEIRAAL 168
G I +P+Y+ IR+ +
Sbjct 458 VGPIGELPNYDRIRSGM 474
> xla:432215 pmpcb, MGC78954; peptidase (mitochondrial processing)
beta (EC:3.4.24.64); K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=479
Score = 95.1 bits (235), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 52/137 (37%), Positives = 77/137 (56%), Gaps = 2/137 (1%)
Query 34 ESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGM-TRVTAEELQRAKNSIKSS 92
S +FNT YTDTGL+GLYM+ PN + + ++ + T VT E+ RAKN +K++
Sbjct 338 HSFQSFNTCYTDTGLWGLYMVCEPNTVEDMMHFVQREWIRLCTSVTENEVARAKNLLKTN 397
Query 93 IYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEK-MFSKPPTVVA 151
+ + L+ + EDIGRQ+L R I E ID + IR V K +++K P V A
Sbjct 398 MLLQLDGSTPICEDIGRQMLCYNRRIPLPELEARIDLISAETIREVCTKYIYNKSPAVAA 457
Query 152 YGDICSVPHYEEIRAAL 168
G I +P Y+ IR+ +
Sbjct 458 VGPIGQLPDYDRIRSGM 474
> ath:AT3G02090 MPPBETA; mitochondrial processing peptidase beta
subunit, putative; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=531
Score = 92.0 bits (227), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 59/165 (35%), Positives = 88/165 (53%), Gaps = 12/165 (7%)
Query 3 MGGGGAFSTGGPGKGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASS 62
+GGG G + R+ +N + ES MAFNT Y DTGLFG+Y +A +
Sbjct 369 VGGGKHV-----GSDLTQRVAINEI-----AESIMAFNTNYKDTGLFGVYAVAKADCLDD 418
Query 63 -AVKVMAEQFAGMTRVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPA 121
+ +M E RV+ ++ RA+N +KSS+ ++++ + EDIGRQLL GR I A
Sbjct 419 LSYAIMYEVTKLAYRVSDADVTRARNQLKSSLLLHMDGTSPIAEDIGRQLLTYGRRIPTA 478
Query 122 EFCRAIDNVKEADIRRVAEK-MFSKPPTVVAYGDICSVPHYEEIR 165
E ID V + ++RVA K ++ K + A G I +P Y + R
Sbjct 479 ELFARIDAVDASTVKRVANKYIYDKDIAISAIGPIQDLPDYNKFR 523
> hsa:9512 PMPCB, Beta-MPP, MPP11, MPPB, MPPP52, P-52; peptidase
(mitochondrial processing) beta (EC:3.4.24.64); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=489
Score = 90.1 bits (222), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 48/136 (35%), Positives = 79/136 (58%), Gaps = 2/136 (1%)
Query 33 VESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGM-TRVTAEELQRAKNSIKS 91
S +FNT YTDTGL+GLYM+ + + + V+ +++ + T VT E+ RA+N +K+
Sbjct 347 CHSFQSFNTSYTDTGLWGLYMVCESSTVADMLHVVQKEWMRLCTSVTESEVARARNLLKT 406
Query 92 SIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEK-MFSKPPTVV 150
++ + L+ + EDIGRQ+L R I E ID V IR V K ++++ P +
Sbjct 407 NMLLQLDGSTPICEDIGRQMLCYNRRIPIPELEARIDAVNAETIREVCTKYIYNRSPAIA 466
Query 151 AYGDICSVPHYEEIRA 166
A G I +P +++IR+
Sbjct 467 AVGPIKQLPDFKQIRS 482
> ath:AT1G51980 mitochondrial processing peptidase alpha subunit,
putative; K01412 mitochondrial processing peptidase [EC:3.4.24.64]
Length=451
Score = 89.7 bits (221), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 45/102 (44%), Positives = 67/102 (65%), Gaps = 2/102 (1%)
Query 16 KGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMT 75
KGM++ LY VLN+Y V+S AF + + DTGLFG+Y ++P A+ A+++ A++ +
Sbjct 348 KGMHSWLYRRVLNEYQEVQSCTAFTSIFNDTGLFGIYGCSSPQFAAKAIELAAKELKDVA 407
Query 76 --RVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSG 115
+V L RAK + KS++ MNLE+R I EDIGRQ+L G
Sbjct 408 GGKVNQAHLDRAKAATKSAVLMNLESRMIAAEDIGRQILTYG 449
> mmu:73078 Pmpcb, 3110004O18Rik, MPP11, MPPB, MPPP52; peptidase
(mitochondrial processing) beta (EC:3.4.24.64); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=489
Score = 88.6 bits (218), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 50/139 (35%), Positives = 76/139 (54%), Gaps = 2/139 (1%)
Query 33 VESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGM-TRVTAEELQRAKNSIKS 91
S +FNT YTDTGL+GLYM+ + + V+ ++ + T VT E+ RAKN +K+
Sbjct 347 CHSFQSFNTSYTDTGLWGLYMVCEQATVADMLHVVQNEWKRLCTDVTESEVARAKNLLKT 406
Query 92 SIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEK-MFSKPPTVV 150
++ + L+ + EDIGRQ+L R I E ID V +RRV K + K P +
Sbjct 407 NMLLQLDGSTPICEDIGRQMLCYNRRIPIPELEARIDAVDAETVRRVCTKYIHDKSPAIA 466
Query 151 AYGDICSVPHYEEIRAALR 169
A G I +P + +I + +R
Sbjct 467 ALGPIERLPDFNQICSNMR 485
> xla:379401 uqcrc1, MGC53748; ubiquinol-cytochrome c reductase
core protein I (EC:1.10.2.2); K00414 ubiquinol-cytochrome
c reductase core subunit 1 [EC:1.10.2.2]
Length=478
Score = 87.0 bits (214), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 51/160 (31%), Positives = 86/160 (53%), Gaps = 3/160 (1%)
Query 11 TGGPGKGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKV-MAE 69
T G GK + +R+ +V ++ +S FN +Y+DTGLFG++ + + + + + E
Sbjct 315 TYGGGKNLSSRV-ASVAAEHKLCQSYQTFNIRYSDTGLFGMHFVTDKHNIEDMLHIAQGE 373
Query 70 QFAGMTRVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDN 129
+ T VT E+ +AKN++K+++ L+ V EDIGRQ+L G+ +S E ID
Sbjct 374 WMSLCTSVTDSEVAQAKNALKTALVAQLDGTTPVCEDIGRQILSYGQRVSLEELNARIDA 433
Query 130 VKEADIRRVAEK-MFSKPPTVVAYGDICSVPHYEEIRAAL 168
V + + K ++ K P V G I +P Y IR+A+
Sbjct 434 VDAKKVSEICSKYLYDKCPAVAGVGPIEQIPDYNRIRSAM 473
> dre:503532 pmpcb, zgc:110738; peptidase (mitochondrial processing)
beta (EC:3.4.24.64); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=470
Score = 85.9 bits (211), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 48/135 (35%), Positives = 73/135 (54%), Gaps = 2/135 (1%)
Query 33 VESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGM-TRVTAEELQRAKNSIKS 91
S +FNT YTDTGL+GLYM+ P ++ ++ + T VT E+ RAKN +K+
Sbjct 331 CHSFQSFNTCYTDTGLWGLYMVCEPGTVHDMIRFTQLEWKSLCTSVTESEVNRAKNLLKT 390
Query 92 SIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEK-MFSKPPTVV 150
++ ++L+ + EDIGRQ+L R I E ID + I+ V K +++K P +
Sbjct 391 NMLLHLDGSTPICEDIGRQMLCYSRRIPLHELEARIDAINATTIKDVCLKYIYNKAPAIA 450
Query 151 AYGDICSVPHYEEIR 165
A G I + Y IR
Sbjct 451 AVGPIEQLLDYNSIR 465
> dre:436930 uqcrc2a, fb33d01, wu:fb33d01, wu:fc43c05, zgc:92453;
ubiquinol-cytochrome c reductase core protein IIa (EC:1.10.2.2);
K00415 ubiquinol-cytochrome c reductase core subunit
2 [EC:1.10.2.2]
Length=460
Score = 82.8 bits (203), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 44/134 (32%), Positives = 82/134 (61%), Gaps = 8/134 (5%)
Query 36 AMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMT-----RVTAEELQRAKNSIK 90
A AF+T Y+D+GLFGLY++ ++A S +V++ A +T ++T ++L RAKN +K
Sbjct 330 ATAFSTTYSDSGLFGLYVI---SQADSTREVISSAVAQVTAVAEGKLTTDDLTRAKNQLK 386
Query 91 SSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEKMFSKPPTVV 150
+ M+LE+ +++E++G QLL SG SP ++ID+V +D+ + A + ++
Sbjct 387 ADYLMSLESSDVLLEELGVQLLNSGVYSSPQTVTQSIDSVTSSDVLKAARRFVEGQKSMS 446
Query 151 AYGDICSVPHYEEI 164
+ G + + P +E+
Sbjct 447 SCGYLENTPFLDEL 460
> xla:446522 uqcrc2, MGC130698, MGC80228; ubiquinol-cytochrome
c reductase core protein II (EC:1.10.2.2); K00415 ubiquinol-cytochrome
c reductase core subunit 2 [EC:1.10.2.2]
Length=451
Score = 81.6 bits (200), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 47/152 (30%), Positives = 78/152 (51%), Gaps = 2/152 (1%)
Query 15 GKGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGM 74
G ++L+ V + AFN Y+D+GLFG+Y ++ AS + Q +
Sbjct 300 GSNTSSKLFQAVNKATNQPFDVSAFNASYSDSGLFGIYTVSQAAAASEVINAALNQVKAV 359
Query 75 TR--VTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKE 132
+ VT ++ RAKN +KS M LE+ ++ +IG Q L SG I+P E + ID+V
Sbjct 360 AQGNVTEADVTRAKNQLKSQYLMTLESSCGLIGEIGSQALASGTYITPTETIQQIDSVTS 419
Query 133 ADIRRVAEKMFSKPPTVVAYGDICSVPHYEEI 164
AD+ A+K S ++ A G++ + P ++
Sbjct 420 ADVVSAAKKFASGKKSMAATGNLENTPFVSDL 451
> xla:380093 uqcrc2, MGC53943; Ubiquinol-cytochrome C reductase
complex; K00415 ubiquinol-cytochrome c reductase core subunit
2 [EC:1.10.2.2]
Length=451
Score = 80.1 bits (196), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 44/129 (34%), Positives = 70/129 (54%), Gaps = 2/129 (1%)
Query 38 AFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMTR--VTAEELQRAKNSIKSSIYM 95
AFN Y+D+GLFG+Y ++ AS + Q + + VT ++ RAKN +KS M
Sbjct 323 AFNASYSDSGLFGVYTVSQAAAASEVINAALNQVKAVAQGNVTEADVTRAKNQLKSQYLM 382
Query 96 NLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEKMFSKPPTVVAYGDI 155
LE+ ++ DIG Q L SG +P E + ID+V AD+ A+K S ++ A G++
Sbjct 383 PLESSCGLIGDIGSQALASGTYTTPTETIQQIDSVTSADVVSAAKKFASGKKSMAATGNL 442
Query 156 CSVPHYEEI 164
+ P ++
Sbjct 443 ENTPFVSDL 451
> dre:322549 uqcrc2b, fi06a11, uqcrc2, wu:fb64g04, wu:fi06a11;
ubiquinol-cytochrome c reductase core protein IIb (EC:1.10.2.2);
K00415 ubiquinol-cytochrome c reductase core subunit 2
[EC:1.10.2.2]
Length=454
Score = 78.6 bits (192), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 42/131 (32%), Positives = 70/131 (53%), Gaps = 2/131 (1%)
Query 36 AMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMTR--VTAEELQRAKNSIKSSI 93
A AFN YTD+GLFGLY + N + +K Q + + + A +L +AKN + +
Sbjct 324 ASAFNANYTDSGLFGLYTICQANAVNDVIKAAVGQVNAIAQGNLAAADLSKAKNQLTADY 383
Query 94 YMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEKMFSKPPTVVAYG 153
M++E+ +M+ IG +L G +P + I+ V AD+ VA+K S T+ + G
Sbjct 384 LMSIESSEGLMDVIGTHVLSEGTYHTPEAVTQKINAVSSADVVNVAKKFMSGKKTMASSG 443
Query 154 DICSVPHYEEI 164
++ + P +EI
Sbjct 444 NLVNTPFVDEI 454
> hsa:7385 UQCRC2, QCR2, UQCR2; ubiquinol-cytochrome c reductase
core protein II (EC:1.10.2.2); K00415 ubiquinol-cytochrome
c reductase core subunit 2 [EC:1.10.2.2]
Length=453
Score = 78.2 bits (191), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 39/129 (30%), Positives = 72/129 (55%), Gaps = 2/129 (1%)
Query 38 AFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMTR--VTAEELQRAKNSIKSSIYM 95
AFN Y+D+GLFG+Y ++ A +K Q + + ++ ++Q AKN +K+ M
Sbjct 325 AFNASYSDSGLFGIYTISQATAAGDVIKAAYNQVKTIAQGNLSNTDVQAAKNKLKAGYLM 384
Query 96 NLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEKMFSKPPTVVAYGDI 155
++E+ +E++G Q L++G + P+ + ID+V ADI A+K S ++ A G++
Sbjct 385 SVESSECFLEEVGSQALVAGSYMPPSTVLQQIDSVANADIINAAKKFVSGQKSMAASGNL 444
Query 156 CSVPHYEEI 164
P +E+
Sbjct 445 GHTPFVDEL 453
> sce:YLR163C MAS1, MIF1; Mas1p (EC:3.4.24.64); K01412 mitochondrial
processing peptidase [EC:3.4.24.64]
Length=462
Score = 77.8 bits (190), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 41/139 (29%), Positives = 86/139 (61%), Gaps = 5/139 (3%)
Query 35 SAMAFNTQYTDTGLFGLYMLANPNKASSAVKV--MAEQFAGMT--RVTAEELQRAKNSIK 90
S M+F+T Y D+GL+G+Y++ + N+ + + V + +++ + +++ E+ RAK +K
Sbjct 322 SYMSFSTSYADSGLWGMYIVTDSNEHNVQLIVNEILKEWKRIKSGKISDAEVNRAKAQLK 381
Query 91 SSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAE-KMFSKPPTV 149
+++ ++L+ ++EDIGRQ++ +G+ +SP E +D + + DI A ++ +KP ++
Sbjct 382 AALLLSLDGSTAIVEDIGRQVVTTGKRLSPEEVFEQVDKITKDDIIMWANYRLQNKPVSM 441
Query 150 VAYGDICSVPHYEEIRAAL 168
VA G+ +VP+ I L
Sbjct 442 VALGNTSTVPNVSYIEEKL 460
> mmu:67003 Uqcrc2, 1500004O06Rik; ubiquinol cytochrome c reductase
core protein 2 (EC:1.10.2.2); K00415 ubiquinol-cytochrome
c reductase core subunit 2 [EC:1.10.2.2]
Length=453
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 37/129 (28%), Positives = 71/129 (55%), Gaps = 2/129 (1%)
Query 38 AFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMTR--VTAEELQRAKNSIKSSIYM 95
AFN Y+D+GLFG+Y ++ A + Q + + +++ ++Q AKN +K+ M
Sbjct 325 AFNASYSDSGLFGIYTISQAAAAGEVINAAYNQVKAVAQGNLSSADVQAAKNKLKAGYLM 384
Query 96 NLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEKMFSKPPTVVAYGDI 155
++E + +IG Q L +G + P+ + ID+V +AD+ + A+K S ++ A G++
Sbjct 385 SVETSEGFLSEIGSQALAAGSYMPPSTVLQQIDSVADADVVKAAKKFVSGKKSMAASGNL 444
Query 156 CSVPHYEEI 164
P +E+
Sbjct 445 GHTPFLDEL 453
> xla:100505438 hypothetical protein LOC100505438; K00414 ubiquinol-cytochrome
c reductase core subunit 1 [EC:1.10.2.2]
Length=480
Score = 73.9 bits (180), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 45/138 (32%), Positives = 68/138 (49%), Gaps = 2/138 (1%)
Query 33 VESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGM-TRVTAEELQRAKNSIKS 91
+S FN Y+DTGL G + + + V + Q+ + T T E+ R KN +++
Sbjct 338 CQSFQTFNISYSDTGLLGAHFVCDAMSIDDMVFFLQGQWMRLCTSATESEVTRGKNILRN 397
Query 92 SIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEKMF-SKPPTVV 150
++ +L+ V EDIGR LL GR I AE+ I V +R + K F + P V
Sbjct 398 ALVSHLDGTTPVCEDIGRSLLTYGRRIPLAEWESRIQEVDAQMLRDICSKYFYDQCPAVA 457
Query 151 AYGDICSVPHYEEIRAAL 168
YG I +P Y IR+ +
Sbjct 458 GYGPIEQLPDYNRIRSGM 475
> mmu:22273 Uqcrc1, 1110032G10Rik, MGC97899; ubiquinol-cytochrome
c reductase core protein 1 (EC:1.10.2.2); K00414 ubiquinol-cytochrome
c reductase core subunit 1 [EC:1.10.2.2]
Length=480
Score = 73.9 bits (180), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 45/138 (32%), Positives = 68/138 (49%), Gaps = 2/138 (1%)
Query 33 VESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGM-TRVTAEELQRAKNSIKS 91
+S FN Y+DTGL G + + + V + Q+ + T T E+ R KN +++
Sbjct 338 CQSFQTFNISYSDTGLLGAHFVCDAMSIDDMVFFLQGQWMRLCTSATESEVTRGKNILRN 397
Query 92 SIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEKMF-SKPPTVV 150
++ +L+ V EDIGR LL GR I AE+ I V +R + K F + P V
Sbjct 398 ALVSHLDGTTPVCEDIGRSLLTYGRRIPLAEWESRIQEVDAQMLRDICSKYFYDQCPAVA 457
Query 151 AYGDICSVPHYEEIRAAL 168
YG I +P Y IR+ +
Sbjct 458 GYGPIEQLPDYNRIRSGM 475
> dre:393793 uqcrc1, MGC73404, zgc:73404, zgc:85750; ubiquinol-cytochrome
c reductase core protein I (EC:1.10.2.2); K00414
ubiquinol-cytochrome c reductase core subunit 1 [EC:1.10.2.2]
Length=474
Score = 72.8 bits (177), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 50/160 (31%), Positives = 81/160 (50%), Gaps = 3/160 (1%)
Query 11 TGGPGKGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQ 70
T G GK + +RL + + S F + Y+DTGL G+Y + K +
Sbjct 311 TFGGGKHLSSRLAQRA-AELNLCHSFQTFYSSYSDTGLLGIYFVTEKLKIEDMMHWAQNA 369
Query 71 FAGM-TRVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDN 129
+ + T VT ++ RAKN++++S+ L V ++IGR +L GR I AE+ I+
Sbjct 370 WINVCTTVTESDVARAKNALRASLVGQLNGTTPVCDEIGRHILNYGRRIPLAEWDARIEA 429
Query 130 VKEADIRRVAEK-MFSKPPTVVAYGDICSVPHYEEIRAAL 168
V + +R V K ++ K P V A G I +P Y +R+A+
Sbjct 430 VTPSVVRDVCSKYIYDKCPAVSAVGPIEQLPDYNRMRSAM 469
> tgo:TGME49_036210 mitochondrial-processing peptidase beta subunit,
putative (EC:3.4.24.64); K01412 mitochondrial processing
peptidase [EC:3.4.24.64]
Length=524
Score = 72.4 bits (176), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 56/148 (37%), Positives = 79/148 (53%), Gaps = 11/148 (7%)
Query 14 PGKGMYTRLYLNVLNKYDWVESAM--AFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQF 71
PGK NV NK + M AFNT Y+DTGLFG Y A ++ + VM F
Sbjct 346 PGKVSANATVRNVCNKMTVGCADMFSAFNTCYSDTGLFGFY--AQCDEVALEHCVMEIMF 403
Query 72 AGMTR----VTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAI 127
G+T VT EE++RAK +K+ + +L++ V EDIGRQ+L GR + AEF + +
Sbjct 404 -GITSLSYAVTDEEVERAKAQLKTQLLGHLDSTTAVAEDIGRQMLAYGRRMPLAEFLKRL 462
Query 128 DNVKEADIRRVAEKMFSKP--PTVVAYG 153
+ + +++RVA K P V A G
Sbjct 463 EVIDAEEVKRVAWKYLHDAVRPKVGALG 490
> pfa:PFI1625c organelle processing peptidase, putative; K01412
mitochondrial processing peptidase [EC:3.4.24.64]
Length=484
Score = 72.0 bits (175), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 50/159 (31%), Positives = 81/159 (50%), Gaps = 10/159 (6%)
Query 14 PGKGMYTRLYLNVLNKYD--WVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQF 71
PGK R N+ NK + +FNT Y +TGLFG Y+ + A + E
Sbjct 321 PGKLSANRTVNNICNKMTVGCADYFTSFNTCYNNTGLFGFYVQCDEIAVEHA---LGELM 377
Query 72 AGMTR----VTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAI 127
G+T +T EE++ AK +K+ + E+ + E++ RQLL+ GR IS AEF +
Sbjct 378 FGVTSLSYSITDEEVELAKIHLKTQLISMFESSSTLAEEVSRQLLVYGRKISLAEFILRL 437
Query 128 DNVKEADIRRVAEK-MFSKPPTVVAYGDICSVPHYEEIR 165
+ + +++RVA K + + V A G + +P Y ++R
Sbjct 438 NEIDTEEVKRVAWKYLHDRDIAVAAIGALHGMPQYIDLR 476
> hsa:7384 UQCRC1, D3S3191, QCR1, UQCR1; ubiquinol-cytochrome
c reductase core protein I (EC:1.10.2.2); K00414 ubiquinol-cytochrome
c reductase core subunit 1 [EC:1.10.2.2]
Length=480
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 43/138 (31%), Positives = 71/138 (51%), Gaps = 2/138 (1%)
Query 33 VESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGM-TRVTAEELQRAKNSIKS 91
+S F+ Y +TGL G + + + K + V+ Q+ + T T E+ R KN +++
Sbjct 338 CQSFQTFSICYAETGLLGAHFVCDRMKIDDMMFVLQGQWMRLCTSATESEVARGKNILRN 397
Query 92 SIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEK-MFSKPPTVV 150
++ +L+ V EDIGR LL GR I AE+ I V + +R + K ++ + P V
Sbjct 398 ALVSHLDGTTPVCEDIGRSLLTYGRRIPLAEWESRIAEVDASVVREICSKYIYDQCPAVA 457
Query 151 AYGDICSVPHYEEIRAAL 168
YG I +P Y IR+ +
Sbjct 458 GYGPIEQLPDYNRIRSGM 475
> cel:ZC410.2 mppb-1; Mitochondrial Processing Peptidase Beta
family member (mppb-1); K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=458
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 35/112 (31%), Positives = 59/112 (52%), Gaps = 1/112 (0%)
Query 33 VESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGM-TRVTAEELQRAKNSIKS 91
+E +FNT Y +TGL G Y +A P + + + +Q+ + + + RAK S+ +
Sbjct 317 IEVFQSFNTCYKETGLVGTYFVAAPESIDNLIDSVLQQWVWLANNIDEAAVDRAKRSLHT 376
Query 92 SIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEKMF 143
++ + L+ V EDIGRQLL GR I E I+++ +R V ++F
Sbjct 377 NLLLMLDGSTPVCEDIGRQLLCYGRRIPTPELHARIESITVQQLRDVCRRVF 428
> tpv:TP01_0151 biquinol-cytochrome C reductase complex core protein
I (EC:1.10.2.2); K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=518
Score = 63.5 bits (153), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 44/133 (33%), Positives = 65/133 (48%), Gaps = 8/133 (6%)
Query 38 AFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMTR----VTAEELQRAKNSIKSSI 93
AFNT Y DTGLFG Y A+ V E G+T VT EE++RAK +
Sbjct 381 AFNTCYKDTGLFGFYAKADEVAVDHCV---GELLFGITSLSYSVTDEEVERAKRQLMLQF 437
Query 94 YMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEKMFSKPPTVV-AY 152
E+ V E++ RQ+L+ GR + AEF ++ + +++RVA K V A
Sbjct 438 LSMTESTSSVAEEVARQILVYGRRMPVAEFLLRLEKIDAEEVKRVAWKYLHDSEVAVSAM 497
Query 153 GDICSVPHYEEIR 165
G + +P ++R
Sbjct 498 GPLHGMPSLVDLR 510
> bbo:BBOV_IV001260 21.m02910; mitochondrial processing peptidase
beta subunit; K01412 mitochondrial processing peptidase
[EC:3.4.24.64]
Length=514
Score = 60.5 bits (145), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 39/134 (29%), Positives = 65/134 (48%), Gaps = 2/134 (1%)
Query 34 ESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMT-RVTAEELQRAKNSIKSS 92
E+ AFNT Y DTGLFG Y + V + M+ +T EE++RAK +
Sbjct 373 EAFSAFNTCYKDTGLFGFYAQCDEVAVDHCVGELMFGVTSMSYSITDEEVERAKRQLMLQ 432
Query 93 IYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEK-MFSKPPTVVA 151
++ V E++ RQ+++ GR + EF ++ + +++RVA K + V A
Sbjct 433 FLSMNDSTSTVAEEVARQIIVYGRRMPVTEFLLRLEQIDAEEVKRVAWKYLHDHEVAVTA 492
Query 152 YGDICSVPHYEEIR 165
G + +P +IR
Sbjct 493 MGPLHGMPSLIDIR 506
> cel:F56D2.1 ucr-1; Ubiquinol-Cytochrome c oxidoReductase complex
family member (ucr-1)
Length=471
Score = 59.3 bits (142), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 39/141 (27%), Positives = 68/141 (48%), Gaps = 5/141 (3%)
Query 33 VESAMAFNTQYTDTGLFGLYMLANP---NKASSAVKVMAEQFAGM-TRVTAEELQRAKNS 88
V + FN Y DTGLFG+Y +A+ N S +K +A ++ + + T EE+ AKN
Sbjct 326 VHNLQHFNINYKDTGLFGIYFVADAHDLNDTSGIMKSVAHEWKHLASAATEEEVAMAKNQ 385
Query 89 IKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRR-VAEKMFSKPP 147
++++Y NLE ++LL +G + +E I V +R ++ ++ +
Sbjct 386 FRTNLYQNLETNTQKAGFNAKELLYTGNLRQLSELEAQIQKVDAGAVREAISRHVYDRDL 445
Query 148 TVVAYGDICSVPHYEEIRAAL 168
V G + P+Y RA +
Sbjct 446 AAVGVGRTEAFPNYALTRAGM 466
> cel:VW06B3R.1 ucr-2.1; Ubiquinol-Cytochrome c oxidoReductase
complex family member (ucr-2.1); K00415 ubiquinol-cytochrome
c reductase core subunit 2 [EC:1.10.2.2]
Length=424
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 37/149 (24%), Positives = 69/149 (46%), Gaps = 7/149 (4%)
Query 16 KGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMT 75
+ + ++ LN K A++ N Y D+GL G+ A + + K +A + +
Sbjct 283 QAVVAQILLNAAQKV--TSEAISVNVNYQDSGLVGVQFAACNTQITQVTKSIA---SAIK 337
Query 76 RVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADI 135
A+ L AKN+ + + ++ V + Q+L +G +SP EF AI V D+
Sbjct 338 SAKADGLDNAKNTAAVQVLSDAQHASEVALEKATQVL-AGVEVSPREFADAIRAVTAQDV 396
Query 136 RRVAEKMFSKPPTVVAYGDICSVPHYEEI 164
+ ++ K ++ AYG VP+ +E+
Sbjct 397 TQALSRVNGK-LSLAAYGSTSLVPYLDEL 424
> cel:T24C4.1 ucr-2.3; Ubiquinol-Cytochrome c oxidoReductase complex
family member (ucr-2.3); K00415 ubiquinol-cytochrome
c reductase core subunit 2 [EC:1.10.2.2]
Length=427
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 33/128 (25%), Positives = 61/128 (47%), Gaps = 8/128 (6%)
Query 38 AFNTQYTDTGLFGLYMLANPNKASSAVKVMAEQFAGMTRVT-AEELQRAKNSIKSSIYMN 96
AF Y +GL G+Y+LA A SAV+ + + R T ++++ K + I N
Sbjct 307 AFQAPYDGSGLVGVYLLATGANADSAVRATVK----VLRTTQVQDIEGCKRRAIADILFN 362
Query 97 LENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEKMFSKPPTVVAYGDIC 156
EN D+ L +G +E I ++E+D+++ + F + + AYG+
Sbjct 363 AENNVYSAYDLATNALYNGP--EQSELIAEIQKIQESDVQKYTKAAFER-LAISAYGNYG 419
Query 157 SVPHYEEI 164
+P+ +E+
Sbjct 420 RIPYADEL 427
> cel:T10B10.2 ucr-2.2; Ubiquinol-Cytochrome c oxidoReductase
complex family member (ucr-2.2); K00415 ubiquinol-cytochrome
c reductase core subunit 2 [EC:1.10.2.2]
Length=422
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 34/132 (25%), Positives = 68/132 (51%), Gaps = 13/132 (9%)
Query 35 SAMAFNTQYTDTGLFGLYMLANPNKASSAVK--VMAEQFAGMTRVTAEELQRAKNSIKSS 92
SA AF + D+GL G+Y++ ++A+ AV V A + + + A + Q N++++S
Sbjct 302 SASAFQAVHADSGLAGVYLVVEGSQANQAVSNVVGALKSLKVADIEAVKKQAFNNALRAS 361
Query 93 IYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEKMFSKPPTVVAY 152
+ +N I + QL S + + I NV +D+ A+K+ +K ++ +Y
Sbjct 362 AHS--DNFAI---ERASQLFQ-----SQDNYIQQIPNVSASDVEVAAKKLTTK-LSLASY 410
Query 153 GDICSVPHYEEI 164
G++ VP+ + +
Sbjct 411 GNVSEVPYVDTL 422
> sce:YBL045C COR1, QCR1; Cor1p; K00414 ubiquinol-cytochrome c
reductase core subunit 1 [EC:1.10.2.2]
Length=457
Score = 37.7 bits (86), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 36/165 (21%), Positives = 76/165 (46%), Gaps = 5/165 (3%)
Query 7 GAFSTGGPGKGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLA-NPNKASSAVK 65
G+++ P + L+ + +Y ++ F+ Y D+GL+G N +
Sbjct 286 GSYNAFEPASRLQGIKLLDNIQEYQLCDNFNHFSLSYKDSGLWGFSTATRNVTMIDDLIH 345
Query 66 VMAEQFAGMT-RVTAEELQRAKNSIKSSIYMNLENRGIVMED--IGRQLLMSGRVISPAE 122
+Q+ +T VT E++RAK+ +K + E+ V + +G ++L+ G +S E
Sbjct 346 FTLKQWNRLTISVTDTEVERAKSLLKLQLGQLYESGNPVNDANLLGAEVLIKGSKLSLGE 405
Query 123 FCRAIDNVKEADIRRVA-EKMFSKPPTVVAYGDICSVPHYEEIRA 166
+ ID + D++ A ++++ + + G I + Y IR+
Sbjct 406 AFKKIDAITVKDVKAWAGKRLWDQDIAIAGTGQIEGLLDYMRIRS 450
> mmu:19011 Endou, Pp11r, Tcl-30; endonuclease, polyU-specific;
K14648 poly(U)-specific endoribonuclease [EC:3.1.-.-]
Length=412
Score = 34.7 bits (78), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 35/70 (50%), Gaps = 8/70 (11%)
Query 77 VTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIR 136
VT EEL+ SI +IY N+ EDI +L S ISP+E +D E
Sbjct 137 VTKEELE----SISETIYRVDTNKA-QKEDI---VLNSQNRISPSETGNQVDRSPEPLFT 188
Query 137 RVAEKMFSKP 146
V EK+FSKP
Sbjct 189 YVNEKLFSKP 198
> hsa:8909 ENDOU, MGC133268, P11, PP11, PRSS26; endonuclease,
polyU-specific; K14648 poly(U)-specific endoribonuclease [EC:3.1.-.-]
Length=410
Score = 34.3 bits (77), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 34/70 (48%), Gaps = 8/70 (11%)
Query 77 VTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIR 136
+T EE+Q SI IY N+ EDI +L S ISP+E +D +
Sbjct 136 ITKEEIQ----SISEKIYRADTNKA-QKEDI---VLNSQNCISPSETRNQVDRCPKPLFT 187
Query 137 RVAEKMFSKP 146
V EK+FSKP
Sbjct 188 YVNEKLFSKP 197
> ath:AT5G18510 hypothetical protein
Length=702
Score = 33.1 bits (74), Expect = 0.53, Method: Composition-based stats.
Identities = 26/106 (24%), Positives = 45/106 (42%), Gaps = 1/106 (0%)
Query 48 LFGLYMLANPNKASSAVKVMAEQFAGMTRVTAEELQRAKNSIKSSIYMNLENRGIVMEDI 107
L G +L +P + M + + +V + L +AK + S + RG ME
Sbjct 129 LLGFSVLGSPVFSPLECSEMRDSAEKLEKVRRDSLGKAKRVSQRSWTSSFMGRGGQMEHE 188
Query 108 GRQLLMSGRVISPAEFCRAID-NVKEADIRRVAEKMFSKPPTVVAY 152
+L + P +FCR+I NV +R + + P V+A+
Sbjct 189 AFLVLWLSLFVFPGKFCRSISTNVIPIAVRLARGERIALAPAVLAF 234
> ath:AT2G41220 GLU2; GLU2 (GLUTAMATE SYNTHASE 2); glutamate synthase
(ferredoxin); K00284 glutamate synthase (ferredoxin)
[EC:1.4.7.1]
Length=1629
Score = 31.6 bits (70), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 35/68 (51%), Gaps = 2/68 (2%)
Query 4 GGGGAFSTGGPGKGMYTRLYLNVLNKYDWVESAMAFNTQYTDTGLFGLYMLANPNKASSA 63
GG G+ +T G G G+ T + ++ N++ + +F+ +T G+ L++ + N A
Sbjct 139 GGCGSDNTSGDGSGLMTSIPWDLFNEWAEKQGIASFDRTHTGVGM--LFLPRDDNIRKEA 196
Query 64 VKVMAEQF 71
KV+ F
Sbjct 197 KKVITSIF 204
> cel:F21F3.5 unc-38; UNCoordinated family member (unc-38); K05312
nicotinic acetylcholine receptor, invertebrate
Length=511
Score = 30.4 bits (67), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 128 DNVKEADIRRVAEKMFSKPPTVVAYGDICSV 158
+N++ A I +KM+ PP V A+ +IC +
Sbjct 408 NNIRNATIDDTIQKMYYSPPVVKAFENICFI 438
> bbo:BBOV_I000420 16.m00758; hypothetical protein
Length=1736
Score = 30.4 bits (67), Expect = 3.4, Method: Composition-based stats.
Identities = 27/111 (24%), Positives = 50/111 (45%), Gaps = 8/111 (7%)
Query 42 QYTDTGLFGLYM------LAN-PNKASSAVKVMAEQFAGMTRVTAEELQRAKNSIKSSIY 94
+YTD + + + L N P KA + ++ E F M + EE+ + N KSS Y
Sbjct 1499 EYTDEPILHVLLQVLMLCLGNLPVKAEQNMNLVKELFYRMAKTFPEEVNQVWNVCKSS-Y 1557
Query 95 MNLENRGIVMEDIGRQLLMSGRVISPAEFCRAIDNVKEADIRRVAEKMFSK 145
+ + R +I ++L+ ++ + I +AD R V+ + +K
Sbjct 1558 IKKQIREFTKAEITQKLIADELRMAKYDTSTTIRITHDADYRNVSASLDTK 1608
> tgo:TGME49_111860 hypothetical protein
Length=3213
Score = 29.6 bits (65), Expect = 5.3, Method: Composition-based stats.
Identities = 20/83 (24%), Positives = 36/83 (43%), Gaps = 2/83 (2%)
Query 68 AEQFAGMTRVTAEEL--QRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRVISPAEFCR 125
AE G TR +++ K + + +E + +E +GR++ G V SP + C+
Sbjct 2163 AEGGMGATRCGTDDMLGWEVKGKAPRAGKLQVEGQKATLETVGREVAEPGAVTSPRDECK 2222
Query 126 AIDNVKEADIRRVAEKMFSKPPT 148
D + A +A S P +
Sbjct 2223 GRDGRRRAGTVHLAPSEASHPES 2245
> mmu:108000 Cenpf, 6530404A22Rik, AI325968, CENF, Lek1, PRO1779,
mitosin; centromere protein F; K11499 centromere protein
F
Length=2997
Score = 29.6 bits (65), Expect = 6.0, Method: Composition-based stats.
Identities = 20/85 (23%), Positives = 41/85 (48%), Gaps = 1/85 (1%)
Query 58 NKASSAVKVMAEQFAGMTRVTAEELQRAKNSIKSSIYMNLENRGIVMEDIGRQLLMSGRV 117
++ + + + ++ M +ELQ+ K+ + S++ + LE V + + R L
Sbjct 394 SRQHQSFQALDSEYTQMKTRLTQELQQVKH-LHSTLQLELEKVTSVKQQLERNLEEIRLK 452
Query 118 ISPAEFCRAIDNVKEADIRRVAEKM 142
+S AE V E ++RR +E+M
Sbjct 453 LSRAEQALQASQVAENELRRSSEEM 477
Lambda K H
0.318 0.134 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4535951560
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40