bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1760_orf1
Length=110
Score E
Sequences producing significant alignments: (Bits) Value
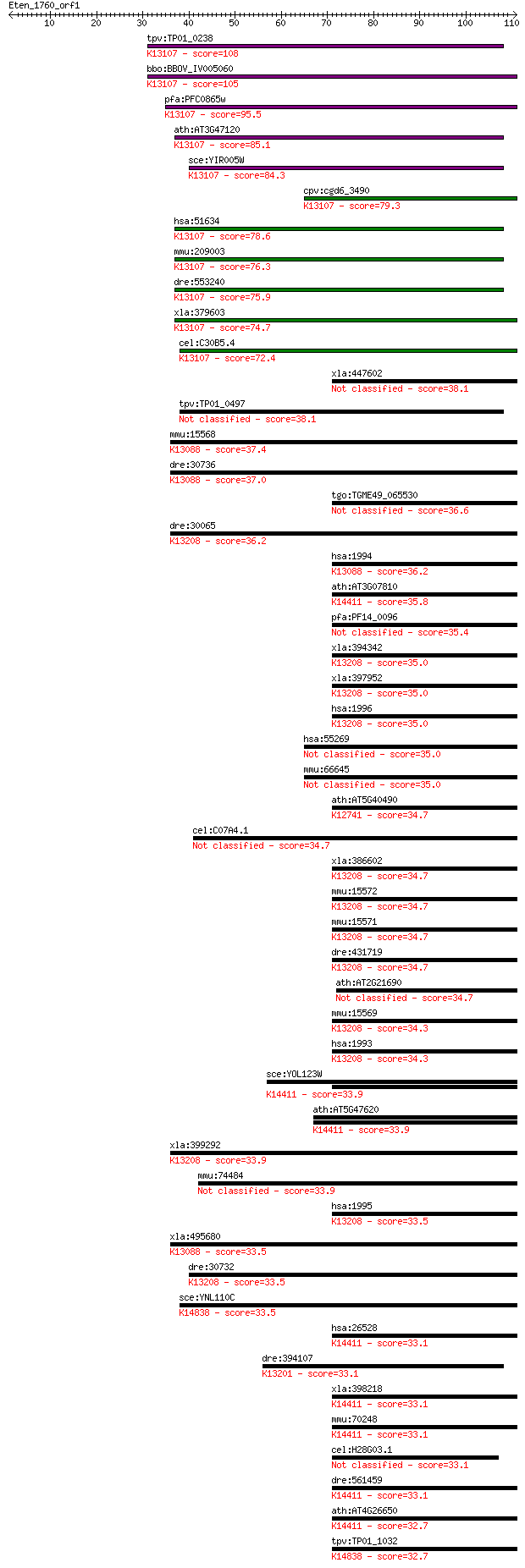
tpv:TP01_0238 hypothetical protein; K13107 RNA-binding motif p... 108 6e-24
bbo:BBOV_IV005060 23.m06028; RNA-binding protein; K13107 RNA-b... 105 4e-23
pfa:PFC0865w RNA binding protein, putative; K13107 RNA-binding... 95.5 3e-20
ath:AT3G47120 RNA recognition motif (RRM)-containing protein; ... 85.1 5e-17
sce:YIR005W IST3, SNU17; Component of the U2 snRNP, required f... 84.3 1e-16
cpv:cgd6_3490 RNA-binding protein ; K13107 RNA-binding motif p... 79.3 2e-15
hsa:51634 RBMX2, CGI-79; RNA binding motif protein, X-linked 2... 78.6 5e-15
mmu:209003 Rbmx2, 2700089A17Rik, 2810411A03Rik, CGI-79; RNA bi... 76.3 3e-14
dre:553240 rbmx2, im:7141316, zgc:113884; RNA binding motif pr... 75.9 3e-14
xla:379603 rbmx2, MGC64376; RNA binding motif protein, X-linke... 74.7 6e-14
cel:C30B5.4 hypothetical protein; K13107 RNA-binding motif pro... 72.4 3e-13
xla:447602 MGC84815 protein 38.1 0.007
tpv:TP01_0497 hypothetical protein 38.1 0.008
mmu:15568 Elavl1, 2410055N02Rik, HUR, Hua, W91709; ELAV (embry... 37.4 0.012
dre:30736 elavl1, elra, fa25f08, fb59a02, fc60g02, hua, wu:fa2... 37.0 0.015
tgo:TGME49_065530 RNA binding motif-containing protein 36.6 0.019
dre:30065 hug, fb72d01, fk65g02, wu:fb72d01, wu:fk65g02, zHuG;... 36.2 0.024
hsa:1994 ELAVL1, ELAV1, HUR, Hua, MelG; ELAV (embryonic lethal... 36.2 0.025
ath:AT3G07810 heterogeneous nuclear ribonucleoprotein, putativ... 35.8 0.032
pfa:PF14_0096 RNA binding protein, putative 35.4 0.042
xla:394342 elavl2-a, Xel-1, el-1, elavl2, elrB, hel-n1, heln1,... 35.0 0.060
xla:397952 elavl2-b, elavl2, elrB, hel-n1, heln1, hub, xel-1; ... 35.0 0.062
hsa:1996 ELAVL4, HUD, PNEM; ELAV (embryonic lethal, abnormal v... 35.0 0.062
hsa:55269 PSPC1, DKFZp566B1447, FLJ10955, PSP1; paraspeckle co... 35.0 0.063
mmu:66645 Pspc1, 5730470C09Rik, AI327109, AI449052; paraspeckl... 35.0 0.066
ath:AT5G40490 RNA recognition motif (RRM)-containing protein; ... 34.7 0.069
cel:C07A4.1 hypothetical protein 34.7 0.073
xla:386602 elavl4, HuD, elrD, elrD1, elrD2; ELAV (embryonic le... 34.7 0.073
mmu:15572 Elavl4, Elav, Hud, PNEM; ELAV (embryonic lethal, abn... 34.7 0.075
mmu:15571 Elavl3, 2600009P04Rik, Huc, PLE21, mHuC; ELAV (embry... 34.7 0.075
dre:431719 elavl2, zgc:91918; ELAV (embryonic lethal, abnormal... 34.7 0.085
ath:AT2G21690 RNA-binding protein, putative 34.7 0.087
mmu:15569 Elavl2, Hub, mel-N1; ELAV (embryonic lethal, abnorma... 34.3 0.093
hsa:1993 ELAVL2, HEL-N1, HELN1, HUB; ELAV (embryonic lethal, a... 34.3 0.099
sce:YOL123W HRP1, NAB4, NAB5; Hrp1p; K14411 RNA-binding protei... 33.9 0.12
ath:AT5G47620 heterogeneous nuclear ribonucleoprotein, putativ... 33.9 0.12
xla:399292 elavl1-a, Elavl1, MGC132161, Xel-1, elav1, elrA, hu... 33.9 0.13
mmu:74484 Rbm31y, 4933434H11Rik; RNA binding motif 31, Y-linked 33.9 0.14
hsa:1995 ELAVL3, DKFZp547J036, HUC, HUCL, MGC20653, PLE21; ELA... 33.5 0.15
xla:495680 elavl1-b, ElrA, Xel-1, elav1, elavl1, hua, hur, mel... 33.5 0.16
dre:30732 elavl3, HuC, elrc, id:ibd1248, wu:fb77b03, zHuC; ELA... 33.5 0.19
sce:YNL110C NOP15; Constituent of 66S pre-ribosomal particles,... 33.5 0.19
hsa:26528 DAZAP1, MGC19907; DAZ associated protein 1; K14411 R... 33.1 0.20
dre:394107 tial1, MGC66130, zgc:66130; TIA1 cytotoxic granule-... 33.1 0.20
xla:398218 dazap1; DAZ associated protein 1; K14411 RNA-bindin... 33.1 0.20
mmu:70248 Dazap1, 2410042M16Rik, mPrrp; DAZ associated protein... 33.1 0.20
cel:H28G03.1 hypothetical protein 33.1 0.25
dre:561459 dazap1; DAZ associated protein 1; K14411 RNA-bindin... 33.1 0.25
ath:AT4G26650 RNA recognition motif (RRM)-containing protein; ... 32.7 0.27
tpv:TP01_1032 hypothetical protein; K14838 nucleolar protein 15 32.7
> tpv:TP01_0238 hypothetical protein; K13107 RNA-binding motif
protein, X-linked 2
Length=310
Score = 108 bits (269), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 49/77 (63%), Positives = 59/77 (76%), Gaps = 0/77 (0%)
Query 31 LGVTGAFSNINAIQRLNAEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFS 90
+ VTG +N NAI RL+ EL+ GI E SWHN YKDSCYI+I GLD R+TEGD+ IVFS
Sbjct 1 MSVTGPAANTNAIHRLSELELKKGIIGEGSWHNQYKDSCYIFIGGLDHRMTEGDIIIVFS 60
Query 91 QWGEPIDVVLVRDQKTG 107
Q+GEPID+ L RD+ TG
Sbjct 61 QFGEPIDINLKRDKDTG 77
> bbo:BBOV_IV005060 23.m06028; RNA-binding protein; K13107 RNA-binding
motif protein, X-linked 2
Length=248
Score = 105 bits (262), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 49/80 (61%), Positives = 61/80 (76%), Gaps = 0/80 (0%)
Query 31 LGVTGAFSNINAIQRLNAEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFS 90
+ VTG +NI+AI RL+ EL GI E+SWHN YKDSCYI+I GL R+TEGD+ IVFS
Sbjct 1 MSVTGPSANISAISRLSEFELEKGIIGESSWHNRYKDSCYIFIGGLHRRMTEGDIIIVFS 60
Query 91 QWGEPIDVVLVRDQKTGVPR 110
Q+G+PID+ L RD+KTG R
Sbjct 61 QFGDPIDIHLKRDKKTGASR 80
> pfa:PFC0865w RNA binding protein, putative; K13107 RNA-binding
motif protein, X-linked 2
Length=309
Score = 95.5 bits (236), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 43/76 (56%), Positives = 55/76 (72%), Gaps = 0/76 (0%)
Query 35 GAFSNINAIQRLNAEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGE 94
G F I I++LN EL+ + ++SWH+ Y+DS YIYI LD RLTEGD+ IVFSQ+GE
Sbjct 2 GLFDKIRNIEKLNEAELKNIGNNDSSWHDQYRDSSYIYIGNLDNRLTEGDIVIVFSQYGE 61
Query 95 PIDVVLVRDQKTGVPR 110
PIDV LVRD +TG +
Sbjct 62 PIDVNLVRDNETGKSK 77
> ath:AT3G47120 RNA recognition motif (RRM)-containing protein;
K13107 RNA-binding motif protein, X-linked 2
Length=352
Score = 85.1 bits (209), Expect = 5e-17, Method: Composition-based stats.
Identities = 37/71 (52%), Positives = 51/71 (71%), Gaps = 0/71 (0%)
Query 37 FSNINAIQRLNAEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEPI 96
+ + +Q++NA E LGIS EASWH YK+S Y+Y+ G+ LTEGD+ VFSQ+GE +
Sbjct 4 LTQVKNLQKINARESDLGISDEASWHAKYKNSAYVYVGGIPFDLTEGDLLAVFSQYGEIV 63
Query 97 DVVLVRDQKTG 107
DV L+RD+ TG
Sbjct 64 DVNLIRDKGTG 74
> sce:YIR005W IST3, SNU17; Component of the U2 snRNP, required
for the first catalytic step of splicing and for spliceosomal
assembly; interacts with Rds3p and is required for Mer1p-activated
splicing; K13107 RNA-binding motif protein, X-linked
2
Length=148
Score = 84.3 bits (207), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 38/69 (55%), Positives = 51/69 (73%), Gaps = 1/69 (1%)
Query 40 INAIQRLNAEELRLGI-SQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEPIDV 98
+N IQ++N +EL+ GI S SWHN YKD+ YIYI L+ LTEGD+ VFS++G P+DV
Sbjct 1 MNKIQQINDKELQSGILSPHQSWHNEYKDNAYIYIGNLNRELTEGDILTVFSEYGVPVDV 60
Query 99 VLVRDQKTG 107
+L RD+ TG
Sbjct 61 ILSRDENTG 69
> cpv:cgd6_3490 RNA-binding protein ; K13107 RNA-binding motif
protein, X-linked 2
Length=153
Score = 79.3 bits (194), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 33/46 (71%), Positives = 41/46 (89%), Gaps = 0/46 (0%)
Query 65 YKDSCYIYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
YK+S YIYISGLD RLTEGD+AI+FSQWGEPID+ L+RD+K G+ +
Sbjct 1 YKNSSYIYISGLDLRLTEGDIAIIFSQWGEPIDINLIRDKKLGISK 46
> hsa:51634 RBMX2, CGI-79; RNA binding motif protein, X-linked
2; K13107 RNA-binding motif protein, X-linked 2
Length=322
Score = 78.6 bits (192), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 34/71 (47%), Positives = 51/71 (71%), Gaps = 0/71 (0%)
Query 37 FSNINAIQRLNAEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEPI 96
+ + I LN E++LG++ + SWH+ YKDS +I++ GL LTEGD+ VFSQ+GE +
Sbjct 4 LTKVKLINELNEREVQLGVADKVSWHSEYKDSAWIFLGGLPYELTEGDIICVFSQYGEIV 63
Query 97 DVVLVRDQKTG 107
++ LVRD+KTG
Sbjct 64 NINLVRDKKTG 74
> mmu:209003 Rbmx2, 2700089A17Rik, 2810411A03Rik, CGI-79; RNA
binding motif protein, X-linked 2; K13107 RNA-binding motif
protein, X-linked 2
Length=326
Score = 76.3 bits (186), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 33/71 (46%), Positives = 51/71 (71%), Gaps = 0/71 (0%)
Query 37 FSNINAIQRLNAEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEPI 96
+ + I LN E++LG++++ SWH+ YK S +I++ GL LTEGD+ VFSQ+GE +
Sbjct 4 LTKVKLINELNEREVQLGVAEKVSWHSEYKHSAWIFVGGLPYELTEGDIICVFSQYGEIV 63
Query 97 DVVLVRDQKTG 107
++ LVRD+KTG
Sbjct 64 NINLVRDKKTG 74
> dre:553240 rbmx2, im:7141316, zgc:113884; RNA binding motif
protein, X-linked 2; K13107 RNA-binding motif protein, X-linked
2
Length=434
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 33/71 (46%), Positives = 47/71 (66%), Gaps = 0/71 (0%)
Query 37 FSNINAIQRLNAEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEPI 96
+ + I LN E +LGI + SWH+ YK+S +++I G LTEGD+ VFSQ+GE
Sbjct 4 LTKVKLINELNEREAQLGIKETVSWHSVYKNSAWVFIGGFPYELTEGDIICVFSQYGEIA 63
Query 97 DVVLVRDQKTG 107
++ LVRD+KTG
Sbjct 64 NINLVRDKKTG 74
> xla:379603 rbmx2, MGC64376; RNA binding motif protein, X-linked
2; K13107 RNA-binding motif protein, X-linked 2
Length=272
Score = 74.7 bits (182), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 35/74 (47%), Positives = 46/74 (62%), Gaps = 0/74 (0%)
Query 37 FSNINAIQRLNAEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEPI 96
+ + I LNA E LG+ SWH YKDS +I+I GL LTEGDV VFSQ+GE +
Sbjct 4 LTKVKLINELNAREASLGVKDSVSWHQDYKDSAWIFIGGLPFELTEGDVICVFSQYGEVV 63
Query 97 DVVLVRDQKTGVPR 110
++ L RD+ +G R
Sbjct 64 NINLARDKSSGRSR 77
> cel:C30B5.4 hypothetical protein; K13107 RNA-binding motif protein,
X-linked 2
Length=302
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 36/75 (48%), Positives = 49/75 (65%), Gaps = 2/75 (2%)
Query 38 SNINAIQRLNAEELRLGISQE--ASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEP 95
+NI R+N EL LG + + SWH YKDS +IYI GL L+EGDV VFSQ+GE
Sbjct 5 TNIKNQNRMNERELSLGYAGDLKKSWHQTYKDSAWIYIGGLSYALSEGDVIAVFSQYGEV 64
Query 96 IDVVLVRDQKTGVPR 110
+++ L+RD+ TG +
Sbjct 65 MNINLIRDKDTGKSK 79
> xla:447602 MGC84815 protein
Length=291
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+++ GL+P+ E + +SQWGE DVV+++D ++ R
Sbjct 15 LFVGGLNPKTNEDSLRAYYSQWGEITDVVVMKDPRSNKSR 54
> tpv:TP01_0497 hypothetical protein
Length=539
Score = 38.1 bits (87), Expect = 0.008, Method: Composition-based stats.
Identities = 23/72 (31%), Positives = 38/72 (52%), Gaps = 4/72 (5%)
Query 38 SNINAIQ--RLNAEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEP 95
S++ A+Q E RLG +Q A D +++ L LTE D+A +F +GEP
Sbjct 206 SSLGAVQIRYATGEVERLGFTQMAGEPG--MDEAKLFVGSLPKSLTEDDLASLFKDFGEP 263
Query 96 IDVVLVRDQKTG 107
++V +++D G
Sbjct 264 LEVFVLKDLTCG 275
> mmu:15568 Elavl1, 2410055N02Rik, HUR, Hua, W91709; ELAV (embryonic
lethal, abnormal vision, Drosophila)-like 1 (Hu antigen
R); K13088 ELAV-like protein 1
Length=326
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 46/75 (61%), Gaps = 4/75 (5%)
Query 36 AFSNINAIQRLNAEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEP 95
A S +N + RL ++ +++ ++ +S KD+ +YISGL +T+ DV +FS++G
Sbjct 77 AISTLNGL-RLQSKTIKVSYARPSS--EVIKDAN-LYISGLPRTMTQKDVEDMFSRFGRI 132
Query 96 IDVVLVRDQKTGVPR 110
I+ ++ DQ TG+ R
Sbjct 133 INSRVLVDQTTGLSR 147
> dre:30736 elavl1, elra, fa25f08, fb59a02, fc60g02, hua, wu:fa25f08,
wu:fb59a02, wu:fc60g02, zHuA; ELAV (embryonic lethal,
abnormal vision, Drosophila)-like 1 (Hu antigen R); K13088
ELAV-like protein 1
Length=324
Score = 37.0 bits (84), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 46/75 (61%), Gaps = 4/75 (5%)
Query 36 AFSNINAIQRLNAEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEP 95
A S +N + RL ++ +++ ++ +S + KD+ +YISGL +T+ DV +F ++G
Sbjct 75 AISTLNGL-RLQSKTIKVSYARPSS--DSIKDAN-LYISGLPKTMTQKDVEEMFGRYGRI 130
Query 96 IDVVLVRDQKTGVPR 110
I+ ++ DQ +G+ R
Sbjct 131 INSRVLVDQASGLSR 145
> tgo:TGME49_065530 RNA binding motif-containing protein
Length=492
Score = 36.6 bits (83), Expect = 0.019, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
I++ G++P L+E DV FS++G V ++RD TG R
Sbjct 195 IFVGGVNPSLSESDVEKCFSKFGTVDKVSIIRDAATGKSR 234
> dre:30065 hug, fb72d01, fk65g02, wu:fb72d01, wu:fk65g02, zHuG;
HuG; K13208 ELAV like protein 2/3/4
Length=322
Score = 36.2 bits (82), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 47/75 (62%), Gaps = 4/75 (5%)
Query 36 AFSNINAIQRLNAEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEP 95
A + +N + RL ++ +++ ++ +S + KD+ +YISGL +T+ +V +F+Q+G
Sbjct 75 AINTLNGL-RLQSKTIKVSYARPSS--DGIKDAN-LYISGLPKTMTQKNVEDMFTQYGRI 130
Query 96 IDVVLVRDQKTGVPR 110
I+ ++ DQ +G+ R
Sbjct 131 INSRILVDQASGLSR 145
> hsa:1994 ELAVL1, ELAV1, HUR, Hua, MelG; ELAV (embryonic lethal,
abnormal vision, Drosophila)-like 1 (Hu antigen R); K13088
ELAV-like protein 1
Length=326
Score = 36.2 bits (82), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 17/40 (42%), Positives = 27/40 (67%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+YISGL +T+ DV +FS++G I+ ++ DQ TG+ R
Sbjct 108 LYISGLPRTMTQKDVEDMFSRFGRIINSRVLVDQTTGLSR 147
> ath:AT3G07810 heterogeneous nuclear ribonucleoprotein, putative
/ hnRNP, putative; K14411 RNA-binding protein Musashi
Length=494
Score = 35.8 bits (81), Expect = 0.032, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
I++ GL +TE D F Q+G DVV++ D T PR
Sbjct 110 IFVGGLPSSVTESDFKTYFEQFGTTTDVVVMYDHNTQRPR 149
> pfa:PF14_0096 RNA binding protein, putative
Length=514
Score = 35.4 bits (80), Expect = 0.042, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+++S + LTE D+ +F ++G DVV ++D+K V R
Sbjct 56 LFVSSIPKNLTEEDIKSIFQEYGNIQDVVFIKDKKPNVNR 95
> xla:394342 elavl2-a, Xel-1, el-1, elavl2, elrB, hel-n1, heln1,
hub, p45; ELAV (embryonic lethal, abnormal vision)-like 2
(Hu antigen B); K13208 ELAV like protein 2/3/4
Length=389
Score = 35.0 bits (79), Expect = 0.060, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+Y+SGL +T+ ++ +FSQ+G I ++ DQ TGV R
Sbjct 155 LYVSGLPKTMTQKELEQLFSQYGRIITSRILVDQVTGVSR 194
> xla:397952 elavl2-b, elavl2, elrB, hel-n1, heln1, hub, xel-1;
ELAV (embryonic lethal, abnormal vision)-like 2 (Hu antigen
B); K13208 ELAV like protein 2/3/4
Length=389
Score = 35.0 bits (79), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+Y+SGL +T+ ++ +FSQ+G I ++ DQ TGV R
Sbjct 155 LYVSGLPKTMTQKELEQLFSQYGRIITSRILVDQVTGVSR 194
> hsa:1996 ELAVL4, HUD, PNEM; ELAV (embryonic lethal, abnormal
vision, Drosophila)-like 4 (Hu antigen D); K13208 ELAV like
protein 2/3/4
Length=366
Score = 35.0 bits (79), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+Y+SGL +T+ ++ +FSQ+G I ++ DQ TGV R
Sbjct 134 LYVSGLPKTMTQKELEQLFSQYGRIITSRILVDQVTGVSR 173
> hsa:55269 PSPC1, DKFZp566B1447, FLJ10955, PSP1; paraspeckle
component 1
Length=523
Score = 35.0 bits (79), Expect = 0.063, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 25/46 (54%), Gaps = 0/46 (0%)
Query 65 YKDSCYIYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
Y C +++ L +TE D +F ++GEP +V + RD+ G R
Sbjct 78 YTQRCRLFVGNLPTDITEEDFKRLFERYGEPSEVFINRDRGFGFIR 123
> mmu:66645 Pspc1, 5730470C09Rik, AI327109, AI449052; paraspeckle
protein 1
Length=523
Score = 35.0 bits (79), Expect = 0.066, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 25/46 (54%), Gaps = 0/46 (0%)
Query 65 YKDSCYIYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
Y C +++ L +TE D +F ++GEP +V + RD+ G R
Sbjct 77 YTQRCRLFVGNLPTDITEEDFKRLFERYGEPSEVFINRDRGFGFIR 122
> ath:AT5G40490 RNA recognition motif (RRM)-containing protein;
K12741 heterogeneous nuclear ribonucleoprotein A1/A3
Length=423
Score = 34.7 bits (78), Expect = 0.069, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 24/40 (60%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
I++ GL T + F ++GE D V+++D+KTG PR
Sbjct 44 IFVGGLARETTSAEFLKHFGKYGEITDSVIMKDRKTGQPR 83
> cel:C07A4.1 hypothetical protein
Length=360
Score = 34.7 bits (78), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 17/70 (24%), Positives = 37/70 (52%), Gaps = 0/70 (0%)
Query 41 NAIQRLNAEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEPIDVVL 100
N R N+EE R ++ E +++ D+ +Y+ + + T+ D+ +FS +G+ +V +
Sbjct 209 NWAARKNSEENRDKLTFEQVFNSTKADNTSVYVGNISQQTTDADLRDLFSTYGDIAEVRI 268
Query 101 VRDQKTGVPR 110
+ Q+ R
Sbjct 269 FKTQRYAFVR 278
> xla:386602 elavl4, HuD, elrD, elrD1, elrD2; ELAV (embryonic
lethal, abnormal vision)-like 4 (Hu antigen D); K13208 ELAV
like protein 2/3/4
Length=400
Score = 34.7 bits (78), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+Y+SGL +T+ ++ +FSQ+G I ++ DQ TGV R
Sbjct 168 LYVSGLPKTMTQKELEQLFSQYGRIITSRILVDQVTGVSR 207
> mmu:15572 Elavl4, Elav, Hud, PNEM; ELAV (embryonic lethal, abnormal
vision, Drosophila)-like 4 (Hu antigen D); K13208 ELAV
like protein 2/3/4
Length=380
Score = 34.7 bits (78), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+Y+SGL +T+ ++ +FSQ+G I ++ DQ TGV R
Sbjct 134 LYVSGLPKTMTQKELEQLFSQYGRIITSRILVDQVTGVSR 173
> mmu:15571 Elavl3, 2600009P04Rik, Huc, PLE21, mHuC; ELAV (embryonic
lethal, abnormal vision, Drosophila)-like 3 (Hu antigen
C); K13208 ELAV like protein 2/3/4
Length=367
Score = 34.7 bits (78), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+Y+SGL +++ ++ +FSQ+G I ++ DQ TGV R
Sbjct 127 LYVSGLPKTMSQKEMEQLFSQYGRIITSRILLDQATGVSR 166
> dre:431719 elavl2, zgc:91918; ELAV (embryonic lethal, abnormal
vision, Drosophila)-like 2 (Hu antigen B); K13208 ELAV like
protein 2/3/4
Length=360
Score = 34.7 bits (78), Expect = 0.085, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+Y+SGL +T+ ++ +FSQ+G I ++ DQ TGV R
Sbjct 127 LYVSGLPKTMTQKELEQLFSQFGRIITSRILVDQVTGVSR 166
> ath:AT2G21690 RNA-binding protein, putative
Length=117
Score = 34.7 bits (78), Expect = 0.087, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 72 YISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
++ GLD E D+ +FS++G ID ++ D+ TG R
Sbjct 10 FVRGLDQDTDEKDLTDIFSKFGNVIDSKIIYDRDTGKSR 48
> mmu:15569 Elavl2, Hub, mel-N1; ELAV (embryonic lethal, abnormal
vision, Drosophila)-like 2 (Hu antigen B); K13208 ELAV like
protein 2/3/4
Length=359
Score = 34.3 bits (77), Expect = 0.093, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+Y+SGL +T+ ++ +FSQ+G I ++ DQ TG+ R
Sbjct 127 LYVSGLPKTMTQKELEQLFSQYGRIITSRILVDQVTGISR 166
> hsa:1993 ELAVL2, HEL-N1, HELN1, HUB; ELAV (embryonic lethal,
abnormal vision, Drosophila)-like 2 (Hu antigen B); K13208
ELAV like protein 2/3/4
Length=346
Score = 34.3 bits (77), Expect = 0.099, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+Y+SGL +T+ ++ +FSQ+G I ++ DQ TG+ R
Sbjct 127 LYVSGLPKTMTQKELEQLFSQYGRIITSRILVDQVTGISR 166
> sce:YOL123W HRP1, NAB4, NAB5; Hrp1p; K14411 RNA-binding protein
Musashi
Length=534
Score = 33.9 bits (76), Expect = 0.12, Method: Composition-based stats.
Identities = 17/54 (31%), Positives = 31/54 (57%), Gaps = 0/54 (0%)
Query 57 QEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+E S + K+SC ++I GL+ TE ++ F ++G D+ +++D TG R
Sbjct 147 EERSKADLSKESCKMFIGGLNWDTTEDNLREYFGKYGTVTDLKIMKDPATGRSR 200
Score = 33.9 bits (76), Expect = 0.12, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
I++ G+ P + + FSQWG ID L+ D+ TG R
Sbjct 245 IFVGGIGPDVRPKEFEEFFSQWGTIIDAQLMLDKDTGQSR 284
> ath:AT5G47620 heterogeneous nuclear ribonucleoprotein, putative
/ hnRNP, putative; K14411 RNA-binding protein Musashi
Length=431
Score = 33.9 bits (76), Expect = 0.12, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 67 DSCYIYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+S I++ GL +TE + F+Q+G DVV++ D +T PR
Sbjct 104 NSKKIFVGGLASSVTEAEFKKYFAQFGMITDVVVMYDHRTQRPR 147
Score = 31.6 bits (70), Expect = 0.72, Method: Composition-based stats.
Identities = 13/44 (29%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 67 DSCYIYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+SC ++I G+ +E + F +GE ++ V+++D+ TG R
Sbjct 4 ESCKLFIGGISWETSEDRLRDYFHSFGEVLEAVIMKDRATGRAR 47
> xla:399292 elavl1-a, Elavl1, MGC132161, Xel-1, elav1, elrA,
hua, hur, melg, p36; ELAV (embryonic lethal, abnormal vision)-like
1 (Hu antigen R); K13208 ELAV like protein 2/3/4
Length=337
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 44/75 (58%), Gaps = 4/75 (5%)
Query 36 AFSNINAIQRLNAEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEP 95
A + +N + RL ++ +++ ++ +S KD+ +YISGL +T+ DV +F +G
Sbjct 88 AINTLNGL-RLQSKTIKVSFARPSS--ESIKDAN-LYISGLPRTMTQKDVEDMFLPFGHI 143
Query 96 IDVVLVRDQKTGVPR 110
I+ ++ DQ TG+ R
Sbjct 144 INSRVLVDQATGLSR 158
> mmu:74484 Rbm31y, 4933434H11Rik; RNA binding motif 31, Y-linked
Length=565
Score = 33.9 bits (76), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 19/70 (27%), Positives = 39/70 (55%), Gaps = 3/70 (4%)
Query 42 AIQRLN-AEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEPIDVVL 100
A+++ N E+ R G +A+ + +D+ ++I GL +++ + S++GE ID ++
Sbjct 7 AVEKDNETEQFREGFKIDATKNQ--QDASKMFIGGLSQEMSKQVLLEYLSKFGEIIDFII 64
Query 101 VRDQKTGVPR 110
D TG+ R
Sbjct 65 KTDPNTGLSR 74
> hsa:1995 ELAVL3, DKFZp547J036, HUC, HUCL, MGC20653, PLE21; ELAV
(embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu
antigen C); K13208 ELAV like protein 2/3/4
Length=367
Score = 33.5 bits (75), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 26/40 (65%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+Y+SGL +++ ++ +FSQ+G I ++ DQ TGV R
Sbjct 127 LYVSGLPKTMSQKEMEQLFSQYGRIITSRILVDQVTGVSR 166
> xla:495680 elavl1-b, ElrA, Xel-1, elav1, elavl1, hua, hur, melg;
ELAV (embryonic lethal, abnormal vision)-like 1 (Hu antigen
R); K13088 ELAV-like protein 1
Length=326
Score = 33.5 bits (75), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 44/75 (58%), Gaps = 4/75 (5%)
Query 36 AFSNINAIQRLNAEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEP 95
A + +N + RL ++ +++ ++ +S KD+ +YISGL +T+ DV +F +G
Sbjct 77 AINTLNGL-RLQSKTIKVSFARPSS--ETIKDAN-LYISGLPRTMTQKDVEDMFLPFGHI 132
Query 96 IDVVLVRDQKTGVPR 110
I+ ++ DQ TG+ R
Sbjct 133 INSRVLVDQATGLSR 147
> dre:30732 elavl3, HuC, elrc, id:ibd1248, wu:fb77b03, zHuC; ELAV
(embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu
antigen C); K13208 ELAV like protein 2/3/4
Length=345
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 37/71 (52%), Gaps = 1/71 (1%)
Query 40 INAIQRLNAEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEPIDVV 99
IN + L + + +S +D+ +Y+SGL +++ D+ +FSQ+G I
Sbjct 96 INTLNGLKLQTKTIKVSYARPSSASIRDAN-LYVSGLPKTMSQKDMEQLFSQYGRIITSR 154
Query 100 LVRDQKTGVPR 110
++ +Q TG+ R
Sbjct 155 ILVNQVTGISR 165
> sce:YNL110C NOP15; Constituent of 66S pre-ribosomal particles,
involved in 60S ribosomal subunit biogenesis; localizes to
both nucleolus and cytoplasm; K14838 nucleolar protein 15
Length=220
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 39/74 (52%), Gaps = 3/74 (4%)
Query 38 SNINAIQRLN-AEELRLGISQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEPI 96
S + I+RLN ++ S++ Y S IY+S L E +++ F+Q+G+
Sbjct 61 SGTHKIKRLNPKKQANEKKSKDKKTLEEY--SGIIYVSRLPHGFHEKELSKYFAQFGDLK 118
Query 97 DVVLVRDQKTGVPR 110
+V L R++KTG R
Sbjct 119 EVRLARNKKTGNSR 132
> hsa:26528 DAZAP1, MGC19907; DAZ associated protein 1; K14411
RNA-binding protein Musashi
Length=407
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+++ GLD T+ + FSQ+GE +D V+++D+ T R
Sbjct 12 LFVGGLDWSTTQETLRSYFSQYGEVVDCVIMKDKTTNQSR 51
> dre:394107 tial1, MGC66130, zgc:66130; TIA1 cytotoxic granule-associated
RNA binding protein-like 1; K13201 nucleolysin
TIA-1/TIAR
Length=370
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 28/52 (53%), Gaps = 4/52 (7%)
Query 56 SQEASWHNHYKDSCYIYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTG 107
SQ+ NH+ ++++ L P +T D+ F+ +G+ D +V+D TG
Sbjct 87 SQKKDTSNHF----HVFVGDLSPEITTDDIRAAFAPFGKISDARVVKDMTTG 134
> xla:398218 dazap1; DAZ associated protein 1; K14411 RNA-binding
protein Musashi
Length=360
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+++ GLD T+ + FSQ+GE +D V+++D+ T R
Sbjct 12 LFVGGLDWSTTQETLRSYFSQYGEVVDCVIMKDKTTNQSR 51
> mmu:70248 Dazap1, 2410042M16Rik, mPrrp; DAZ associated protein
1; K14411 RNA-binding protein Musashi
Length=406
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+++ GLD T+ + FSQ+GE +D V+++D+ T R
Sbjct 12 LFVGGLDWSTTQETLRSYFSQYGEVVDCVIMKDKTTNQSR 51
> cel:H28G03.1 hypothetical protein
Length=336
Score = 33.1 bits (74), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKT 106
++I GL T+ + FSQWG +D +++RD T
Sbjct 72 LFIGGLSHDTTDEQLGNYFSQWGPVVDAIVIRDPNT 107
> dre:561459 dazap1; DAZ associated protein 1; K14411 RNA-binding
protein Musashi
Length=418
Score = 33.1 bits (74), Expect = 0.25, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
+++ GLD T+ + FSQ+GE +D V+++D+ T R
Sbjct 25 LFVGGLDWSTTQETLRNYFSQYGEVVDCVIMKDKSTNQSR 64
> ath:AT4G26650 RNA recognition motif (RRM)-containing protein;
K14411 RNA-binding protein Musashi
Length=452
Score = 32.7 bits (73), Expect = 0.27, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
I++ GL +TE + F Q+G DVV++ D T PR
Sbjct 121 IFVGGLPSSITEAEFKNYFDQFGTIADVVVMYDHNTQRPR 160
> tpv:TP01_1032 hypothetical protein; K14838 nucleolar protein
15
Length=195
Score = 32.7 bits (73), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 71 IYISGLDPRLTEGDVAIVFSQWGEPIDVVLVRDQKTGVPR 110
IY+ L +LTE + F+Q+G+ I + L + +KT R
Sbjct 15 IYVGNLPKQLTEEQLKTYFNQFGDVIKIRLFKSRKTNRSR 54
Lambda K H
0.322 0.138 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067351240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40