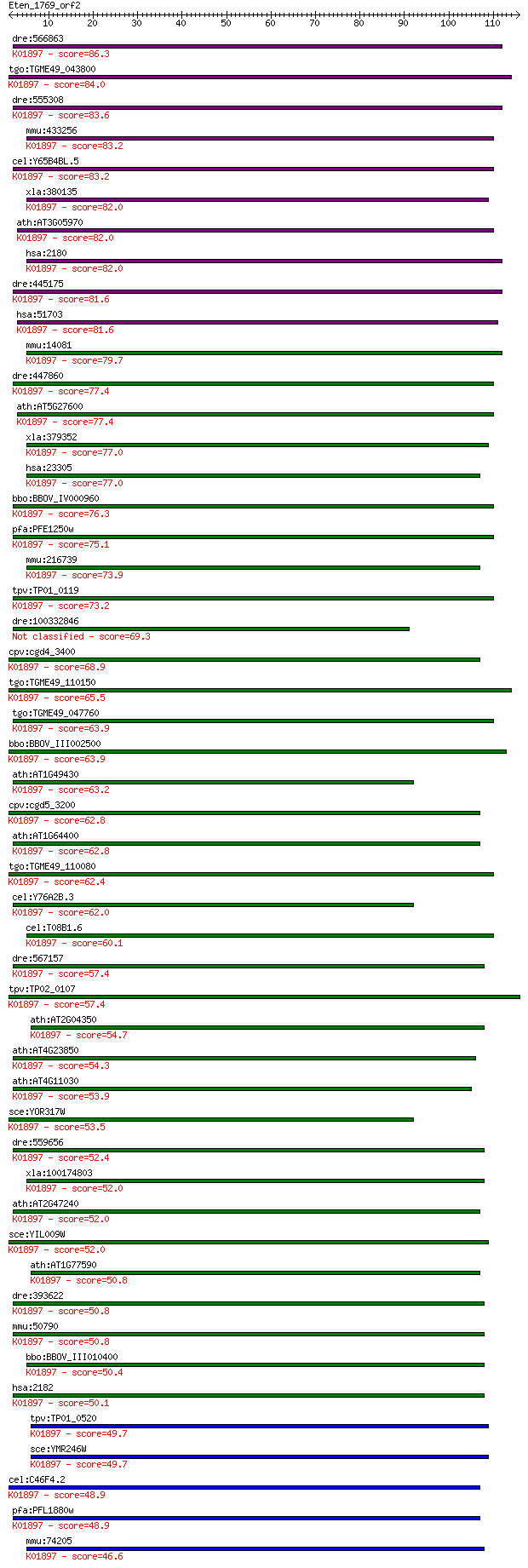
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1769_orf2
Length=115
Score E
Sequences producing significant alignments: (Bits) Value
dre:566863 fa04h02, wu:fc05h07; wu:fa04h02; K01897 long-chain ... 86.3 2e-17
tgo:TGME49_043800 long-chain fatty acid CoA ligase, putative (... 84.0 1e-16
dre:555308 acsl1, MGC110081, zgc:110081; acyl-CoA synthetase l... 83.6 2e-16
mmu:433256 Acsl5, 1700030F05Rik, ACS2, ACS5, Facl5; acyl-CoA s... 83.2 2e-16
cel:Y65B4BL.5 acs-13; fatty Acid CoA Synthetase family member ... 83.2 2e-16
xla:380135 acsl1, MGC132122, MGC52902, facl2; acyl-CoA synthet... 82.0 4e-16
ath:AT3G05970 LACS6; LACS6 (long-chain acyl-CoA synthetase 6);... 82.0 4e-16
hsa:2180 ACSL1, ACS1, FACL1, FACL2, LACS, LACS1, LACS2; acyl-C... 82.0 4e-16
dre:445175 zgc:101071 (EC:6.2.1.3); K01897 long-chain acyl-CoA... 81.6 5e-16
hsa:51703 ACSL5, ACS2, ACS5, FACL5; acyl-CoA synthetase long-c... 81.6 6e-16
mmu:14081 Acsl1, Acas, Acas1, Acs, FACS, Facl2; acyl-CoA synth... 79.7 2e-15
dre:447860 acsl5, zgc:92083; acyl-CoA synthetase long-chain fa... 77.4 9e-15
ath:AT5G27600 LACS7; LACS7 (LONG-CHAIN ACYL-COA SYNTHETASE 7);... 77.4 1e-14
xla:379352 MGC53832; similar to fatty acid Coenzyme A ligase; ... 77.0 1e-14
hsa:23305 ACSL6, ACS2, FACL6, FLJ16173, KIAA0837, LACS_6, LACS... 77.0 1e-14
bbo:BBOV_IV000960 21.m02785; fatty acyl-CoA synthetase 3 (EC:6... 76.3 2e-14
pfa:PFE1250w PfACS10; acyl-CoA synthetase, PfACS10 (EC:6.2.1.3... 75.1 6e-14
mmu:216739 Acsl6, A330035H04Rik, AW050338, Facl6, LACS, Lacsl,... 73.9 1e-13
tpv:TP01_0119 long-chain fatty acid CoA ligase; K01897 long-ch... 73.2 2e-13
dre:100332846 acyl-CoA synthetase long-chain family member 6-like 69.3 3e-12
cpv:cgd4_3400 long chain fatty acyl CoA synthetase having a si... 68.9 4e-12
tgo:TGME49_110150 long-chain-fatty-acid-CoA ligase, putative (... 65.5 4e-11
tgo:TGME49_047760 long chain acyl-CoA synthetase, putative (EC... 63.9 1e-10
bbo:BBOV_III002500 17.m07241; fatty acyl-CoA synthetase family... 63.9 1e-10
ath:AT1G49430 LACS2; LACS2 (LONG-CHAIN ACYL-COA SYNTHETASE 2);... 63.2 2e-10
cpv:cgd5_3200 acyl-CoA synthetase ; K01897 long-chain acyl-CoA... 62.8 3e-10
ath:AT1G64400 long-chain-fatty-acid--CoA ligase, putative / lo... 62.8 3e-10
tgo:TGME49_110080 long-chain-fatty-acid-CoA ligase, putative (... 62.4 3e-10
cel:Y76A2B.3 acs-14; fatty Acid CoA Synthetase family member (... 62.0 5e-10
cel:T08B1.6 hypothetical protein; K01897 long-chain acyl-CoA s... 60.1 2e-09
dre:567157 acsl3a, si:dkeyp-109h9.2; acyl-CoA synthetase long-... 57.4 1e-08
tpv:TP02_0107 long-chain fatty acid CoA ligase; K01897 long-ch... 57.4 1e-08
ath:AT2G04350 long-chain-fatty-acid--CoA ligase family protein... 54.7 6e-08
ath:AT4G23850 long-chain-fatty-acid--CoA ligase / long-chain a... 54.3 1e-07
ath:AT4G11030 long-chain-fatty-acid--CoA ligase, putative / lo... 53.9 1e-07
sce:YOR317W FAA1; Faa1p (EC:6.2.1.3); K01897 long-chain acyl-C... 53.5 2e-07
dre:559656 acsl3b, acsl3, fb34c03, im:7155129, si:dkey-20f20.5... 52.4 3e-07
xla:100174803 acsl4, acs4, acsl3, facl4, lacs4, mrx63, mrx68; ... 52.0 4e-07
ath:AT2G47240 long-chain-fatty-acid--CoA ligase family protein... 52.0 5e-07
sce:YIL009W FAA3; Faa3p (EC:6.2.1.3); K01897 long-chain acyl-C... 52.0 5e-07
ath:AT1G77590 LACS9; LACS9 (LONG CHAIN ACYL-COA SYNTHETASE 9);... 50.8 1e-06
dre:393622 acsl4a, MGC66186, acsl4, zgc:66186; acyl-CoA synthe... 50.8 1e-06
mmu:50790 Acsl4, 9430020A05Rik, ACS4, AU018108, Facl4, Lacs4; ... 50.8 1e-06
bbo:BBOV_III010400 17.m07898; long-chain acyl-CoA synthetase (... 50.4 1e-06
hsa:2182 ACSL4, ACS4, FACL4, LACS4, MRX63, MRX68; acyl-CoA syn... 50.1 2e-06
tpv:TP01_0520 long-chain fatty acid CoA ligase; K01897 long-ch... 49.7 2e-06
sce:YMR246W FAA4; Faa4p (EC:6.2.1.3); K01897 long-chain acyl-C... 49.7 2e-06
cel:C46F4.2 acs-17; fatty Acid CoA Synthetase family member (a... 48.9 4e-06
pfa:PFL1880w ACS11; acyl-CoA synthetase, PfACS11 (EC:6.2.1.3);... 48.9 4e-06
mmu:74205 Acsl3, 2610510B12Rik, Acs3, C85929, Facl3, Pro2194; ... 46.6 2e-05
> dre:566863 fa04h02, wu:fc05h07; wu:fa04h02; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=633
Score = 86.3 bits (212), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 45/110 (40%), Positives = 67/110 (60%), Gaps = 5/110 (4%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLK 61
++ LV I+VPD E W ++ +G D F E ++ ++KA+ +DM ++GK GL
Sbjct 529 QSCLVGIIVPDPEVFPSWAQK-KGFDGG----FHELCENKELKKAILEDMVRLGKASGLH 583
Query 62 GFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEIHM 111
FE + I++H E F++ENGLLTPT K KR +E F EEI+ +Y I M
Sbjct 584 SFEQVKDIYIHKEMFSIENGLLTPTLKAKRPELKEYFKEEIQDLYSSISM 633
> tgo:TGME49_043800 long-chain fatty acid CoA ligase, putative
(EC:6.2.1.3); K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=748
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 45/113 (39%), Positives = 66/113 (58%), Gaps = 4/113 (3%)
Query 1 SETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGL 60
S++ LVAI+VPD E+ +W + + AD + E +AV + M V K L
Sbjct 636 SQSCLVAIIVPDAEKAEKWAVQRKLADTS----LETVCTLPEFHRAVTESMAAVAKEHQL 691
Query 61 KGFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEIHMEQ 113
KGFE+ + L SE F++EN LLTPT K+KR+ A+E F +EI +Y E+ E+
Sbjct 692 KGFEVVKHFRLVSEPFSIENELLTPTMKIKRYVAKEFFAKEIDALYSEMETER 744
> dre:555308 acsl1, MGC110081, zgc:110081; acyl-CoA synthetase
long-chain family member 1 (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=697
Score = 83.6 bits (205), Expect = 2e-16, Method: Composition-based stats.
Identities = 46/110 (41%), Positives = 63/110 (57%), Gaps = 5/110 (4%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLK 61
+ LV +VVPD + W + RG + + F + K V+ A+ +DM Q+GK GLK
Sbjct 593 QACLVGVVVPDPDFLPGWAKN-RGIEGS----FNDLCKSKEVKNAILEDMIQLGKEAGLK 647
Query 62 GFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEIHM 111
FE R I LH E F+V+NGLLTPT K KR + F E+I +Y +I M
Sbjct 648 SFEQVRDIALHLEMFSVQNGLLTPTLKAKRTELKSRFREQIDQLYAKIKM 697
> mmu:433256 Acsl5, 1700030F05Rik, ACS2, ACS5, Facl5; acyl-CoA
synthetase long-chain family member 5 (EC:6.2.1.3); K01897
long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=683
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 42/105 (40%), Positives = 62/105 (59%), Gaps = 5/105 (4%)
Query 5 LVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLKGFE 64
L+ +VVPD + + A + FEE K+ V++A+ +D+Q++GK GLK FE
Sbjct 582 LIGVVVPDPDSLPSF-----AAKIGVKGSFEELCKNQCVKEAILEDLQKIGKEGGLKSFE 636
Query 65 LARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEI 109
+ I +H E FT+ENGLLTPT K KR + F +IK++Y I
Sbjct 637 QVKSIFVHPEPFTIENGLLTPTLKAKRVELAKFFQTQIKSLYESI 681
> cel:Y65B4BL.5 acs-13; fatty Acid CoA Synthetase family member
(acs-13); K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=719
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 47/108 (43%), Positives = 63/108 (58%), Gaps = 5/108 (4%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLK 61
+T L+AIVVPD E + + AD FEE + AV+KA+ DDM VGK GL
Sbjct 616 KTCLIAIVVPDAEVLVPAM-----ADQGIKGTFEELCNNDAVKKAILDDMVAVGKKAGLF 670
Query 62 GFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEI 109
FE R I L +EQF+VEN LLTPT K KR + + +++ MY ++
Sbjct 671 SFEQVRDISLSAEQFSVENDLLTPTLKSKRPKLKAHYSKQLDAMYSKL 718
> xla:380135 acsl1, MGC132122, MGC52902, facl2; acyl-CoA synthetase
long-chain family member 1 protein (EC:6.2.1.3); K01897
long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=698
Score = 82.0 bits (201), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 44/106 (41%), Positives = 63/106 (59%), Gaps = 9/106 (8%)
Query 5 LVAIVVPDQEQTLRWL--REVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLKG 62
LV IVVPD + + W R+ G+ +EE K+ + AV +D+ ++GK GLK
Sbjct 597 LVGIVVPDPDNVMNWAKKRKFEGS-------YEELCKNKDFKNAVLEDLLKLGKEAGLKT 649
Query 63 FELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGE 108
FE + I LHSE F++ENGLLTPT K KR ++ F +EI+ +Y
Sbjct 650 FEQVKDIALHSEMFSIENGLLTPTLKAKRPELRKYFKDEIEELYAN 695
> ath:AT3G05970 LACS6; LACS6 (long-chain acyl-CoA synthetase 6);
long-chain-fatty-acid-CoA ligase (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=701
Score = 82.0 bits (201), Expect = 4e-16, Method: Composition-based stats.
Identities = 46/109 (42%), Positives = 62/109 (56%), Gaps = 8/109 (7%)
Query 3 TSLVAIVVPDQEQTLRWLRE--VRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGL 60
+SLVA+V D + W ++G D+ E + V+ AV DM VG+ L
Sbjct 588 SSLVAVVSVDPDVLKSWAASEGIKGGDL------RELCNNPRVKAAVLSDMDTVGREAQL 641
Query 61 KGFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEI 109
+GFE A+ + L E FT+ENGLLTPTFK+KR A+E F E I MY E+
Sbjct 642 RGFEFAKAVTLVLEPFTLENGLLTPTFKIKRPQAKEYFAEAITNMYKEL 690
> hsa:2180 ACSL1, ACS1, FACL1, FACL2, LACS, LACS1, LACS2; acyl-CoA
synthetase long-chain family member 1 (EC:6.2.1.3); K01897
long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=698
Score = 82.0 bits (201), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 45/107 (42%), Positives = 65/107 (60%), Gaps = 5/107 (4%)
Query 5 LVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLKGFE 64
L+AIVVPD E W ++ RG + + FEE ++ V+KA+ +DM ++GK GLK FE
Sbjct 597 LIAIVVPDVETLCSWAQK-RGFEGS----FEELCRNKDVKKAILEDMVRLGKDSGLKPFE 651
Query 65 LARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEIHM 111
+ I LH E F+++NGLLTPT K KR + F +I +Y I +
Sbjct 652 QVKGITLHPELFSIDNGLLTPTMKAKRPELRNYFRSQIDDLYSTIKV 698
> dre:445175 zgc:101071 (EC:6.2.1.3); K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=697
Score = 81.6 bits (200), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 44/110 (40%), Positives = 65/110 (59%), Gaps = 5/110 (4%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLK 61
+ LVAI+VPD + W ++ RG + +E+ K+ ++KA+ +D+ ++GK GLK
Sbjct 593 QACLVAIIVPDPDFLPGWAKK-RGIEGT----YEDLCKNKDIKKAILEDIIKLGKESGLK 647
Query 62 GFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEIHM 111
FE + I LHSE FT++NGLLTPT K KR + F I +Y I M
Sbjct 648 SFEQVKDIALHSEMFTIQNGLLTPTLKAKRADLRNHFRSHIDELYANIKM 697
> hsa:51703 ACSL5, ACS2, ACS5, FACL5; acyl-CoA synthetase long-chain
family member 5 (EC:6.2.1.3); K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=739
Score = 81.6 bits (200), Expect = 6e-16, Method: Composition-based stats.
Identities = 42/108 (38%), Positives = 63/108 (58%), Gaps = 5/108 (4%)
Query 3 TSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLKG 62
+SLV +VVPD + + A + FEE ++ V++A+ +D+Q++GK GLK
Sbjct 636 SSLVGVVVPDTDVLPSF-----AAKLGVKGSFEELCQNQVVREAILEDLQKIGKESGLKT 690
Query 63 FELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEIH 110
FE + I LH E F++ENGLLTPT K KR + F +I ++Y I
Sbjct 691 FEQVKAIFLHPEPFSIENGLLTPTLKAKRGELSKYFRTQIDSLYEHIQ 738
> mmu:14081 Acsl1, Acas, Acas1, Acs, FACS, Facl2; acyl-CoA synthetase
long-chain family member 1 (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=699
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 42/107 (39%), Positives = 63/107 (58%), Gaps = 5/107 (4%)
Query 5 LVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLKGFE 64
L+A+VVPD E W ++ RG + FEE ++ + KA+ DD+ ++GK GLK FE
Sbjct 598 LIAVVVPDVESLPSWAQK-RGLQGS----FEELCRNKDINKAILDDLLKLGKEAGLKPFE 652
Query 65 LARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEIHM 111
+ I +H E F+++NGLLTPT K KR + F +I +Y I +
Sbjct 653 QVKGIAVHPELFSIDNGLLTPTLKAKRPELRNYFRSQIDELYATIKI 699
> dre:447860 acsl5, zgc:92083; acyl-CoA synthetase long-chain
family member 5 (EC:6.2.1.3); K01897 long-chain acyl-CoA synthetase
[EC:6.2.1.3]
Length=681
Score = 77.4 bits (189), Expect = 9e-15, Method: Composition-based stats.
Identities = 42/108 (38%), Positives = 65/108 (60%), Gaps = 5/108 (4%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLK 61
++SLVAIVVPD E L E G + EE ++ ++KA+ D+ ++G+ GLK
Sbjct 579 QSSLVAIVVPDPE-VLPGFAEKLGLKGS----LEELCRNQELKKAIISDLNKLGREAGLK 633
Query 62 GFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEI 109
FE + ++LH + FT+ENGLLTPT K KR + F +I+++Y +
Sbjct 634 SFEQVKDLYLHPDMFTIENGLLTPTLKAKRADLTKYFRVQIESLYANM 681
> ath:AT5G27600 LACS7; LACS7 (LONG-CHAIN ACYL-COA SYNTHETASE 7);
long-chain-fatty-acid-CoA ligase/ protein binding (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=700
Score = 77.4 bits (189), Expect = 1e-14, Method: Composition-based stats.
Identities = 44/107 (41%), Positives = 58/107 (54%), Gaps = 4/107 (3%)
Query 3 TSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLKG 62
+SLVAIV D E W ++ + D V+K V +M +G+ L+G
Sbjct 588 SSLVAIVSVDPEVMKDW----AASEGIKYEHLGQLCNDPRVRKTVLAEMDDLGREAQLRG 643
Query 63 FELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEI 109
FE A+ + L E FT+ENGLLTPTFK+KR A+ F E I MY EI
Sbjct 644 FEFAKAVTLVPEPFTLENGLLTPTFKIKRPQAKAYFAEAISKMYAEI 690
> xla:379352 MGC53832; similar to fatty acid Coenzyme A ligase;
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=698
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 41/106 (38%), Positives = 62/106 (58%), Gaps = 9/106 (8%)
Query 5 LVAIVVPDQEQTLRWL--REVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLKG 62
LV I+VPD E + W R+ G+ +EE K+ + A+ +D+ ++GK GLK
Sbjct 597 LVGIIVPDPENVINWAKKRKFEGS-------YEELCKNKDFKNAILEDLLKLGKEAGLKT 649
Query 63 FELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGE 108
FE + I L SE F++ENGLLTPT K KR ++ F ++I+ +Y
Sbjct 650 FEQVKDIALCSEMFSIENGLLTPTLKAKRPELRKSFKDQIEELYAN 695
> hsa:23305 ACSL6, ACS2, FACL6, FLJ16173, KIAA0837, LACS_6, LACS2,
LACS5; acyl-CoA synthetase long-chain family member 6 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=722
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 41/102 (40%), Positives = 63/102 (61%), Gaps = 5/102 (4%)
Query 5 LVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLKGFE 64
LV IVVPD E W ++ RG + + + + ++KA+ +DM ++GK GL FE
Sbjct 622 LVGIVVPDPEVMPSWAQK-RGIEGT----YADLCTNKDLKKAILEDMVRLGKESGLHSFE 676
Query 65 LARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMY 106
+ IH+HS+ F+V+NGLLTPT K KR +E F ++I+ +Y
Sbjct 677 QVKAIHIHSDMFSVQNGLLTPTLKAKRPELREYFKKQIEELY 718
> bbo:BBOV_IV000960 21.m02785; fatty acyl-CoA synthetase 3 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=681
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 38/108 (35%), Positives = 66/108 (61%), Gaps = 4/108 (3%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLK 61
+ + VAIV+PD++ RW +G + +S F++ + ++KA+ +D+ ++ L+
Sbjct 572 QVAPVAIVIPDEDVIKRW----KGENGMESMSFKDVCRTPELRKAILEDITRLFDENRLR 627
Query 62 GFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEI 109
GFE + I++ E F VEN LLT T KL+RH +E + E+IK +Y E+
Sbjct 628 GFEKVKAIYVDHELFAVENDLLTVTAKLRRHKVRERYAEQIKELYAEL 675
> pfa:PFE1250w PfACS10; acyl-CoA synthetase, PfACS10 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=673
Score = 75.1 bits (183), Expect = 6e-14, Method: Composition-based stats.
Identities = 40/110 (36%), Positives = 62/110 (56%), Gaps = 9/110 (8%)
Query 2 ETSLVAIVVPDQEQTLRW--LREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQG 59
E+ LV ++ P + W ++++ D EE +K + V +D+ +GK G
Sbjct 569 ESVLVCVICPSTDSIDIWRTQKKIKATD-------EEVIKLPEFKADVINDLTSIGKKDG 621
Query 60 LKGFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEI 109
LKGFE + IH E FT+EN L+TPT K+KRH A++ F +EI MY ++
Sbjct 622 LKGFEQIKDIHFTLEAFTIENDLMTPTGKIKRHEAKKRFKKEIDEMYEKL 671
> mmu:216739 Acsl6, A330035H04Rik, AW050338, Facl6, LACS, Lacsl,
MGC36650, mKIAA0837; acyl-CoA synthetase long-chain family
member 6 (EC:6.2.1.3); K01897 long-chain acyl-CoA synthetase
[EC:6.2.1.3]
Length=722
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 40/102 (39%), Positives = 61/102 (59%), Gaps = 5/102 (4%)
Query 5 LVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLKGFE 64
LV IVVPD E W ++ +G + ++E ++KA+ DDM +GK GL FE
Sbjct 622 LVGIVVPDPEVMPSWAQK-KGIEGT----YQELCMKKELKKAILDDMVMLGKESGLHSFE 676
Query 65 LARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMY 106
+ I++H + F+V+NGLLTPT K KR +E F ++I+ +Y
Sbjct 677 QVKAIYIHCDMFSVQNGLLTPTLKAKRPELREYFKKQIEELY 718
> tpv:TP01_0119 long-chain fatty acid CoA ligase; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=691
Score = 73.2 bits (178), Expect = 2e-13, Method: Composition-based stats.
Identities = 37/108 (34%), Positives = 64/108 (59%), Gaps = 4/108 (3%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLK 61
E VAI+VPD+E W ++ G A+ + KD +++ ++++++ V +K
Sbjct 582 ENYPVAIIVPDEEALQSW-KKTHGLTKAN---LTQLAKDPLLKQFLFNELESVMTSSNVK 637
Query 62 GFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEI 109
G+ ++I++H E F+VEN LLT TFKLKR++ ++ F EI MY +
Sbjct 638 GYNRVKEIYIHDELFSVENNLLTVTFKLKRNFLRQKFENEITQMYASL 685
> dre:100332846 acyl-CoA synthetase long-chain family member 6-like
Length=1391
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 40/89 (44%), Positives = 55/89 (61%), Gaps = 5/89 (5%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLK 61
++ L+ IVVPD E + W +E RG + S +EE ++ V+KAV +DM VGK GLK
Sbjct 1308 QSFLIGIVVPDPEVFVNWAKE-RG--IVGS--YEELCQNPDVKKAVLEDMTAVGKEAGLK 1362
Query 62 GFELARKIHLHSEQFTVENGLLTPTFKLK 90
FE + ++LH E F+V GL TPT K K
Sbjct 1363 SFEQVKDLYLHPELFSVSIGLRTPTLKSK 1391
> cpv:cgd4_3400 long chain fatty acyl CoA synthetase having a
signal peptide ; K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=766
Score = 68.9 bits (167), Expect = 4e-12, Method: Composition-based stats.
Identities = 38/119 (31%), Positives = 69/119 (57%), Gaps = 14/119 (11%)
Query 1 SETSLVAIVVPDQEQTLRWLRE---VRGAD----------VADSRPFEEWVKDSAVQKAV 47
+E+ +VA++VPD E W ++ + +D A+++ FEE ++ +++ +
Sbjct 638 TESVIVAVIVPDSEFVKDWSKKNGVIISSDWQAQFKQICTEANTKYFEE-NTNTPLREEI 696
Query 48 YDDMQQVGKMQGLKGFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMY 106
++ V QG+KGF+ +L + FTVENGLLTPT KL RH A +V+ ++++ +Y
Sbjct 697 IASLKHVEDQQGIKGFKKVNDFYLECDGFTVENGLLTPTNKLMRHKANKVYLDKVEKLY 755
> tgo:TGME49_110150 long-chain-fatty-acid-CoA ligase, putative
(EC:6.2.1.3); K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=866
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 39/122 (31%), Positives = 64/122 (52%), Gaps = 9/122 (7%)
Query 1 SETSLVAIVVPDQEQTLRWLREVRG---------ADVADSRPFEEWVKDSAVQKAVYDDM 51
S++ VA+VVP++++ L W R G A +D F+ + D ++K V D
Sbjct 715 SQSYPVAVVVPNRQKVLAWARTAYGRRAEAERESAAQSDETLFKTALADFELKKEVLADF 774
Query 52 QQVGKMQGLKGFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEIHM 111
Q+ + L GFE + + L ++ +T ENG+LTPTFK KR + EI +Y ++
Sbjct 775 DQLAREADLLGFEKVKNVFLTADVWTPENGMLTPTFKSKRTLLVRKYTPEIDELYRQLVN 834
Query 112 EQ 113
+Q
Sbjct 835 KQ 836
> tgo:TGME49_047760 long chain acyl-CoA synthetase, putative (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=793
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 35/108 (32%), Positives = 64/108 (59%), Gaps = 5/108 (4%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLK 61
+ S+VAIVVP ++ TL+W +E V+ + + +K + ++++V M++ G+ + +
Sbjct 683 QNSVVAIVVPCKDVTLKWAKEHNMGQVS----YTDLLKSTDLRQSVLHSMKEAGRGR-VH 737
Query 62 GFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEI 109
GFE + + L FT ENGL TPT K+ R ++ + ++I MY E+
Sbjct 738 GFEEPKNVFLTPHAFTAENGLATPTMKVVRKQVEKEYRKQIDEMYAEL 785
> bbo:BBOV_III002500 17.m07241; fatty acyl-CoA synthetase family
protein; K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=663
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 33/112 (29%), Positives = 61/112 (54%), Gaps = 4/112 (3%)
Query 1 SETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGL 60
+E VAIVVPD+ + W D+ ++ + ++A+ + M Q +
Sbjct 556 TEVYPVAIVVPDETEAQYWAESQGHGDMT----LQDICQHPVFKEAIMEQMAQAYDEANV 611
Query 61 KGFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEIHME 112
KGFE ++I++ +E F++ N +LT T KL+RH+A++ + + I ++Y H E
Sbjct 612 KGFERCKQIYIEAEPFSIGNNMLTTTNKLRRHHAKKRYEDIIDSLYRMSHTE 663
> ath:AT1G49430 LACS2; LACS2 (LONG-CHAIN ACYL-COA SYNTHETASE 2);
long-chain-fatty-acid-CoA ligase (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=665
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 33/90 (36%), Positives = 47/90 (52%), Gaps = 4/90 (4%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLK 61
E+ LV +VVPD++ W + + FE ++ QK D++ K LK
Sbjct 557 ESFLVGVVVPDRKAIEDWAK----LNYQSPNDFESLCQNLKAQKYFLDELNSTAKQYQLK 612
Query 62 GFELARKIHLHSEQFTVENGLLTPTFKLKR 91
GFE+ + IHL F +E L+TPTFKLKR
Sbjct 613 GFEMLKAIHLEPNPFDIERDLITPTFKLKR 642
> cpv:cgd5_3200 acyl-CoA synthetase ; K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=683
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 37/106 (34%), Positives = 58/106 (54%), Gaps = 3/106 (2%)
Query 1 SETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGL 60
+E+ LVAI V D+E T+++ VR + D F++ V + + + D+++ +
Sbjct 571 TESFLVAIFVLDEEFTMKY---VRTNQLGDDLKFKDAVTHPKIIEKIKLDIEKAELQHKI 627
Query 61 KGFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMY 106
G+E R S F+VEN LLTPTFK+ RH A + + E I MY
Sbjct 628 LGYEKIRAFKCISTPFSVENELLTPTFKVVRHKAIKFYEESINEMY 673
> ath:AT1G64400 long-chain-fatty-acid--CoA ligase, putative /
long-chain acyl-CoA synthetase, putative; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=665
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 35/105 (33%), Positives = 58/105 (55%), Gaps = 5/105 (4%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLK 61
E+ LVA+V P + Q W +E + S FE ++ ++ V + +V K + LK
Sbjct 556 ESYLVAVVCPSKIQIEHWAKEHKV-----SGDFESICRNQKTKEFVLGEFNRVAKDKKLK 610
Query 62 GFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMY 106
GFEL + +HL + F +E L+TP++K+KR + + +EI MY
Sbjct 611 GFELIKGVHLDTVPFDMERDLITPSYKMKRPQLLKYYQKEIDEMY 655
> tgo:TGME49_110080 long-chain-fatty-acid-CoA ligase, putative
(EC:6.2.1.3); K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=841
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 36/114 (31%), Positives = 67/114 (58%), Gaps = 7/114 (6%)
Query 1 SETSLVAIVVPDQEQTLRWLREVRGA-----DVADSRPFEEWVKDSAVQKAVYDDMQQVG 55
S++ VA+VVP+++ W E RGA ++ D + F E ++++ ++K V D+ ++
Sbjct 726 SQSYPVAVVVPNRQTVALWA-EKRGATPKAAELTDEQ-FAELLREAELKKDVLLDLNRLA 783
Query 56 KMQGLKGFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEI 109
L GFE + +HL ++ +T +NG+LTPTFK KR + + ++ +Y E+
Sbjct 784 VDANLLGFEKVKNVHLTTDVWTPDNGMLTPTFKTKRAVMAKTYKADMDRLYDEL 837
> cel:Y76A2B.3 acs-14; fatty Acid CoA Synthetase family member
(acs-14); K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=687
Score = 62.0 bits (149), Expect = 5e-10, Method: Composition-based stats.
Identities = 35/90 (38%), Positives = 49/90 (54%), Gaps = 4/90 (4%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLK 61
E L+A+VVP+ + L+E + SR EE D ++ V ++ +GK L
Sbjct 581 ERWLIAVVVPEPDV----LKEWNDKQGSGSRKIEEICNDEKAKEFVLSELHAIGKANKLN 636
Query 62 GFELARKIHLHSEQFTVENGLLTPTFKLKR 91
E +K+ L S+ FTVENGLLTPT K KR
Sbjct 637 SIEQVKKVILTSDTFTVENGLLTPTLKAKR 666
> cel:T08B1.6 hypothetical protein; K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=653
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 37/105 (35%), Positives = 58/105 (55%), Gaps = 3/105 (2%)
Query 5 LVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLKGFE 64
LV IVV D + + L V+ D +S E +++ V+ AV ++ + K GL+ E
Sbjct 549 LVGIVVLDLPR-FKQLPLVKALDGKES--IEMIMENKEVKNAVIAELNKYAKQNGLQTIE 605
Query 65 LARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEI 109
L R +HL ++FT ENGL+T T K +R ++ F +I MY E+
Sbjct 606 LIRNVHLTLQEFTEENGLITSTLKNRRKILEDYFAPQIAKMYKEV 650
> dre:567157 acsl3a, si:dkeyp-109h9.2; acyl-CoA synthetase long-chain
family member 3a (EC:6.2.1.3); K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=713
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 35/108 (32%), Positives = 58/108 (53%), Gaps = 9/108 (8%)
Query 2 ETSLVAIVVPDQEQTLRWL--REVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQG 59
E+ ++ VVP+Q+Q L + VRG +EE + +++AV + +
Sbjct 611 ESYVIGFVVPNQKQLLALAERKRVRGL-------WEELCNNPVIEEAVLTVLTEAALTAQ 663
Query 60 LKGFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYG 107
L+ FE+ RKI L E +T + GL+T +FKLKR + + +I+ MYG
Sbjct 664 LERFEIPRKIRLSVEPWTPDTGLVTDSFKLKRKELKTHYQNDIERMYG 711
> tpv:TP02_0107 long-chain fatty acid CoA ligase; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=697
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 31/115 (26%), Positives = 57/115 (49%), Gaps = 4/115 (3%)
Query 1 SETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGL 60
+E LV IVV +E W + A+ + E + + ++ + +D +++ K Q
Sbjct 586 TENHLVGIVVVSKENVELWAQ----ANGLAGKTVRELLTNEKLKTRIMEDFEKIAKSQNF 641
Query 61 KGFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGEIHMEQDA 115
E + L + F+VE+G+LTPTFK R+ + + EIK MY ++ ++
Sbjct 642 NSLERLKVFTLVDQPFSVEDGMLTPTFKSVRYKIKARYNAEIKQMYNDVKQTNNS 696
> ath:AT2G04350 long-chain-fatty-acid--CoA ligase family protein
/ long-chain acyl-CoA synthetase family protein (LACS8) (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=720
Score = 54.7 bits (130), Expect = 6e-08, Method: Composition-based stats.
Identities = 36/102 (35%), Positives = 51/102 (50%), Gaps = 4/102 (3%)
Query 6 VAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLKGFEL 65
VA+VVP + +W E A V S F E + K V + + GK L+ FEL
Sbjct 623 VALVVPSRGALEKWAEE---AGVKHSE-FAELCEKGEAVKEVQQSLTKAGKAAKLEKFEL 678
Query 66 ARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYG 107
KI L SE +T E+GL+T K+KR + F +E+ +Y
Sbjct 679 PAKIKLLSEPWTPESGLVTAALKIKREQIKSKFKDELSKLYA 720
> ath:AT4G23850 long-chain-fatty-acid--CoA ligase / long-chain
acyl-CoA synthetase; K01897 long-chain acyl-CoA synthetase
[EC:6.2.1.3]
Length=666
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 35/108 (32%), Positives = 56/108 (51%), Gaps = 9/108 (8%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLK 61
E+ L+AI P+Q RW A+ S ++ ++ ++ + ++ ++ K + +K
Sbjct 556 ESFLIAIANPNQHILERW-----AAENGVSGDYDALCQNEKAKEFILGELVKMAKEKKMK 610
Query 62 GFELARKIHLHSEQFTVENGLLTPTFKLKR----HYAQEVFCEEIKTM 105
GFE+ + IHL F +E LLTPTFK KR Y Q V E KT+
Sbjct 611 GFEIIKAIHLDPVPFDMERDLLTPTFKKKRPQLLKYYQSVIDEMYKTI 658
> ath:AT4G11030 long-chain-fatty-acid--CoA ligase, putative /
long-chain acyl-CoA synthetase, putative (EC:6.2.1.3); K01897
long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=666
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 37/109 (33%), Positives = 52/109 (47%), Gaps = 13/109 (11%)
Query 2 ETSLVAIVVPDQEQTLRWLRE--VRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQG 59
E+ LVAI P Q+ RW E V G F +++ + + ++ + K
Sbjct 556 ESFLVAIANPAQQTLERWAVENGVNG-------DFNSICQNAKAKAFILGELVKTAKENK 608
Query 60 LKGFELARKIHLHSEQFTVENGLLTPTFKLKR----HYAQEVFCEEIKT 104
LKGFE+ + +HL F +E LLTPT+K KR Y Q V E KT
Sbjct 609 LKGFEIIKDVHLEPVAFDMERDLLTPTYKKKRPQLLKYYQNVIHEMYKT 657
> sce:YOR317W FAA1; Faa1p (EC:6.2.1.3); K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=700
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 32/92 (34%), Positives = 51/92 (55%), Gaps = 1/92 (1%)
Query 1 SETSLVAIVVPDQEQTLRWLREVRGADVADSR-PFEEWVKDSAVQKAVYDDMQQVGKMQG 59
S+T V I+VP+ + +++ + DS E +++D+ + KAVY D+ + GK QG
Sbjct 591 SKTKPVGIIVPNHAPLTKLAKKLGIMEQKDSSINIENYLEDAKLIKAVYSDLLKTGKDQG 650
Query 60 LKGFELARKIHLHSEQFTVENGLLTPTFKLKR 91
L G EL I ++T +NG +T KLKR
Sbjct 651 LVGIELLAGIVFFDGEWTPQNGFVTSAQKLKR 682
> dre:559656 acsl3b, acsl3, fb34c03, im:7155129, si:dkey-20f20.5,
wu:fa07a08, wu:fb34c03; acyl-CoA synthetase long-chain family
member 3b; K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=711
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 34/108 (31%), Positives = 58/108 (53%), Gaps = 9/108 (8%)
Query 2 ETSLVAIVVPDQEQ--TLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQG 59
E+ ++ VVP+Q+Q L + +RG+ +EE + ++K V +
Sbjct 609 ESYVIGFVVPNQKQLTALAGQKGIRGS-------WEEICNHAEMEKEVLRIITDAAISGK 661
Query 60 LKGFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYG 107
L+ FE+ +KI L +E +T E GL+T FKLKR + + ++I+ MYG
Sbjct 662 LERFEIPKKICLSAEPWTPETGLVTDAFKLKRKELKTHYQDDIERMYG 709
> xla:100174803 acsl4, acs4, acsl3, facl4, lacs4, mrx63, mrx68;
acyl-CoA synthetase long-chain family member 4 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=670
Score = 52.0 bits (123), Expect = 4e-07, Method: Composition-based stats.
Identities = 32/103 (31%), Positives = 56/103 (54%), Gaps = 5/103 (4%)
Query 5 LVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLKGFE 64
+++ VVP+Q++ L L E +G D +EE ++ V ++++V L+ FE
Sbjct 571 VISFVVPNQKK-LTSLAEQKGIDGT----WEEICNSPVMETEVLKEIKEVANSLKLERFE 625
Query 65 LARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYG 107
+ K+ L + +T E GL+T FKLKR + + +I+ MYG
Sbjct 626 IPIKVRLSPDPWTPETGLVTDAFKLKRKELKNHYITDIERMYG 668
> ath:AT2G47240 long-chain-fatty-acid--CoA ligase family protein
/ long-chain acyl-CoA synthetase family protein (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=660
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 28/105 (26%), Positives = 56/105 (53%), Gaps = 5/105 (4%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLK 61
++ LVA+VVP+ E RW + D+ ++PFEE +++ + +++ + L+
Sbjct 553 KSMLVAVVVPNPETVNRWAK-----DLGFTKPFEELCSFPELKEHIISELKSTAEKNKLR 607
Query 62 GFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMY 106
FE + + + ++ F VE L+T T K +R+ + + +I MY
Sbjct 608 KFEYIKAVTVETKPFDVERDLVTATLKNRRNNLLKYYQVQIDEMY 652
> sce:YIL009W FAA3; Faa3p (EC:6.2.1.3); K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=694
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 36/112 (32%), Positives = 54/112 (48%), Gaps = 10/112 (8%)
Query 1 SETSLVAIVVPDQEQTLRWLREVR----GADVADSRPFEEWVKDSAVQKAVYDDMQQVGK 56
S V IVVP+ ++ ++R G D+ E ++ D A++ AV+ +M K
Sbjct 587 SRVKPVGIVVPNPGPLSKFAVKLRIMKKGEDI------ENYIHDKALRNAVFKEMIATAK 640
Query 57 MQGLKGFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGE 108
QGL G EL I E++T ENG +T KLKR E++ +Y E
Sbjct 641 SQGLVGIELLCGIVFFDEEWTPENGFVTSAQKLKRREILAAVKSEVERVYKE 692
> ath:AT1G77590 LACS9; LACS9 (LONG CHAIN ACYL-COA SYNTHETASE 9);
long-chain-fatty-acid-CoA ligase (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=691
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/101 (33%), Positives = 50/101 (49%), Gaps = 4/101 (3%)
Query 6 VAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLKGFEL 65
VA+VV Q W + +G D A+ FEE K VY + + K L+ FE+
Sbjct 594 VALVVASQHTVEGWASK-QGIDFAN---FEELCTKEQAVKEVYASLVKAAKQSRLEKFEI 649
Query 66 ARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMY 106
KI L + +T E+GL+T KLKR + F E++ +Y
Sbjct 650 PAKIKLLASPWTPESGLVTAALKLKRDVIRREFSEDLTKLY 690
> dre:393622 acsl4a, MGC66186, acsl4, zgc:66186; acyl-CoA synthetase
long-chain family member 4a (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=706
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 30/106 (28%), Positives = 57/106 (53%), Gaps = 5/106 (4%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLK 61
+ +++ +VP+Q++ L L +G + +EE +++ V ++++V L+
Sbjct 604 QNYVISFIVPNQKK-LTALANQKGIEGT----WEEICNHPVMEREVLREIKEVANSIKLQ 658
Query 62 GFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYG 107
FE+ +HL E +T E GL+T FKLKR + + +I+ MYG
Sbjct 659 RFEIPVMVHLSPEPWTPETGLVTDAFKLKRKELKNHYLNDIERMYG 704
> mmu:50790 Acsl4, 9430020A05Rik, ACS4, AU018108, Facl4, Lacs4;
acyl-CoA synthetase long-chain family member 4 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=670
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 30/109 (27%), Positives = 60/109 (55%), Gaps = 11/109 (10%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWV---KDSAVQKAVYDDMQQVGKMQ 58
++ +++ VVP+Q++ L L + +G + + WV + A++ + ++++
Sbjct 568 QSYVISFVVPNQKK-LTLLAQQKGVEGS-------WVDICNNPAMEAEILKEIREAANAM 619
Query 59 GLKGFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYG 107
L+ FE+ K+ L E +T E GL+T FKLKR + + ++I+ MYG
Sbjct 620 KLERFEIPIKVRLSPEPWTPETGLVTDAFKLKRKELKNHYLKDIERMYG 668
> bbo:BBOV_III010400 17.m07898; long-chain acyl-CoA synthetase
(EC:6.2.1.3); K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=687
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 34/107 (31%), Positives = 51/107 (47%), Gaps = 12/107 (11%)
Query 5 LVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQ----QVGKMQGL 60
+VAIV + E + W A + VKD + + D++Q + + L
Sbjct 588 IVAIVNINAEYVMEW--------AASHNLGGKSVKDLLTNQQLIDEIQASFNNITEQNKL 639
Query 61 KGFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYG 107
G E +K L E F+V+NG+LTPTFK R + + +EI MYG
Sbjct 640 NGLERLQKFILSDELFSVDNGMLTPTFKTVRKKVRSRYEKEINAMYG 686
> hsa:2182 ACSL4, ACS4, FACL4, LACS4, MRX63, MRX68; acyl-CoA synthetase
long-chain family member 4 (EC:6.2.1.3); K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=670
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 30/109 (27%), Positives = 59/109 (54%), Gaps = 11/109 (10%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWV---KDSAVQKAVYDDMQQVGKMQ 58
++ +++ VVP+Q++ L L + +G + WV + A++ + ++++
Sbjct 568 QSYVISFVVPNQKR-LTLLAQQKGVE-------GTWVDICNNPAMEAEILKEIREAANAM 619
Query 59 GLKGFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYG 107
L+ FE+ K+ L E +T E GL+T FKLKR + + ++I+ MYG
Sbjct 620 KLERFEIPIKVRLSPEPWTPETGLVTDAFKLKRKELRNHYLKDIERMYG 668
> tpv:TP01_0520 long-chain fatty acid CoA ligase; K01897 long-chain
acyl-CoA synthetase [EC:6.2.1.3]
Length=669
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 29/103 (28%), Positives = 57/103 (55%), Gaps = 4/103 (3%)
Query 6 VAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLKGFEL 65
VA+VVPD+ + W + A++ +E + +++ + ++++V + +KGFE
Sbjct 570 VAVVVPDELELELWSKRYGLANLDR----KEQCEVKQLKEFIAKEIEKVFENSDVKGFEK 625
Query 66 ARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGE 108
+ ++ E F++EN LLT T KLKR+ + + E+I + E
Sbjct 626 IKNFYVEHELFSIENNLLTTTSKLKRYNLSKKYAEQIDRLNDE 668
> sce:YMR246W FAA4; Faa4p (EC:6.2.1.3); K01897 long-chain acyl-CoA
synthetase [EC:6.2.1.3]
Length=694
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 37/107 (34%), Positives = 50/107 (46%), Gaps = 10/107 (9%)
Query 6 VAIVVPDQEQTLRWLRE----VRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLK 61
V IVVP+ + E V G DV E ++ + +Q AV DM K QGL
Sbjct 592 VGIVVPNLGHLSKLAIELGIMVPGEDV------ESYIHEKKLQDAVCKDMLSTAKSQGLN 645
Query 62 GFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYGE 108
G EL I E++T ENGL+T KLKR +++ +Y E
Sbjct 646 GIELLCGIVFFEEEWTPENGLVTSAQKLKRRDILAAVKPDVERVYKE 692
> cel:C46F4.2 acs-17; fatty Acid CoA Synthetase family member
(acs-17); K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=718
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 33/106 (31%), Positives = 60/106 (56%), Gaps = 5/106 (4%)
Query 1 SETSLVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGL 60
+++ LVA+VVP+Q+ L ++ A + FE+ +D V +A+ ++ Q G + L
Sbjct 617 NKSFLVALVVPNQKN----LTQLASKAGASNDNFEKLCEDKKVVEALQKELAQYGASK-L 671
Query 61 KGFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMY 106
+ E+ +KI + E +T +GLLT KLKR ++ F +E+ +Y
Sbjct 672 QRVEIPQKIFICHEPWTPASGLLTEALKLKRKNIEKAFRKELDELY 717
> pfa:PFL1880w ACS11; acyl-CoA synthetase, PfACS11 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=792
Score = 48.9 bits (115), Expect = 4e-06, Method: Composition-based stats.
Identities = 32/109 (29%), Positives = 51/109 (46%), Gaps = 13/109 (11%)
Query 2 ETSLVAIVVPDQEQTLRWLREVRGADVADSR----PFEEWVKDSAVQKAVYDDMQQVGKM 57
E LV+++VP+Q + +D A P+E+ +K + K +++ + K
Sbjct 688 ENELVSVIVPNQ---------IMLSDYAKKHNLNLPYEDLLKHDIIIKLFQEEILNISKT 738
Query 58 QGLKGFELARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMY 106
L G E HL S F+VEN LTPT K+ RH + + I +Y
Sbjct 739 YKLNGIEKIHLFHLTSTPFSVENKQLTPTHKIVRHVILADYKKVINDLY 787
> mmu:74205 Acsl3, 2610510B12Rik, Acs3, C85929, Facl3, Pro2194;
acyl-CoA synthetase long-chain family member 3 (EC:6.2.1.3);
K01897 long-chain acyl-CoA synthetase [EC:6.2.1.3]
Length=720
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 31/103 (30%), Positives = 51/103 (49%), Gaps = 5/103 (4%)
Query 5 LVAIVVPDQEQTLRWLREVRGADVADSRPFEEWVKDSAVQKAVYDDMQQVGKMQGLKGFE 64
++ VVP+Q++ L L +G +EE S ++ V + + L+ FE
Sbjct 621 VIGFVVPNQKE-LTELARTKGF----KGTWEELCNSSEMENEVLKVLSEAAISASLEKFE 675
Query 65 LARKIHLHSEQFTVENGLLTPTFKLKRHYAQEVFCEEIKTMYG 107
+ KI L + +T E GL+T FKLKR + + +I+ MYG
Sbjct 676 IPLKIRLSPDPWTPETGLVTDAFKLKRKELKTHYQADIERMYG 718
Lambda K H
0.318 0.132 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2042676840
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40