bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1817_orf2
Length=162
Score E
Sequences producing significant alignments: (Bits) Value
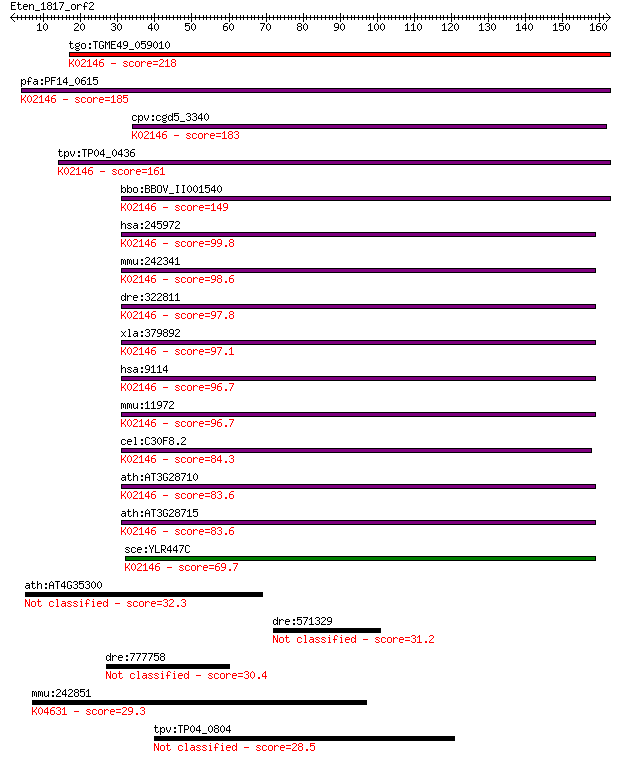
tgo:TGME49_059010 vacuolar ATP synthase subunit D, putative (E... 218 6e-57
pfa:PF14_0615 ATP synthase (C/AC39) subunit, putative; K02146 ... 185 5e-47
cpv:cgd5_3340 vacuolar ATP synthase subunit d ; K02146 V-type ... 183 2e-46
tpv:TP04_0436 vacuolar ATP synthase (C/AC39) subunit (EC:3.6.3... 161 1e-39
bbo:BBOV_II001540 18.m06119; vacuolar ATP synthase subunit d; ... 149 4e-36
hsa:245972 ATP6V0D2, ATP6D2, FLJ38708, VMA6; ATPase, H+ transp... 99.8 3e-21
mmu:242341 Atp6v0d2, 1620401A02Rik, AI324824, V-ATPase; ATPase... 98.6 7e-21
dre:322811 atp6v0d1, fb73h07, wu:fb73h07, zgc:63769; ATPase, H... 97.8 1e-20
xla:379892 atp6v0d1, MGC53957, atp6d; ATPase, H+ transporting,... 97.1 2e-20
hsa:9114 ATP6V0D1, ATP6D, ATP6DV, FLJ43534, P39, VATX, VMA6, V... 96.7 3e-20
mmu:11972 Atp6v0d1, AI267038, Ac39, Atp6d, P39, VATX, Vma6; AT... 96.7 3e-20
cel:C30F8.2 vha-16; Vacuolar H ATPase family member (vha-16); ... 84.3 2e-16
ath:AT3G28710 H+-transporting two-sector ATPase, putative; K02... 83.6 3e-16
ath:AT3G28715 H+-transporting two-sector ATPase, putative; K02... 83.6 3e-16
sce:YLR447C VMA6; Subunit d of the five-subunit V0 integral me... 69.7 4e-12
ath:AT4G35300 TMT2; TMT2 (TONOPLAST MONOSACCHARIDE TRANSPORTER... 32.3 0.73
dre:571329 NLR family, pyrin domain containing 1-like 31.2 1.7
dre:777758 hps3, zgc:152969; Hermansky-Pudlak syndrome 3 30.4
mmu:242851 Gnat3, Ggust, Gtn; guanine nucleotide binding prote... 29.3 6.6
tpv:TP04_0804 hypothetical protein 28.5 9.2
> tgo:TGME49_059010 vacuolar ATP synthase subunit D, putative
(EC:3.6.3.14); K02146 V-type H+-transporting ATPase subunit
AC39 [EC:3.6.3.14]
Length=396
Score = 218 bits (556), Expect = 6e-57, Method: Compositional matrix adjust.
Identities = 100/153 (65%), Positives = 121/153 (79%), Gaps = 7/153 (4%)
Query 17 LLQQQQQQQQQQQLDRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQC 76
L+ QQQQ+DR AL+P+FGYLYP+GTD +RKAWND T+RAAL P+ SYLN+Y+QC
Sbjct 244 LVVNSLSSNQQQQMDRQALYPSFGYLYPEGTDGLRKAWNDTTVRAALAPFSSYLNLYEQC 303
Query 77 RAFYIGDERDKADL------TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKE 130
++FY+G E + TS+FK+LEDLLY E MCEL FEQQFHYG++YAWVKLKE
Sbjct 304 KSFYVGQEGQGNEAAINMASTSKFKSLEDLLYSETATMCELAFEQQFHYGIYYAWVKLKE 363
Query 131 QEIRNIVWIAEMILMKRKEHI-DAIVPLFEPRI 162
QEIRNIVWIA+MILMKRKE+I D IVPLF PR+
Sbjct 364 QEIRNIVWIADMILMKRKEYISDQIVPLFPPRV 396
> pfa:PF14_0615 ATP synthase (C/AC39) subunit, putative; K02146
V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=382
Score = 185 bits (470), Expect = 5e-47, Method: Compositional matrix adjust.
Identities = 85/164 (51%), Positives = 118/164 (71%), Gaps = 5/164 (3%)
Query 4 GRRLSSV---KALLQVLLQQQQQQQQQQQLDRHALFPNFGYLYPDGTDRIRKAWNDPTLR 60
G L SV + L L + Q DR+ +FP FGYLYP+GTDRIRK WN+ T++
Sbjct 219 GHILKSVADFRVLSVTLNTMNSSLSLELQKDRNDMFPCFGYLYPEGTDRIRKCWNNETVQ 278
Query 61 AALEPYPSYLNVYDQCRAFYIGDERDKAD--LTSRFKTLEDLLYREMVQMCELTFEQQFH 118
AALE YP+Y N+Y++C+ FY+ ++ + + + K+LED+LY ++V++CE F+Q H
Sbjct 279 AALEHYPTYYNLYEECKQFYMKNDNANENKIVDHKIKSLEDILYVKLVKLCETAFDQHCH 338
Query 119 YGVFYAWVKLKEQEIRNIVWIAEMILMKRKEHIDAIVPLFEPRI 162
+G+FYAWVKLKEQEIRNIVWI++MILM RK+ ID+IVP+FEP I
Sbjct 339 FGIFYAWVKLKEQEIRNIVWISDMILMNRKDCIDSIVPIFEPHI 382
> cpv:cgd5_3340 vacuolar ATP synthase subunit d ; K02146 V-type
H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=412
Score = 183 bits (465), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 86/148 (58%), Positives = 105/148 (70%), Gaps = 20/148 (13%)
Query 34 ALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDER-------D 86
L+P+FGYLYP+GT++IRKAWND +RAALEPY Y +YDQC+AFY+ D D
Sbjct 263 TLYPSFGYLYPEGTEQIRKAWNDTRVRAALEPYSKYSALYDQCKAFYVNDTANDFGLADD 322
Query 87 KADLTSR-------------FKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEI 133
K + T + FK+LEDLLY E V MCEL F+QQ +YGVFYAW +LKEQEI
Sbjct 323 KDESTDKKKSSSHKNLADRQFKSLEDLLYAETVSMCELVFDQQMNYGVFYAWARLKEQEI 382
Query 134 RNIVWIAEMILMKRKEHIDAIVPLFEPR 161
RN+ WIAEMILM RK+ +DAIVP+F PR
Sbjct 383 RNLTWIAEMILMNRKDQVDAIVPIFAPR 410
> tpv:TP04_0436 vacuolar ATP synthase (C/AC39) subunit (EC:3.6.3.14);
K02146 V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=383
Score = 161 bits (407), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 75/150 (50%), Positives = 106/150 (70%), Gaps = 3/150 (2%)
Query 14 LQVLLQQQQQQQQQQQLDRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVY 73
L + L Q Q DR+ L+P+ GYLYP GTD++ KA+N+ T++AAL PYP Y +Y
Sbjct 236 LSLTLNCLNMTQTAVQQDRNKLYPSIGYLYPYGTDKLCKAFNETTVQAALVPYPRYAELY 295
Query 74 DQCRAFYIGDER-DKADLTSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQE 132
+ ++ + + R K D + K+LEDL Y E V +CE++FEQQ H+G+FYAWVKLKEQE
Sbjct 296 ESSKSNFRAEARVTKYDASE--KSLEDLFYAESVHLCEMSFEQQLHFGIFYAWVKLKEQE 353
Query 133 IRNIVWIAEMILMKRKEHIDAIVPLFEPRI 162
IRNI WIA+MIL+KR+E I ++P+F+ R+
Sbjct 354 IRNITWIADMILLKRREEISRVLPIFKSRV 383
> bbo:BBOV_II001540 18.m06119; vacuolar ATP synthase subunit d;
K02146 V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=374
Score = 149 bits (376), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 67/132 (50%), Positives = 92/132 (69%), Gaps = 0/132 (0%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
DR+ L+ + GYLYP GT+R+ KA+N+ TL+ AL PYP Y +YD C+ E +
Sbjct 243 DRNKLYSSIGYLYPYGTERLCKAFNEATLQQALAPYPKYARLYDMCKGSLGRAEGRSSKF 302
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
++LED Y E V++CE FEQQ H+G+FYAW+KLK+QEIRNI WIA+MIL+KR E
Sbjct 303 DVGERSLEDHFYAESVKLCEQCFEQQLHFGIFYAWLKLKQQEIRNIAWIADMILLKRPEQ 362
Query 151 IDAIVPLFEPRI 162
++P+FEPR+
Sbjct 363 FARVLPIFEPRV 374
> hsa:245972 ATP6V0D2, ATP6D2, FLJ38708, VMA6; ATPase, H+ transporting,
lysosomal 38kDa, V0 subunit d2 (EC:3.6.3.14); K02146
V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=350
Score = 99.8 bits (247), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 53/128 (41%), Positives = 73/128 (57%), Gaps = 13/128 (10%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
DR L+P FG LYP+G + +A + ++ + Y Y +++
Sbjct 236 DRETLYPTFGKLYPEGLRLLAQAEDFDQMKNVADHYGVYKPLFEAVGG------------ 283
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
S KTLED+ Y VQM L F +QFHYGVFYA+VKLKEQEIRNIVWIAE I + +
Sbjct 284 -SGGKTLEDVFYEREVQMNVLAFNRQFHYGVFYAYVKLKEQEIRNIVWIAECISQRHRTK 342
Query 151 IDAIVPLF 158
I++ +P+
Sbjct 343 INSYIPIL 350
> mmu:242341 Atp6v0d2, 1620401A02Rik, AI324824, V-ATPase; ATPase,
H+ transporting, lysosomal V0 subunit D2 (EC:3.6.3.14);
K02146 V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=350
Score = 98.6 bits (244), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 54/128 (42%), Positives = 74/128 (57%), Gaps = 13/128 (10%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
DR LFP G LYP+G + +A + ++ + Y Y ++D +G
Sbjct 236 DRETLFPTCGRLYPEGLRLLAQAEDFEQMKRVADNYGVYKPLFDA-----VGG------- 283
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
S KTLED+ Y VQM L F +QFHYGVFYA+VKLKEQE+RNIVWIAE I + +
Sbjct 284 -SGGKTLEDVFYEREVQMNVLAFNRQFHYGVFYAYVKLKEQEMRNIVWIAECISQRHRTK 342
Query 151 IDAIVPLF 158
I++ +P+
Sbjct 343 INSYIPIL 350
> dre:322811 atp6v0d1, fb73h07, wu:fb73h07, zgc:63769; ATPase,
H+ transporting, V0 subunit D isoform 1 (EC:3.6.3.14); K02146
V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=350
Score = 97.8 bits (242), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 53/128 (41%), Positives = 74/128 (57%), Gaps = 12/128 (9%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
DR LFP+ G LYP+G ++ +A + ++A E YP Y + + G + D
Sbjct 235 DRAKLFPHCGKLYPEGLAQLARADDYEQVKAVAEYYPEY-------KLLFEGAGSNPGD- 286
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
KTLED + V++ +L F QFH+ VFYA+VKLKEQE RNIVWIAE I + +
Sbjct 287 ----KTLEDRFFEHEVKLNKLAFLNQFHFSVFYAYVKLKEQECRNIVWIAECIAQRHRAK 342
Query 151 IDAIVPLF 158
ID +P+F
Sbjct 343 IDNYIPIF 350
> xla:379892 atp6v0d1, MGC53957, atp6d; ATPase, H+ transporting,
lysosomal 38kDa, V0 subunit d1 (EC:3.6.3.14); K02146 V-type
H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=351
Score = 97.1 bits (240), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 51/128 (39%), Positives = 74/128 (57%), Gaps = 12/128 (9%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
DR LFP+ G LYP+G ++ +A + ++ + YP Y + + G + D
Sbjct 236 DRAKLFPHCGKLYPEGLAQLARADDYEQVKTVADYYPEY-------KLLFEGAGNNPGD- 287
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
KTLED + V++ +L F QFH+GVFYA+VKLKEQE RN+VWIAE I + +
Sbjct 288 ----KTLEDRFFEHEVKLNKLAFLNQFHFGVFYAFVKLKEQECRNVVWIAECIAQRHRAK 343
Query 151 IDAIVPLF 158
ID +P+F
Sbjct 344 IDNYIPIF 351
> hsa:9114 ATP6V0D1, ATP6D, ATP6DV, FLJ43534, P39, VATX, VMA6,
VPATPD; ATPase, H+ transporting, lysosomal 38kDa, V0 subunit
d1 (EC:3.6.3.14); K02146 V-type H+-transporting ATPase subunit
AC39 [EC:3.6.3.14]
Length=351
Score = 96.7 bits (239), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 52/128 (40%), Positives = 74/128 (57%), Gaps = 12/128 (9%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
DR LFP+ G LYP+G ++ +A + ++ + YP Y + + G + D
Sbjct 236 DRAKLFPHCGRLYPEGLAQLARADDYEQVKNVADYYPEY-------KLLFEGAGSNPGD- 287
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
KTLED + V++ +L F QFH+GVFYA+VKLKEQE RNIVWIAE I + +
Sbjct 288 ----KTLEDRFFEHEVKLNKLAFLNQFHFGVFYAFVKLKEQECRNIVWIAECIAQRHRAK 343
Query 151 IDAIVPLF 158
ID +P+F
Sbjct 344 IDNYIPIF 351
> mmu:11972 Atp6v0d1, AI267038, Ac39, Atp6d, P39, VATX, Vma6;
ATPase, H+ transporting, lysosomal V0 subunit D1 (EC:3.6.3.14);
K02146 V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=351
Score = 96.7 bits (239), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 52/128 (40%), Positives = 74/128 (57%), Gaps = 12/128 (9%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
DR LFP+ G LYP+G ++ +A + ++ + YP Y + + G + D
Sbjct 236 DRAKLFPHCGRLYPEGLAQLARADDYEQVKNVADYYPEY-------KLLFEGAGSNPGD- 287
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
KTLED + V++ +L F QFH+GVFYA+VKLKEQE RNIVWIAE I + +
Sbjct 288 ----KTLEDRFFEHEVKLNKLAFLNQFHFGVFYAFVKLKEQECRNIVWIAECIAQRHRAK 343
Query 151 IDAIVPLF 158
ID +P+F
Sbjct 344 IDNYIPIF 351
> cel:C30F8.2 vha-16; Vacuolar H ATPase family member (vha-16);
K02146 V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=348
Score = 84.3 bits (207), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 45/127 (35%), Positives = 69/127 (54%), Gaps = 12/127 (9%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
DR L+P G L+PDG + +A + ++ E Y Y + + G +
Sbjct 233 DRQKLYPRCGKLFPDGLTGLSRADDYDQVKQVCEFYSDY-------KPLFEGSGNGPGE- 284
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
KTLED + V++ ++ QFH+GVFYA++KLKEQE+RNI+WIAE I + +
Sbjct 285 ----KTLEDKFFEHEVKLNVHSYLHQFHFGVFYAFIKLKEQEMRNIIWIAECISQRHRTK 340
Query 151 IDAIVPL 157
ID +P+
Sbjct 341 IDNYIPI 347
> ath:AT3G28710 H+-transporting two-sector ATPase, putative; K02146
V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=351
Score = 83.6 bits (205), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 46/129 (35%), Positives = 72/129 (55%), Gaps = 14/129 (10%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
DR L+ NFG LYP G + + + +R +E YP Y ++ + G+
Sbjct 236 DRKKLYSNFGLLYPYGHEELAICEDIDQVRGVMEKYPPYQAIFSK---MSYGES------ 286
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
+ L+ Y E V+ L FEQQFHY VF+A+++L+EQEIRN++WI+E + +K
Sbjct 287 ----QMLDKAFYEEEVRRLCLAFEQQFHYAVFFAYMRLREQEIRNLMWISECVAQNQKSR 342
Query 151 I-DAIVPLF 158
I D++V +F
Sbjct 343 IHDSVVYMF 351
> ath:AT3G28715 H+-transporting two-sector ATPase, putative; K02146
V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=351
Score = 83.6 bits (205), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 45/129 (34%), Positives = 70/129 (54%), Gaps = 14/129 (10%)
Query 31 DRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADL 90
DR L+ NFG LYP G + + + +R +E YP Y ++ +
Sbjct 236 DRKKLYSNFGLLYPYGHEELAICEDIDQVRGVMEKYPPYQAIFSKMSY------------ 283
Query 91 TSRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEH 150
+ L+ Y E V+ L FEQQFHY VF+A+++L+EQEIRN++WI+E + +K
Sbjct 284 -GESQMLDKAFYEEEVRRLCLAFEQQFHYAVFFAYMRLREQEIRNLMWISECVAQNQKSR 342
Query 151 I-DAIVPLF 158
I D++V +F
Sbjct 343 IHDSVVYMF 351
> sce:YLR447C VMA6; Subunit d of the five-subunit V0 integral
membrane domain of vacuolar H+-ATPase (V-ATPase), an electrogenic
proton pump found in the endomembrane system; stabilizes
VO subunits; required for V1 domain assembly on the vacuolar
membrane (EC:3.6.3.14); K02146 V-type H+-transporting ATPase
subunit AC39 [EC:3.6.3.14]
Length=345
Score = 69.7 bits (169), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 44/127 (34%), Positives = 63/127 (49%), Gaps = 19/127 (14%)
Query 32 RHALFPNFGYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDERDKADLT 91
+ L PN G LYP T + +A + +RAAL NVY+ R F
Sbjct 238 KSDLLPNIGKLYPLATFHLAQAQDFEGVRAALA------NVYEY-RGFL----------- 279
Query 92 SRFKTLEDLLYREMVQMCELTFEQQFHYGVFYAWVKLKEQEIRNIVWIAEMILMKRKEHI 151
LED Y+ +++C F QQF +AW+K KEQE+RNI WIAE I ++E I
Sbjct 280 -ETGNLEDHFYQLEMELCRDAFTQQFAISTVWAWMKSKEQEVRNITWIAECIAQNQRERI 338
Query 152 DAIVPLF 158
+ + ++
Sbjct 339 NNYISVY 345
> ath:AT4G35300 TMT2; TMT2 (TONOPLAST MONOSACCHARIDE TRANSPORTER2);
carbohydrate transmembrane transporter/ nucleoside transmembrane
transporter/ sugar:hydrogen symporter
Length=729
Score = 32.3 bits (72), Expect = 0.73, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 31/65 (47%), Gaps = 1/65 (1%)
Query 5 RRLSSVKALLQVLLQQQQQQQQQQQLDRHALFPNFGYLYPDGTDRIR-KAWNDPTLRAAL 63
RR S+ L L ++ R ALFP+FG ++ G ++ R + W++ L
Sbjct 291 RRQGSLIDPLVTLFGSVHEKMPDTGSMRSALFPHFGSMFSVGGNQPRHEDWDEENLVGEG 350
Query 64 EPYPS 68
E YPS
Sbjct 351 EDYPS 355
> dre:571329 NLR family, pyrin domain containing 1-like
Length=1011
Score = 31.2 bits (69), Expect = 1.7, Method: Composition-based stats.
Identities = 11/29 (37%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 72 VYDQCRAFYIGDERDKADLTSRFKTLEDL 100
+Y+QCR +I D D++ +T RF +E++
Sbjct 153 IYEQCRVVFIFDGLDESRITLRFSNIENV 181
> dre:777758 hps3, zgc:152969; Hermansky-Pudlak syndrome 3
Length=1003
Score = 30.4 bits (67), Expect = 2.7, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 27 QQQLDRHALFPNFGYLYPDGTDRIRKAWNDPTL 59
Q QL H L + L+P+ DRIRK ND T+
Sbjct 928 QHQLHHHTLCLWWQKLFPEVCDRIRKTGNDNTI 960
> mmu:242851 Gnat3, Ggust, Gtn; guanine nucleotide binding protein,
alpha transducing 3; K04631 guanine nucleotide binding
protein (G protein), alpha transducing activity polypeptide
Length=354
Score = 29.3 bits (64), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 30/102 (29%), Positives = 47/102 (46%), Gaps = 14/102 (13%)
Query 7 LSSVKAL--LQVLLQQQQQQQQQQQLDRHALFPNFGYLYPDGTDRIRKAWNDPTLRAALE 64
L+ VKA+ L + + ++ Q+QL A G + P + I++ W DP ++A E
Sbjct 82 LAIVKAMATLGIDYVNPRSREDQEQLHSMANTLEDGDMTPQLAEIIKRLWGDPGIQACFE 141
Query 65 ---------PYPSYLNVYDQCRA-FYIGDERDKADLTSRFKT 96
YLN D+ A Y+ +E+D L SR KT
Sbjct 142 RASEYQLNDSAAYYLNDLDRLTAPGYVPNEQDV--LHSRVKT 181
> tpv:TP04_0804 hypothetical protein
Length=3588
Score = 28.5 bits (62), Expect = 9.2, Method: Composition-based stats.
Identities = 21/87 (24%), Positives = 35/87 (40%), Gaps = 6/87 (6%)
Query 40 GYLYPDGTDRIRKAWNDPTLRAALEPYPSYLNVYDQCRAFYIGDER--DKA----DLTSR 93
G LYP G +++ WN R E + LN +++ + ++ D A LT
Sbjct 3379 GTLYPIGFTSVKRFWNTNYFRQKNEEAQANLNYFERQKKYFTNSHAPDDTAGCSNGLTGN 3438
Query 94 FKTLEDLLYREMVQMCELTFEQQFHYG 120
K+ D L+ F+ + H G
Sbjct 3439 HKSCLDQLFLNHNSSFNTAFDHRKHMG 3465
Lambda K H
0.325 0.139 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3767900632
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40