bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1891_orf1
Length=186
Score E
Sequences producing significant alignments: (Bits) Value
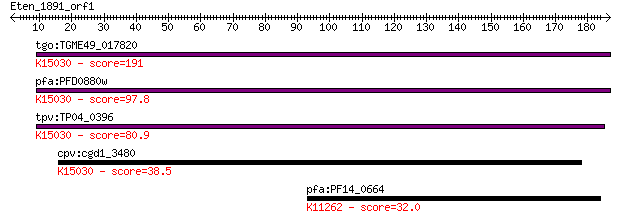
tgo:TGME49_017820 proteasome PCI domain-containing protein ; K... 191 1e-48
pfa:PFD0880w proteasome regulatory component, putative; K15030... 97.8 2e-20
tpv:TP04_0396 hypothetical protein; K15030 translation initiat... 80.9 2e-15
cpv:cgd1_3480 proteasome regulatory complex component with a P... 38.5 0.013
pfa:PF14_0664 ACC1; biotin carboxylase subunit of acetyl CoA c... 32.0 1.3
> tgo:TGME49_017820 proteasome PCI domain-containing protein ;
K15030 translation initiation factor 3 subunit M
Length=463
Score = 191 bits (485), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 97/180 (53%), Positives = 124/180 (68%), Gaps = 13/180 (7%)
Query 9 MTTFVPLAADTDGTASAVAVGEWLLHLITLYDSEDTQRTKDYYREFMSCFTRDEAAGETK 68
MTTFVPLA D DGTASAVAVG+WLL +I L + TQ YY +F+ F +DE GE K
Sbjct 1 MTTFVPLATDGDGTASAVAVGDWLLQIINLKNPSQTQ---SYYTQFLEQFDKDEETGEQK 57
Query 69 ITNPINLLSLLLSQNELVFRWLGEIRSAQDAQGQPPAAAET--RPELKKSYGDALREVQD 126
I + L LLLSQ++LVF + Q + PAAAE +P+ +K++ +A+ EV++
Sbjct 58 IRDHFQLFELLLSQHQLVFNY--------ATQARQPAAAEKGEKPQNRKTFLEAVHEVEE 109
Query 127 FFMLLMSLVVLRITDVEAAGQMAGNFCAVFRASKDQPEFRLKLLMGLYNVFPPSFPYRFP 186
FF +L+++VVLRI +VE AGQ AG C+VFRAS D EFRL+LL LYN FPPSFPYRFP
Sbjct 110 FFTVLIAMVVLRIENVEQAGQAAGTLCSVFRASTDMAEFRLRLLQSLYNAFPPSFPYRFP 169
> pfa:PFD0880w proteasome regulatory component, putative; K15030
translation initiation factor 3 subunit M
Length=429
Score = 97.8 bits (242), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 51/178 (28%), Positives = 91/178 (51%), Gaps = 10/178 (5%)
Query 9 MTTFVPLAADTDGTASAVAVGEWLLHLITLYDSEDTQRTKDYYREFMSCFTRDEAAGETK 68
M+ ++ L D GT +AV +G+W+L ++ ++ Q + ++++F+ + + + E
Sbjct 1 MSIYIQLTHDLSGTVTAVTIGDWILGILK---EKNIQEYEVFFKKFLHIYQKQDKDSEC- 56
Query 69 ITNPINLLSLLLSQNELVFRWLGEIRSAQDAQGQPPAAAETRPELKKSYGDALREVQDFF 128
N + SLLLS ++ VF L E + + E KSY + +EV+++F
Sbjct 57 -NNRSEIFSLLLSASDFVFETLVETK-----KNNKIKVIIDNKESVKSYNEFYKEVEEYF 110
Query 129 MLLMSLVVLRITDVEAAGQMAGNFCAVFRASKDQPEFRLKLLMGLYNVFPPSFPYRFP 186
++L+S++ L VE NF + + PE RLK+L LYN F +F +RFP
Sbjct 111 VILISILQLEFKSVEELNNATNNFIKAIKNYNEFPELRLKILQLLYNSFNVNFSFRFP 168
> tpv:TP04_0396 hypothetical protein; K15030 translation initiation
factor 3 subunit M
Length=450
Score = 80.9 bits (198), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 58/196 (29%), Positives = 94/196 (47%), Gaps = 28/196 (14%)
Query 9 MTTFVPLAADTDGTASAVAVGEWLLHLITLYDSEDTQRTKDYYREFMSCFTRDEAAGETK 68
MTTFVPL+ DT+G S+ ++G+W+L ++ + +T YY + + F E GE
Sbjct 1 MTTFVPLSQDTEGAVSSASLGDWVLGILKV---RCPSKTATYYAQLLDQF--QEVEGEQI 55
Query 69 ITNPINLLSLLLSQNELVFRWLGEIRSAQDA---QGQPPAAAETRP-------------- 111
I L LLLS++ VF +L E ++ P + AE+
Sbjct 56 IKESFQLFELLLSEHLTVFEFLEESKTNGTTVMLSLDPTSKAESEVFFTLLNYLPIFYYI 115
Query 112 ELKKSYGDALREVQDFFMLLMSLVVLRITD---VEAAGQMAGNFCAVFRASKDQPEFRLK 168
++ Y DAL++V+++F +LM ++ LR T +E AG + + + E RL+
Sbjct 116 PVRVKYSDALKQVEEYFTVLMYMLQLRFTSSGQIEKAGHL---LLKAIQGGESFLELRLR 172
Query 169 LLMGLYNVFPPSFPYR 184
LL LYN + P R
Sbjct 173 LLQMLYNSVESTLPLR 188
> cpv:cgd1_3480 proteasome regulatory complex component with a
PINT domain at the C-terminus ; K15030 translation initiation
factor 3 subunit M
Length=456
Score = 38.5 bits (88), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 32/162 (19%), Positives = 78/162 (48%), Gaps = 16/162 (9%)
Query 16 AADTDGTASAVAVGEWLLHLITLYDSEDTQRTKDYYREFMSCFTRDEAAGETKITNPINL 75
++ +G ++++ + +WL T+ D D +++ +Y E + RD+ + + ++
Sbjct 7 TSNGNGRSTSILLCDWLF---TILDYLDKKKSNSFY-EKVCKLVRDDISE----VDMASV 58
Query 76 LSLLLSQNELVFRWLGEIRSAQDAQGQPPAAAETRPELKKSYGDALREVQDFFMLLMSLV 135
LL+ + +F L +DA+ + R ++K+S+G+ +E ++FF+ L+S++
Sbjct 59 FELLVENLDGIFEIL------KDAEVKKEIV--IRDDVKRSFGELYKESEEFFIPLISML 110
Query 136 VLRITDVEAAGQMAGNFCAVFRASKDQPEFRLKLLMGLYNVF 177
+ D F + + P+ R+ +L Y++F
Sbjct 111 LHPKFDKSRIDTCKKIFLEKLMSKGEIPKLRINVLQVYYDIF 152
> pfa:PF14_0664 ACC1; biotin carboxylase subunit of acetyl CoA
carboxylase, putative (EC:6.4.1.2); K11262 acetyl-CoA carboxylase
/ biotin carboxylase [EC:6.4.1.2 6.3.4.14]
Length=3367
Score = 32.0 bits (71), Expect = 1.3, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 44/94 (46%), Gaps = 8/94 (8%)
Query 93 IRSAQDAQGQPPAAAETRPELKKSYGDALREVQDFFMLLMSLV-VLRITDVEAAGQMAGN 151
I+++Q G+ E E+KK+Y E+ + + LM + +R +++ G M GN
Sbjct 686 IKASQGGGGKGIRKVENEYEIKKAYEQVQNELPNSPIFLMKVCNNVRHIEIQVVGDMYGN 745
Query 152 FCAVF-RASKDQPEFRLKLLMGLYNVFPPS-FPY 183
C++ R Q F+ ++ PPS PY
Sbjct 746 VCSLSGRDCTTQRRFQ-----KIFEEGPPSVVPY 774
Lambda K H
0.320 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5170784960
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40