bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1909_orf1
Length=175
Score E
Sequences producing significant alignments: (Bits) Value
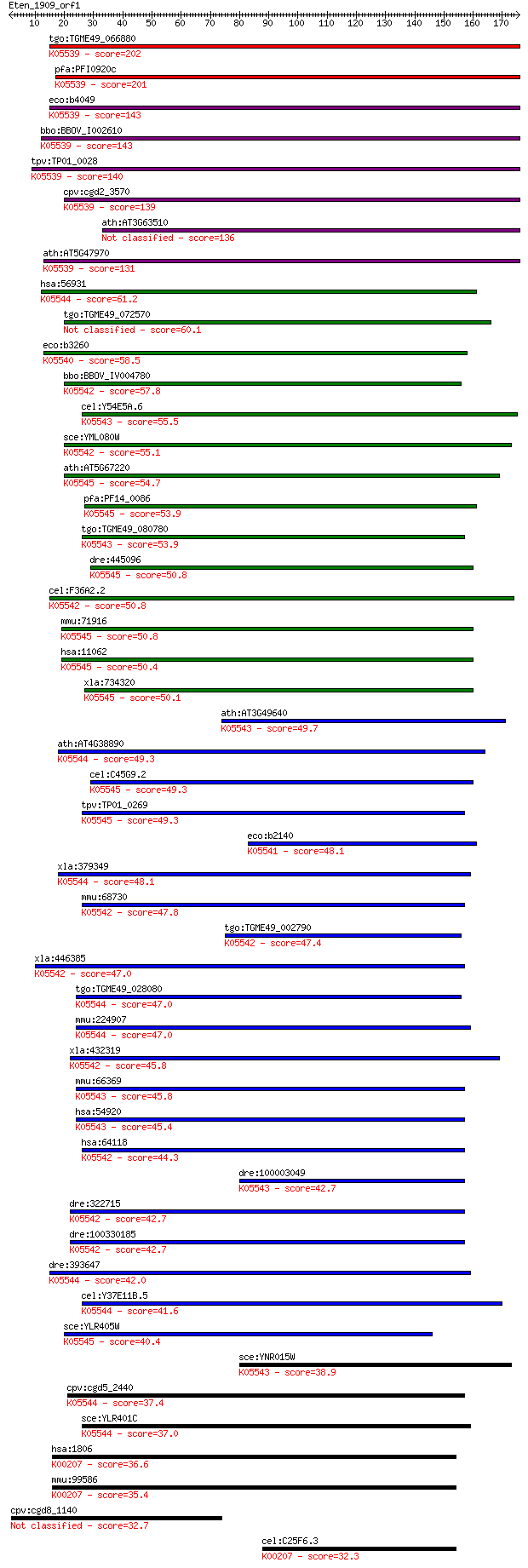
tgo:TGME49_066880 dihydrouridine synthase domain-containing pr... 202 4e-52
pfa:PFI0920c dihydrouridine synthase, putative; K05539 tRNA-di... 201 1e-51
eco:b4049 dusA, ECK4041, JW5950, yjbN; tRNA-dihydrouridine syn... 143 2e-34
bbo:BBOV_I002610 19.m02130; dihydrouridine synthase (Dus) doma... 143 2e-34
tpv:TP01_0028 tRNA-dihydrouridine synthase A; K05539 tRNA-dihy... 140 3e-33
cpv:cgd2_3570 YjbN-like Dus1p tRNA dihydouriding synthase Tim ... 139 4e-33
ath:AT3G63510 FAD binding / catalytic/ tRNA dihydrouridine syn... 136 3e-32
ath:AT5G47970 nitrogen regulation family protein; K05539 tRNA-... 131 1e-30
hsa:56931 DUS3L, DUS3, FLJ13896; dihydrouridine synthase 3-lik... 61.2 2e-09
tgo:TGME49_072570 hypothetical protein 60.1 3e-09
eco:b3260 dusB, ECK3247, JW3228, yhdG; tRNA-dihydrouridine syn... 58.5 1e-08
bbo:BBOV_IV004780 23.m05837; dihydrouridine synthase; K05542 t... 57.8 2e-08
cel:Y54E5A.6 hypothetical protein; K05543 tRNA-dihydrouridine ... 55.5 9e-08
sce:YML080W DUS1; Dihydrouridine synthase, member of a widespr... 55.1 1e-07
ath:AT5G67220 nitrogen regulation family protein; K05545 tRNA-... 54.7 1e-07
pfa:PF14_0086 tRNA-dihydrouridine synthase, putative; K05545 t... 53.9 2e-07
tgo:TGME49_080780 dihydrouridine synthase domain-containing pr... 53.9 3e-07
dre:445096 dus4l, zgc:92033; dihydrouridine synthase 4-like (S... 50.8 2e-06
cel:F36A2.2 hypothetical protein; K05542 tRNA-dihydrouridine s... 50.8 2e-06
mmu:71916 Dus4l, 2310069P03Rik, 2700089B10Rik, AI482040, MGC13... 50.8 2e-06
hsa:11062 DUS4L, DUS4, MGC133233, PP35; dihydrouridine synthas... 50.4 3e-06
xla:734320 dus4l; dihydrouridine synthase 4-like; K05545 tRNA-... 50.1 4e-06
ath:AT3G49640 FAD binding / catalytic/ tRNA dihydrouridine syn... 49.7 4e-06
ath:AT4G38890 dihydrouridine synthase family protein; K05544 t... 49.3 6e-06
cel:C45G9.2 hypothetical protein; K05545 tRNA-dihydrouridine s... 49.3 6e-06
tpv:TP01_0269 hypothetical protein; K05545 tRNA-dihydrouridine... 49.3 6e-06
eco:b2140 dusC, ECK2133, JW2128, yohI; tRNA-dihydrouridine syn... 48.1 1e-05
xla:379349 dus3l, MGC53781; dihydrouridine synthase 3-like; K0... 48.1 2e-05
mmu:68730 Dus1l, 1110032N12Rik; dihydrouridine synthase 1-like... 47.8 2e-05
tgo:TGME49_002790 dihydrouridine synthase domain-containing pr... 47.4 3e-05
xla:446385 dus1l, MGC83715; dihydrouridine synthase 1-like; K0... 47.0 3e-05
tgo:TGME49_028080 hypothetical protein ; K05544 tRNA-dihydrour... 47.0 3e-05
mmu:224907 Dus3l, AI662135, AW557805, MGC56820; dihydrouridine... 47.0 4e-05
xla:432319 MGC132093; hypothetical protein MGC78973; K05542 tR... 45.8 8e-05
mmu:66369 Dus2l, 2310016K04Rik; dihydrouridine synthase 2-like... 45.8 8e-05
hsa:54920 DUS2L, DUS2, FLJ20399, SMM1, URLC8; dihydrouridine s... 45.4 1e-04
hsa:64118 DUS1L, DUS1, PP3111; dihydrouridine synthase 1-like ... 44.3 2e-04
dre:100003049 dus2l; dihydrouridine synthase 2-like, SMM1 homo... 42.7 6e-04
dre:322715 dus1l, wu:fb71f06, zgc:63748; dihydrouridine syntha... 42.7 7e-04
dre:100330185 dihydrouridine synthase 1-like (S. cerevisiae)-l... 42.7 7e-04
dre:393647 MGC63779; zgc:63779; K05544 tRNA-dihydrouridine syn... 42.0 0.001
cel:Y37E11B.5 hypothetical protein; K05544 tRNA-dihydrouridine... 41.6 0.001
sce:YLR405W DUS4; Dihydrouridine synthase, member of a widespr... 40.4 0.003
sce:YNR015W SMM1, DUS2; Dihydrouridine synthase, member of a f... 38.9 0.010
cpv:cgd5_2440 Ylr401cp-like protein with 2 CCCH domains plus D... 37.4 0.027
sce:YLR401C DUS3; Dihydrouridine synthase, member of a widespr... 37.0 0.031
hsa:1806 DPYD, DHP, DHPDHASE, DPD, MGC132008, MGC70799; dihydr... 36.6 0.043
mmu:99586 Dpyd, AI315208, DPD, E330028L06Rik, MGC37940; dihydr... 35.4 0.11
cpv:cgd8_1140 Sec14d domain containing protein 32.7 0.62
cel:C25F6.3 dpyd-1; DihydroPYrimidine Dehydrogenase family mem... 32.3 0.80
> tgo:TGME49_066880 dihydrouridine synthase domain-containing
protein (EC:1.3.1.2); K05539 tRNA-dihydrouridine synthase A
[EC:1.-.-.-]
Length=394
Score = 202 bits (514), Expect = 4e-52, Method: Compositional matrix adjust.
Identities = 93/161 (57%), Positives = 125/161 (77%), Gaps = 0/161 (0%)
Query 15 ETEISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFD 74
++E + +L+VAPM+AV+NTHFRNFMR TR + ++TEMV D IL+N +L+ +L +
Sbjct 6 DSEGPGKPILSVAPMLAVTNTHFRNFMRCFTREAQLWTEMVTDGAILNNMDRLQQNLCLE 65
Query 75 DVEHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTK 134
D+EHPIVCQLGGS+P+ +AEA E G+DE+NLNVGCPS RVV +G FGAALM++P
Sbjct 66 DIEHPIVCQLGGSDPKTLAEAGKLIEKLGFDEINLNVGCPSNRVVSQGCFGAALMKTPET 125
Query 135 VRDIVYEMKRRVQIPVTVKCRLGVDNLDSPEFTKQFVETVS 175
VRDIV+E++R VQIPVTVK R+G D+ DS + + FV+TVS
Sbjct 126 VRDIVHEIRRHVQIPVTVKTRIGYDHCDSRDVLRNFVQTVS 166
> pfa:PFI0920c dihydrouridine synthase, putative; K05539 tRNA-dihydrouridine
synthase A [EC:1.-.-.-]
Length=577
Score = 201 bits (511), Expect = 1e-51, Method: Composition-based stats.
Identities = 95/159 (59%), Positives = 129/159 (81%), Gaps = 0/159 (0%)
Query 17 EISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDV 76
E S+ + VAPMI V+N HFR +R+IT+R+ ++TEM+VD+T+L+N + LE HLGFD+
Sbjct 193 EKSTTPFIQVAPMINVTNRHFRAMVRIITKRAQLWTEMIVDNTLLYNLNNLEEHLGFDNN 252
Query 77 EHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVR 136
EHPIVCQLGG + +++EAA E AGYDE+N+NVGCPS +V +KG+FGA+LM++P +VR
Sbjct 253 EHPIVCQLGGCDMNSMSEAAILVEQAGYDEININVGCPSTKVANKGAFGASLMKNPEQVR 312
Query 137 DIVYEMKRRVQIPVTVKCRLGVDNLDSPEFTKQFVETVS 175
+IVYE+K++VQIPVTVK R GVDN DS +F K F+ETVS
Sbjct 313 NIVYEIKKKVQIPVTVKIRTGVDNYDSFDFLKTFIETVS 351
> eco:b4049 dusA, ECK4041, JW5950, yjbN; tRNA-dihydrouridine synthase
A; K05539 tRNA-dihydrouridine synthase A [EC:1.-.-.-]
Length=330
Score = 143 bits (361), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 74/161 (45%), Positives = 102/161 (63%), Gaps = 4/161 (2%)
Query 15 ETEISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFD 74
+T++ +VAPM+ ++ H R F+RL++R + +YTEMV I+H ++L +
Sbjct 4 KTDVHWSGRFSVAPMLDWTDRHCRYFLRLLSRNTLLYTEMVTTGAIIHGK---GDYLAYS 60
Query 75 DVEHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTK 134
+ EHP+ QLGGS+P +A+ A AE GYDE+NLNVGCPS R V G FGA LM +
Sbjct 61 EEEHPVALQLGGSDPAALAQCAKLAEARGYDEINLNVGCPSDR-VQNGMFGACLMGNAQL 119
Query 135 VRDIVYEMKRRVQIPVTVKCRLGVDNLDSPEFTKQFVETVS 175
V D V M+ V IPVTVK R+G+D+ DS EF F+ TVS
Sbjct 120 VADCVKAMRDVVSIPVTVKTRIGIDDQDSYEFLCDFINTVS 160
> bbo:BBOV_I002610 19.m02130; dihydrouridine synthase (Dus) domain
containing protein; K05539 tRNA-dihydrouridine synthase
A [EC:1.-.-.-]
Length=362
Score = 143 bits (361), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 76/169 (44%), Positives = 111/169 (65%), Gaps = 5/169 (2%)
Query 12 TMGETEISSRA----LLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILH-NTHQ 66
+M + +IS R+ L+ +APM+ ++ FR FMRLIT+R+ ++TEM+ D ++++ ++ +
Sbjct 2 SMEDADISGRSHDRPLVQIAPMLDITYYEFRQFMRLITKRAQLWTEMMADGSLIYGDSDK 61
Query 67 LENHLGFDDVEHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGA 126
+ + L D EHPIV QLGG+N ++++A A GY E NLNVGCPS RV KG FGA
Sbjct 62 VGSMLHCDANEHPIVLQLGGNNLDSLSKAGNIALGYGYTEFNLNVGCPSTRVSGKGCFGA 121
Query 127 ALMRSPTKVRDIVYEMKRRVQIPVTVKCRLGVDNLDSPEFTKQFVETVS 175
ALM V IV ++ + PVTVK RLGVD+ DS EF + F+ TV+
Sbjct 122 ALMNDAPLVGRIVKHLRSELGTPVTVKHRLGVDHNDSYEFVRDFISTVA 170
> tpv:TP01_0028 tRNA-dihydrouridine synthase A; K05539 tRNA-dihydrouridine
synthase A [EC:1.-.-.-]
Length=471
Score = 140 bits (352), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 82/201 (40%), Positives = 116/201 (57%), Gaps = 34/201 (16%)
Query 9 EIDTMGETEIS-SRALLAVAPMIAVSNTHFR----------NFMRLITRRSTVYTEMVVD 57
++TM +S S+ L +APM+ V+ HFR FMRL+TR++ ++TEM V
Sbjct 70 HLNTMRNGTVSHSKPSLQIAPMLDVTYLHFRYFMDFKFDFRQFMRLLTRKTQLWTEMFVA 129
Query 58 STILHNTHQ-LENHLGFDDVEHPIVCQLG----------GSNPQNVAEAATWAELAGYDE 106
S++++ +++ + L F++ EHPIV QLG G+ P+ + EA + GYDE
Sbjct 130 SSLINASNESVSRWLKFEENEHPIVAQLGIFYQHKIVPGGNCPETLVEAGRILKKFGYDE 189
Query 107 VNLNVGCPSCRVVDKG------------SFGAALMRSPTKVRDIVYEMKRRVQIPVTVKC 154
+NLN GCPS RV KG FGA+LM+ VRDI + M R +++PVTVK
Sbjct 190 INLNAGCPSPRVSGKGIYKFMLKLFNLGCFGASLMKEKELVRDIAHHMLRELEMPVTVKT 249
Query 155 RLGVDNLDSPEFTKQFVETVS 175
RLGVD DS EF + FV TVS
Sbjct 250 RLGVDEFDSYEFVRDFVSTVS 270
> cpv:cgd2_3570 YjbN-like Dus1p tRNA dihydouriding synthase Tim
barrel ; K05539 tRNA-dihydrouridine synthase A [EC:1.-.-.-]
Length=519
Score = 139 bits (350), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 70/170 (41%), Positives = 107/170 (62%), Gaps = 15/170 (8%)
Query 20 SRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLEN--------HL 71
S ++++VAPM+ V+N+HFR R+I++++ ++TEMVVD TI+H + EN H
Sbjct 24 SNSIISVAPMLDVTNSHFRMLCRIISKKTELWTEMVVDDTIIH-CYNDENRRKILQDVHF 82
Query 72 GFDDVEHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRS 131
+D+E+P+V QLGG+NPQ + +A A G+ NLNVGCPSC+V KGSFGA+L ++
Sbjct 83 KKNDIENPLVLQLGGNNPQKMEKAIEIAYKYGFQNFNLNVGCPSCKVASKGSFGASLFKN 142
Query 132 PTKVRDIVYEMKRR------VQIPVTVKCRLGVDNLDSPEFTKQFVETVS 175
P +V IV ++ V ++VK R+GVD D+ + F+ VS
Sbjct 143 PLRVAKIVDTCNKKLIHLGLVNKRISVKTRIGVDQYDTYQHLYNFISLVS 192
> ath:AT3G63510 FAD binding / catalytic/ tRNA dihydrouridine synthase
Length=386
Score = 136 bits (343), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 65/143 (45%), Positives = 90/143 (62%), Gaps = 0/143 (0%)
Query 33 SNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHPIVCQLGGSNPQNV 92
S+ H+R RLIT+ + +YTEM+ T++H L+ L F +HPIV QLGGSN +N+
Sbjct 30 SDNHYRTLARLITKHAWLYTEMIAAETLVHQQTNLDRFLAFSPQQHPIVLQLGGSNVENL 89
Query 93 AEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIVYEMKRRVQIPVTV 152
A+AA ++ GYDE+NLN GCPS +V G FG +LM P V + + + +PVTV
Sbjct 90 AKAAKLSDAYGYDEINLNCGCPSPKVAGHGCFGVSLMLKPKLVGEAMSAIAANTNVPVTV 149
Query 153 KCRLGVDNLDSPEFTKQFVETVS 175
KCR+GVDN DS + F+ VS
Sbjct 150 KCRIGVDNHDSYDELCDFIYKVS 172
> ath:AT5G47970 nitrogen regulation family protein; K05539 tRNA-dihydrouridine
synthase A [EC:1.-.-.-]
Length=387
Score = 131 bits (329), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 66/163 (40%), Positives = 95/163 (58%), Gaps = 0/163 (0%)
Query 13 MGETEISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLG 72
M +E S L ++APM+ ++ H+R RLIT+ + +YTEM+ TI++ L++ L
Sbjct 1 MTVSEAYSPPLFSIAPMMGWTDNHYRTLARLITKHAWLYTEMLAAETIVYQEDNLDSFLA 60
Query 73 FDDVEHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSP 132
F +HPIV Q+GG N +N+A+A A YDE+N N GCPS +V +G FGA LM P
Sbjct 61 FSPDQHPIVLQIGGRNLENLAKATRLANAYAYDEINFNCGCPSPKVSGRGCFGALLMLDP 120
Query 133 TKVRDIVYEMKRRVQIPVTVKCRLGVDNLDSPEFTKQFVETVS 175
V + + + VTVKCR+GVD+ DS F+ VS
Sbjct 121 KFVGEAMSVIAANTNAAVTVKCRIGVDDHDSYNELCDFIHIVS 163
> hsa:56931 DUS3L, DUS3, FLJ13896; dihydrouridine synthase 3-like
(S. cerevisiae); K05544 tRNA-dihydrouridine synthase 3 [EC:1.-.-.-]
Length=408
Score = 61.2 bits (147), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 49/150 (32%), Positives = 66/150 (44%), Gaps = 3/150 (2%)
Query 12 TMGETEISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHL 71
T ETE+ R L +AP+ N FR + T EM V + +L L
Sbjct 53 TCRETELDIRGKLYLAPLTTCGNLPFRRICKRFGADVTC-GEMAVCTNLLQGQMSEWALL 111
Query 72 GFDDVEHPIVCQLGGSNPQNVAEAATW-AELAGYDEVNLNVGCPSCRVVDKGSFGAALMR 130
E QL G+ P + + A + D V++NVGCP V KG G ALM
Sbjct 112 KRHQCEDIFGVQLEGAFPDTMTKCAELLSRTVEVDFVDINVGCPIDLVYKKGG-GCALMN 170
Query 131 SPTKVRDIVYEMKRRVQIPVTVKCRLGVDN 160
TK + IV M + + +P+TVK R GV
Sbjct 171 RSTKFQQIVRGMNQVLDVPLTVKIRTGVQE 200
> tgo:TGME49_072570 hypothetical protein
Length=668
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 57/190 (30%), Positives = 76/190 (40%), Gaps = 45/190 (23%)
Query 20 SRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVV-------------DSTILHNTHQ 66
S LL+ APM+ +N H+R + L++RR T+YTEMV D
Sbjct 97 SGGLLSAAPMMDYTNRHWRRLISLMSRRLTLYTEMVPVPPAPCESPDSSQDLPFSVKRKL 156
Query 67 LENHLGFDDVEHPIVCQLGGSNPQNVAEAATWAELAGYDE----------------VNLN 110
LE V + +V Q+G + Q+ A D VNLN
Sbjct 157 LEEAGSLASVPNDVVLQVGVGSAQSATAVARVLNEFSVDRDHAEPVTRMRGKRTFLVNLN 216
Query 111 VGCPSCRVVDKGSFGAALMRSPTKVRDIVY---------EMKRRVQIPVTVKCRLG---- 157
GCPS RV KGSFG LM P +V ++ E ++R + V K G
Sbjct 217 CGCPSPRVA-KGSFGLILMEDPKRVAEMCQILVDGANNREGQKRCSLDVMGKSARGDTKD 275
Query 158 --VDNLDSPE 165
V DSPE
Sbjct 276 RTVHTNDSPE 285
> eco:b3260 dusB, ECK3247, JW3228, yhdG; tRNA-dihydrouridine synthase
B; K05540 tRNA-dihydrouridine synthase B [EC:1.-.-.-]
Length=321
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 40/147 (27%), Positives = 79/147 (53%), Gaps = 8/147 (5%)
Query 13 MGETEISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLG 72
+G+ ++ +R L APM +++ FR + TV M + + + ++ L
Sbjct 3 IGQYQLRNR--LIAAPMAGITDRPFRTLCYEMGAGLTVSEMMSSNPQVWESD---KSRLR 57
Query 73 FDDVEHPIV--CQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMR 130
++ P + Q+ GS+P+ +A+AA +G +++N+GCP+ + V++ G+AL++
Sbjct 58 MVHIDEPGIRTVQIAGSDPKEMADAARINVESGAQIIDINMGCPA-KKVNRKLAGSALLQ 116
Query 131 SPTKVRDIVYEMKRRVQIPVTVKCRLG 157
P V+ I+ E+ V +PVT+K R G
Sbjct 117 YPDVVKSILTEVVNAVDVPVTLKIRTG 143
> bbo:BBOV_IV004780 23.m05837; dihydrouridine synthase; K05542
tRNA-dihydrouridine synthase 1 [EC:1.-.-.-]
Length=355
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 44/140 (31%), Positives = 63/140 (45%), Gaps = 11/140 (7%)
Query 20 SRALLAVAPMIAVSNTHFRNFMRLITRRSTV---YTEMVVDSTILHNTHQLENHLGFDDV 76
R APM+ S FR L+ RR V YT M+ + N H D
Sbjct 10 GRPRYVAAPMVNQSELPFR----LLCRRYNVDLTYTPMLHGRIFVENEKYRAVHFQTSDD 65
Query 77 EHPIVCQLGGSNPQNVAEAATWAELAGY-DEVNLNVGCPSCRVVDKGSFGAALMRSPTKV 135
+ P++ Q+ G + + +AA L G+ V+LN+GCP + G +G+ L+ P V
Sbjct 66 DRPLIAQVCGDDAGTITQAARL--LKGHVSAVDLNLGCPQA-IAKDGHYGSFLLDEPDLV 122
Query 136 RDIVYEMKRRVQIPVTVKCR 155
IV + R V I VT K R
Sbjct 123 TGIVSRVTREVGIAVTCKIR 142
> cel:Y54E5A.6 hypothetical protein; K05543 tRNA-dihydrouridine
synthase 2 [EC:1.-.-.-]
Length=436
Score = 55.5 bits (132), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 47/165 (28%), Positives = 79/165 (47%), Gaps = 25/165 (15%)
Query 26 VAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGF------DDV--- 76
+APM+ T R + L YTE +VD ++ T + LG DD+
Sbjct 11 LAPMVRAGRTPLR-LLCLKYGADLCYTEEIVDKKLIEATRVVNEALGTIDYRNGDDIILR 69
Query 77 -----EHPIVCQLGGSNPQNVAEAATWAELAGYD--EVNLNVGCPSCRVVDKGSFGAALM 129
+ + Q+G ++ + +AA A++ G D +++N+GCP + G GAAL+
Sbjct 70 LAPEEKGRCILQIGTNSGE---KAAKIAQIVGDDVAGIDVNMGCPKPFSIHCG-MGAALL 125
Query 130 RSPTKVRDIVYEMKRRVQIPVTVKCRLGVDNLDSPEFTKQFVETV 174
K+ DI+ +K ++PVT K R+ LD PE T + V+ +
Sbjct 126 TQTEKIVDILTSLKSAAKVPVTCKIRV----LDDPEDTLKLVQEI 166
> sce:YML080W DUS1; Dihydrouridine synthase, member of a widespread
family of conserved proteins including Smm1p, Dus3p, and
Dus4p; modifies pre-tRNA(Phe) at U17 (EC:1.-.-.-); K05542
tRNA-dihydrouridine synthase 1 [EC:1.-.-.-]
Length=423
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 42/159 (26%), Positives = 76/159 (47%), Gaps = 12/159 (7%)
Query 20 SRALLAVAPMIAVSNTHFRNFMRLITRR---STVYTEMVVDSTILHNTHQLENH---LGF 73
R VAPM+ S +R +++RR + YT M+ + E++ L
Sbjct 27 GRPTRIVAPMVDQSELAWR----ILSRRYGATLAYTPMLHAKLFATSKKYREDNWSSLDG 82
Query 74 DDVEHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPT 133
V+ P+V Q ++P+ + AA E D V+LN+GCP + KG +G+ LM
Sbjct 83 SSVDRPLVVQFCANDPEYLLAAAKLVE-DKCDAVDLNLGCPQG-IAKKGHYGSFLMEEWD 140
Query 134 KVRDIVYEMKRRVQIPVTVKCRLGVDNLDSPEFTKQFVE 172
+ +++ + + +++PVT K R+ D S + K ++
Sbjct 141 LIHNLINTLHKNLKVPVTAKIRIFDDCEKSLNYAKMVLD 179
> ath:AT5G67220 nitrogen regulation family protein; K05545 tRNA-dihydrouridine
synthase 4 [EC:1.-.-.-]
Length=423
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 42/150 (28%), Positives = 74/150 (49%), Gaps = 5/150 (3%)
Query 20 SRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENH-LGFDDVEH 78
R VAPM+ S FR + ++ YT M+ S I T + N +
Sbjct 86 GRPKYIVAPMVDNSELPFRLLCQKYGAQA-AYTPML-HSRIFTETEKYRNQEFTTCKEDR 143
Query 79 PIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDI 138
P+ Q ++P + EAA E D V++N+GCP R+ +G++GA LM + V+ +
Sbjct 144 PLFVQFCANDPDTLLEAAKRVE-PYCDYVDINLGCPQ-RIARRGNYGAFLMDNLPLVKSL 201
Query 139 VYEMKRRVQIPVTVKCRLGVDNLDSPEFTK 168
V ++ + + +PV+ K R+ + D+ ++ K
Sbjct 202 VEKLAQNLNVPVSCKIRIFPNLEDTLKYAK 231
> pfa:PF14_0086 tRNA-dihydrouridine synthase, putative; K05545
tRNA-dihydrouridine synthase 4 [EC:1.-.-.-]
Length=340
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 36/140 (25%), Positives = 72/140 (51%), Gaps = 16/140 (11%)
Query 27 APMIAVSNTHFRNFMRLITRRST---VYTEMVVDSTILHNTHQLENHLGFDDVEHPIVCQ 83
APM+ +S FR L+ R+ +T M+ + + + + D++ P++ Q
Sbjct 31 APMVDLSELPFR----LLCRKYNCDLTFTPMLHSKNFVEHEKYRKGYFKSCDMDKPVIAQ 86
Query 84 LGGSNPQNVAEAATWAELAGYDEVN---LNVGCPSCRVVDKGSFGAALMRSPTKVRDIVY 140
G++ + + EA + + D+VN +N+GCP ++ KG++GA L+ +V +++
Sbjct 87 FCGNDSKILLEAINFIK----DDVNAVDINLGCPQ-QIAKKGNYGAFLLHKHDEVVNLIS 141
Query 141 EMKRRVQIPVTVKCRLGVDN 160
++ IP+T K R +DN
Sbjct 142 DITNNCVIPITCKIR-KIDN 160
> tgo:TGME49_080780 dihydrouridine synthase domain-containing
protein ; K05543 tRNA-dihydrouridine synthase 2 [EC:1.-.-.-]
Length=653
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 44/149 (29%), Positives = 66/149 (44%), Gaps = 23/149 (15%)
Query 26 VAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHP------ 79
+APM+ + FR L VYTE +D +L++ D H
Sbjct 258 LAPMVRIGILPFR-LECLKYGADLVYTEETIDHKLLNSVRTFNEDFSTIDFLHKSEKTCV 316
Query 80 ----------IVCQLGGSNPQNVAEAATWAELAGYD--EVNLNVGCPSCRVVDKGSFGAA 127
+V QLG S+ +AA L D V++N+GCP ++ G GAA
Sbjct 317 FSTCLEERGKVVMQLGTSDATRALKAAM---LVAQDVAAVDVNMGCPKSFSIN-GGMGAA 372
Query 128 LMRSPTKVRDIVYEMKRRVQIPVTVKCRL 156
L+++P DI+ ++R + IPVT K RL
Sbjct 373 LLKTPLIATDILKTLRRNLDIPVTCKIRL 401
> dre:445096 dus4l, zgc:92033; dihydrouridine synthase 4-like
(S. cerevisiae); K05545 tRNA-dihydrouridine synthase 4 [EC:1.-.-.-]
Length=285
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/134 (24%), Positives = 65/134 (48%), Gaps = 6/134 (4%)
Query 29 MIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHPIVCQLGGSN 88
M+ S FR+ +R +T M++ + + + ++ + + P++ Q +
Sbjct 1 MVRYSKLAFRSLVRKYDC-DVCFTPMIIAADFMRSAKARDSEFTTNKNDRPLIVQFAAKD 59
Query 89 PQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIVYEMKRRVQI 148
Q +A+AA D V+LN GCP + +G +GA L+ P V+D+V ++ ++
Sbjct 60 AQTLADAACVVSPFS-DGVDLNCGCPQRWAMSEG-YGACLINKPELVKDMVRHVRNQIDN 117
Query 149 P---VTVKCRLGVD 159
P V++K R+ D
Sbjct 118 PNYAVSIKIRIHKD 131
> cel:F36A2.2 hypothetical protein; K05542 tRNA-dihydrouridine
synthase 1 [EC:1.-.-.-]
Length=527
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 40/159 (25%), Positives = 71/159 (44%), Gaps = 3/159 (1%)
Query 15 ETEISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFD 74
ET + R +APM+ S FR F R + T +T M+ +++ N L
Sbjct 66 ETLENQRITKVLAPMVDQSELAFRMFTRKYGAQLT-FTPMIHAHLFVNDGTYRRNSLALV 124
Query 75 DVEHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTK 134
+ P+V Q + A E D V+LN+GCP V +G +G+ L
Sbjct 125 KADRPLVVQFCANKVDTFLAACRLVEDV-CDGVDLNLGCPQM-VAKRGRYGSWLQDEVDL 182
Query 135 VRDIVYEMKRRVQIPVTVKCRLGVDNLDSPEFTKQFVET 173
+ ++V ++ ++P++ K R+ D + E+ K+ V+
Sbjct 183 ICEMVSAVRDYCRLPISCKIRVRDDRQQTVEYAKRLVDA 221
> mmu:71916 Dus4l, 2310069P03Rik, 2700089B10Rik, AI482040, MGC130363,
Pp35; dihydrouridine synthase 4-like (S. cerevisiae);
K05545 tRNA-dihydrouridine synthase 4 [EC:1.-.-.-]
Length=324
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 34/144 (23%), Positives = 70/144 (48%), Gaps = 6/144 (4%)
Query 19 SSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEH 78
S + + APM+ S FR +R + YT M++ + + + ++ + +
Sbjct 24 SGQLVKVCAPMVRYSKLAFRTLVRKYSC-DLCYTPMIIAADFVRSIKARDSEFTTNQGDC 82
Query 79 PIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDI 138
P++ Q ++ + +++AA + +++N GCP R +GA L+ P V D+
Sbjct 83 PLIVQFAANDARLLSDAALLV-CPYANGIDINCGCPQ-RWAMADGYGACLINKPELVHDM 140
Query 139 VYEMKRRVQIP---VTVKCRLGVD 159
V +++ RV+ P V++K R+ D
Sbjct 141 VRQVRNRVESPRFSVSIKIRIHDD 164
> hsa:11062 DUS4L, DUS4, MGC133233, PP35; dihydrouridine synthase
4-like (S. cerevisiae); K05545 tRNA-dihydrouridine synthase
4 [EC:1.-.-.-]
Length=317
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 34/144 (23%), Positives = 73/144 (50%), Gaps = 6/144 (4%)
Query 19 SSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEH 78
S + + APM+ S FR +R + YT M+V + + + ++ + +
Sbjct 24 SGQLVKVCAPMVRYSKLAFRTLVRKYSC-DLCYTPMIVAADFVKSIKARDSEFTTNQGDC 82
Query 79 PIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDI 138
P++ Q ++ + +++AA + +++N GCP + +G +GA L+ P V+D+
Sbjct 83 PLIVQFAANDARLLSDAARIV-CPYANGIDINCGCPQRWAMAEG-YGACLINKPELVQDM 140
Query 139 VYEMKRRVQIP---VTVKCRLGVD 159
V +++ +V+ P V++K R+ D
Sbjct 141 VKQVRNQVETPGFSVSIKIRIHDD 164
> xla:734320 dus4l; dihydrouridine synthase 4-like; K05545 tRNA-dihydrouridine
synthase 4 [EC:1.-.-.-]
Length=308
Score = 50.1 bits (118), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 33/136 (24%), Positives = 66/136 (48%), Gaps = 6/136 (4%)
Query 27 APMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHPIVCQLGG 86
APM+ S FR +R YT M++ + + + ++ + + P++ Q
Sbjct 22 APMVRYSKLAFRTLVRKYDC-DLCYTPMIIAADFVKSVKARDSEFTTNQGDCPLIVQFAA 80
Query 87 SNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIVYEMKRRV 146
Q +A+AA+ ++LN GCP + +G +GA L+ +P V D+V +++ +V
Sbjct 81 KEAQVLADAASLVSPFA-SGIDLNCGCPQRWAMAEG-YGACLINNPELVSDMVRQVRNQV 138
Query 147 ---QIPVTVKCRLGVD 159
+ +++K R+ D
Sbjct 139 GSSEFTISIKIRIHAD 154
> ath:AT3G49640 FAD binding / catalytic/ tRNA dihydrouridine synthase;
K05543 tRNA-dihydrouridine synthase 2 [EC:1.-.-.-]
Length=290
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 53/99 (53%), Gaps = 6/99 (6%)
Query 74 DDVEHPIVCQLGGSNPQNVAEAATWAELAGYD--EVNLNVGCPSCRVVDKGSFGAALMRS 131
D+ + +V Q+G S+ +A+ E+ D +++N+GCP + +G GAAL+
Sbjct 33 DEEKSRVVFQMGTSDAVRALKAS---EIVCNDVATIDINMGCPKAFSI-QGGMGAALLSK 88
Query 132 PTKVRDIVYEMKRRVQIPVTVKCRLGVDNLDSPEFTKQF 170
P + DI+ +KR + +PVT K RL D+ E ++
Sbjct 89 PELIHDILATLKRNLDVPVTCKIRLLKSPADTVELARRI 127
> ath:AT4G38890 dihydrouridine synthase family protein; K05544
tRNA-dihydrouridine synthase 3 [EC:1.-.-.-]
Length=700
Score = 49.3 bits (116), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 43/151 (28%), Positives = 70/151 (46%), Gaps = 7/151 (4%)
Query 18 ISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVE 77
I R L +AP+ V N FR +++ T EM + + +L L E
Sbjct 343 IDFRDKLYLAPLTTVGNLPFRRLCKVLGADVTC-GEMAMCTNLLQGQASEWALLRRHSSE 401
Query 78 HPIVCQLGGSNPQNVAEAATWAEL-AGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVR 136
Q+ GS P V+ + D +++N+GCP VV+K S G+AL+ P +++
Sbjct 402 DLFGVQICGSYPDTVSRVVELIDRECTVDFIDINMGCPIDMVVNK-SAGSALLNKPLRMK 460
Query 137 DIVYEMKRRVQIPVTVKCRL----GVDNLDS 163
+IV V+ P+T+K R G + +DS
Sbjct 461 NIVEVSSSIVETPITIKVRTAFFEGKNRIDS 491
> cel:C45G9.2 hypothetical protein; K05545 tRNA-dihydrouridine
synthase 4 [EC:1.-.-.-]
Length=284
Score = 49.3 bits (116), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 36/134 (26%), Positives = 62/134 (46%), Gaps = 6/134 (4%)
Query 29 MIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHPIVCQLGGSN 88
M+ S FR +R + +T M+ + + + L + + P++ Q +
Sbjct 1 MVRYSKLAFRQLVR-VYDVDVCFTPMIYAKNFIESEKCRSSELSVCEGDSPLIVQFATDD 59
Query 89 PQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIVYEMKRRVQI 148
P ++EAA V+LN GCP V KG FG+AL+ P + D+V + + R+
Sbjct 60 PFVLSEAAEMVYKCSTG-VDLNCGCPKHDVRSKG-FGSALLSKPELLADMVRQTRARIPD 117
Query 149 P---VTVKCRLGVD 159
P V++K R+ D
Sbjct 118 PDFSVSLKIRINHD 131
> tpv:TP01_0269 hypothetical protein; K05545 tRNA-dihydrouridine
synthase 4 [EC:1.-.-.-]
Length=327
Score = 49.3 bits (116), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 34/134 (25%), Positives = 62/134 (46%), Gaps = 12/134 (8%)
Query 26 VAPMIAVSNTHFRNFMRLITRRSTV---YTEMVVDSTILHNTHQLENHLGFDDVEHPIVC 82
VAPM+ S FR L+ RR + YT M+ N + H + P++
Sbjct 30 VAPMVDQSELPFR----LLCRRYSADLAYTPMLHAKIFSENENYRNIHFHTSSDDKPLIA 85
Query 83 QLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIVYEM 142
Q G++P A+++ + +++N+GCP + KG +G+ L+ P + +IV +
Sbjct 86 QFCGNDPNAFILASSYIK-NDVSAIDINLGCPQ-GIARKGRYGSFLLEYPQLINEIVRSL 143
Query 143 KRRVQIPVTVKCRL 156
Q + V C++
Sbjct 144 ---TQTGINVTCKI 154
> eco:b2140 dusC, ECK2133, JW2128, yohI; tRNA-dihydrouridine synthase
C; K05541 tRNA-dihydrouridine synthase C [EC:1.-.-.-]
Length=315
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 32/80 (40%), Positives = 43/80 (53%), Gaps = 3/80 (3%)
Query 83 QLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIVYEM 142
QL G PQ +AE A A G V+LN GCPS + V+ GA L++ P + M
Sbjct 68 QLLGQFPQWLAENAARAVELGSWGVDLNCGCPS-KTVNGSGGGATLLKDPELIYQGAKAM 126
Query 143 KRRV--QIPVTVKCRLGVDN 160
+ V +PV+VK RLG D+
Sbjct 127 REAVPAHLPVSVKVRLGWDS 146
> xla:379349 dus3l, MGC53781; dihydrouridine synthase 3-like;
K05544 tRNA-dihydrouridine synthase 3 [EC:1.-.-.-]
Length=640
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 43/142 (30%), Positives = 58/142 (40%), Gaps = 3/142 (2%)
Query 18 ISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVE 77
I R L +AP+ N FR + + T EM + + +L L E
Sbjct 291 IDFRNKLYLAPLTTCGNLPFRRICKRLGADITC-GEMAMCTNLLQGQPSEWALLKRHHSE 349
Query 78 HPIVCQLGGSNPQNVAEAATWAELA-GYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVR 136
QL G+ P + + A D V++NVGCP V KG G LM K
Sbjct 350 DIFGVQLEGAFPDTMTKCAELLNRTIDVDFVDINVGCPIDLVYKKGG-GCGLMNRTNKFE 408
Query 137 DIVYEMKRRVQIPVTVKCRLGV 158
IV M + +P+TVK R GV
Sbjct 409 QIVKGMNSVLDVPLTVKIRTGV 430
> mmu:68730 Dus1l, 1110032N12Rik; dihydrouridine synthase 1-like
(S. cerevisiae); K05542 tRNA-dihydrouridine synthase 1 [EC:1.-.-.-]
Length=475
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 37/137 (27%), Positives = 68/137 (49%), Gaps = 13/137 (9%)
Query 26 VAPMIAVSNTHFRNFMRLITRR---STVYTEMVVDSTILHNTHQLENHLGFD--DVEHPI 80
VAPM+ S +R L++RR YT M+ + + + + +L D + P+
Sbjct 21 VAPMVDQSELAWR----LLSRRHGAQLCYTPMLHAQVFVRDANYRKENLYCDVCPEDRPL 76
Query 81 VCQLGGSNPQNVAEAATWAELAGY-DEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIV 139
+ Q ++P+ +AA A+ Y D ++LN+GCP + +G +GA L ++ ++
Sbjct 77 IVQFCANDPEVFVQAALLAQ--DYCDAIDLNLGCPQ-MIAKRGHYGAFLQEEWDLLQRMI 133
Query 140 YEMKRRVQIPVTVKCRL 156
R+ +PVT K R+
Sbjct 134 LLAHERLSVPVTCKIRV 150
> tgo:TGME49_002790 dihydrouridine synthase domain-containing
protein (EC:5.1.3.9); K05542 tRNA-dihydrouridine synthase 1
[EC:1.-.-.-]
Length=540
Score = 47.4 bits (111), Expect = 3e-05, Method: Composition-based stats.
Identities = 25/81 (30%), Positives = 42/81 (51%), Gaps = 2/81 (2%)
Query 75 DVEHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTK 134
+ + P+ Q G +P + AA E + V++N GCP + +G +GA L+ P
Sbjct 138 EFDEPVFVQFCGDSPATLLAAAQLVE-DEVEAVDVNFGCPQ-GIARRGHYGAFLLNEPEL 195
Query 135 VRDIVYEMKRRVQIPVTVKCR 155
+ DIV + + ++ PVT K R
Sbjct 196 LVDIVSTLHKHLKTPVTCKMR 216
> xla:446385 dus1l, MGC83715; dihydrouridine synthase 1-like;
K05542 tRNA-dihydrouridine synthase 1 [EC:1.-.-.-]
Length=464
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 40/153 (26%), Positives = 76/153 (49%), Gaps = 15/153 (9%)
Query 10 IDTMGETEISSRALLAVAPMIAVSNTHFRNFMRLITRRSTV---YTEMVVDSTILHNTHQ 66
+D T S+R +LA PM+ S +R L++RR V YT M+ + + +
Sbjct 1 MDFWKNTLRSARYVLA--PMVDQSELAWR----LLSRRHGVQLCYTPMLHAQVFVRDANY 54
Query 67 LENHLGFDDV--EHPIVCQLGGSNPQNVAEAATWAELAGY-DEVNLNVGCPSCRVVDKGS 123
+ +L + + P++ Q ++P+ +AA A+ Y D ++LN+GCP + +G
Sbjct 55 RKENLYCERCSEDRPLIVQFCANDPEVFVQAALLAQ--DYCDAIDLNLGCPQ-MIAKRGH 111
Query 124 FGAALMRSPTKVRDIVYEMKRRVQIPVTVKCRL 156
+GA L + ++ +++ +PVT K R+
Sbjct 112 YGAFLQDEWDLLEKMIQLAHQKLSVPVTCKIRV 144
> tgo:TGME49_028080 hypothetical protein ; K05544 tRNA-dihydrouridine
synthase 3 [EC:1.-.-.-]
Length=1220
Score = 47.0 bits (110), Expect = 3e-05, Method: Composition-based stats.
Identities = 37/135 (27%), Positives = 59/135 (43%), Gaps = 4/135 (2%)
Query 24 LAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHPIVCQ 83
L +AP+ V N FR + TV +EM V +I+ + L E Q
Sbjct 821 LILAPLTTVGNLPFRRLCVELGAEVTV-SEMAVAKSIVDGKQSELSLLKRHKSETFFGVQ 879
Query 84 LGGSNPQ--NVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIVYE 141
+ G P+ N +E D ++LN CP ++ + GA ++ P ++ +V
Sbjct 880 IAGGTPEVLNACTDILASEDVSCDFIDLNAACPLVQLHRRFKAGACMLDHPKRLESLVES 939
Query 142 M-KRRVQIPVTVKCR 155
M R ++PVTVK R
Sbjct 940 MTTRHPEVPVTVKLR 954
> mmu:224907 Dus3l, AI662135, AW557805, MGC56820; dihydrouridine
synthase 3-like (S. cerevisiae); K05544 tRNA-dihydrouridine
synthase 3 [EC:1.-.-.-]
Length=637
Score = 47.0 bits (110), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 42/136 (30%), Positives = 57/136 (41%), Gaps = 3/136 (2%)
Query 24 LAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHPIVCQ 83
L +AP+ N FR + T EM V + +L L E Q
Sbjct 294 LYLAPLTTCGNLPFRRICKRFGADVTC-GEMAVCTNLLQGQMSEWALLKRHPCEDIFGVQ 352
Query 84 LGGSNPQNVAEAATWAELA-GYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIVYEM 142
L G+ P + + A D V++NVGCP V KG G ALM K + IV +
Sbjct 353 LEGAFPDTMTKCAELLNRTIDVDFVDINVGCPIDLVYKKGG-GCALMNRSAKFQQIVRGV 411
Query 143 KRRVQIPVTVKCRLGV 158
+ +P+TVK R GV
Sbjct 412 NEVLDVPLTVKMRTGV 427
> xla:432319 MGC132093; hypothetical protein MGC78973; K05542
tRNA-dihydrouridine synthase 1 [EC:1.-.-.-]
Length=465
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 36/152 (23%), Positives = 74/152 (48%), Gaps = 11/152 (7%)
Query 22 ALLAVAPMIAVSNTHFRNFMRLITRRSTV---YTEMVVDSTILHNTHQLENHLGFD--DV 76
A +APM+ S +R L++RR V YT M+ + + + + +L +
Sbjct 11 ACYVLAPMVDQSELAWR----LLSRRHGVQLCYTPMLHAQVFVRDANYRKENLYCEVCPE 66
Query 77 EHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVR 136
+ P++ Q ++P+ +AA A+ D ++LN+GCP + +G +G+ L +
Sbjct 67 DRPLIVQFCANDPEVFVQAALLAQ-DYCDAIDLNLGCPQ-MIAKRGHYGSFLQDEWDLLE 124
Query 137 DIVYEMKRRVQIPVTVKCRLGVDNLDSPEFTK 168
++ +++ +PVT K R+ + + E+ K
Sbjct 125 KMIQLAHQKLSVPVTCKIRVFPEIEKTVEYAK 156
> mmu:66369 Dus2l, 2310016K04Rik; dihydrouridine synthase 2-like
(SMM1, S. cerevisiae); K05543 tRNA-dihydrouridine synthase
2 [EC:1.-.-.-]
Length=493
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 42/150 (28%), Positives = 62/150 (41%), Gaps = 22/150 (14%)
Query 24 LAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHP---- 79
L +APM+ V R + L VY E ++D +L + L D P
Sbjct 14 LILAPMVRVGTLPMR-LLALDYGADIVYCEELIDLKMLQCKRVVNEVLSTVDFVAPDDRV 72
Query 80 -----------IVCQLGGSNPQNVAEAATWAE--LAGYDEVNLNVGCPSCRVVDKGSFGA 126
+V Q+G S+ + A E +AG D +N+GCP KG GA
Sbjct 73 VFRTCEREQSRVVFQMGTSDAERALAVARLVENDVAGID---VNMGCPK-EYSTKGGMGA 128
Query 127 ALMRSPTKVRDIVYEMKRRVQIPVTVKCRL 156
AL+ P K+ I+ + + PVT K R+
Sbjct 129 ALLSDPDKIEKILSTLVKGTHRPVTCKIRI 158
> hsa:54920 DUS2L, DUS2, FLJ20399, SMM1, URLC8; dihydrouridine
synthase 2-like, SMM1 homolog (S. cerevisiae); K05543 tRNA-dihydrouridine
synthase 2 [EC:1.-.-.-]
Length=493
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 41/150 (27%), Positives = 64/150 (42%), Gaps = 22/150 (14%)
Query 24 LAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHP---- 79
L +APM+ V R + L VY E ++D ++ + L D P
Sbjct 14 LILAPMVRVGTLPMR-LLALDYGADIVYCEELIDLKMIQCKRVVNEVLSTVDFVAPDDRV 72
Query 80 -----------IVCQLGGSNPQNVAEAATWAE--LAGYDEVNLNVGCPSCRVVDKGSFGA 126
+V Q+G S+ + A E +AG D +N+GCP + KG GA
Sbjct 73 VFRTCEREQNRVVFQMGTSDAERALAVARLVENDVAGID---VNMGCPK-QYSTKGGMGA 128
Query 127 ALMRSPTKVRDIVYEMKRRVQIPVTVKCRL 156
AL+ P K+ I+ + + + PVT K R+
Sbjct 129 ALLSDPDKIEKILSTLVKGTRRPVTCKIRI 158
> hsa:64118 DUS1L, DUS1, PP3111; dihydrouridine synthase 1-like
(S. cerevisiae); K05542 tRNA-dihydrouridine synthase 1 [EC:1.-.-.-]
Length=473
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 35/137 (25%), Positives = 68/137 (49%), Gaps = 13/137 (9%)
Query 26 VAPMIAVSNTHFRNFMRLITRR---STVYTEMVVDSTILHNTHQLENHLGFD--DVEHPI 80
VAPM+ S +R L++RR YT M+ + + + + +L + + P+
Sbjct 21 VAPMVDQSELAWR----LLSRRHGAQLCYTPMLHAQVFVRDANYRKENLYCEVCPEDRPL 76
Query 81 VCQLGGSNPQNVAEAATWAELAGY-DEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIV 139
+ Q ++P+ +AA A+ Y D ++LN+GCP + +G +GA L ++ ++
Sbjct 77 IVQFCANDPEVFVQAALLAQ--DYCDAIDLNLGCPQ-MIAKRGHYGAFLQDEWDLLQRMI 133
Query 140 YEMKRRVQIPVTVKCRL 156
++ +PVT K R+
Sbjct 134 LLAHEKLSVPVTCKIRV 150
> dre:100003049 dus2l; dihydrouridine synthase 2-like, SMM1 homolog
(S. cerevisiae); K05543 tRNA-dihydrouridine synthase 2
[EC:1.-.-.-]
Length=504
Score = 42.7 bits (99), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 24/77 (31%), Positives = 40/77 (51%), Gaps = 2/77 (2%)
Query 80 IVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIV 139
+V Q+G ++P+ A E V++N+GCP KG G+AL+ P K+ I+
Sbjct 89 VVFQMGTADPERALAVAKLVE-NDVAAVDVNMGCPK-EYSTKGGMGSALLSDPEKIEAIL 146
Query 140 YEMKRRVQIPVTVKCRL 156
+ + + PVT K R+
Sbjct 147 TTLVKGISKPVTCKIRI 163
> dre:322715 dus1l, wu:fb71f06, zgc:63748; dihydrouridine synthase
1-like (S. cerevisiae); K05542 tRNA-dihydrouridine synthase
1 [EC:1.-.-.-]
Length=479
Score = 42.7 bits (99), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 35/142 (24%), Positives = 68/142 (47%), Gaps = 15/142 (10%)
Query 22 ALLAVAPMIAVSNTHFRNFMRLITRR---STVYTEMVVDSTILHNTHQLENHLGFDDV-- 76
A VAPM+ S +R L++RR YT M+ + + + +L + +V
Sbjct 17 ARFVVAPMVDQSELAWR----LLSRRHGAELCYTPMLHAQVFVRDANYRRENL-YSEVNQ 71
Query 77 -EHPIVCQLGGSNPQNVAEAATWAELAGY-DEVNLNVGCPSCRVVDKGSFGAALMRSPTK 134
+ P++ Q ++P+ +AA A+ Y D ++LN+GCP + +G +G L
Sbjct 72 EDRPLITQFCANDPEVFIQAALLAQ--DYCDAIDLNLGCPQ-MIAKRGHYGVFLQDEWDL 128
Query 135 VRDIVYEMKRRVQIPVTVKCRL 156
+ ++ ++ +P+T K R+
Sbjct 129 LEKMIKLANEKLSVPITCKIRV 150
> dre:100330185 dihydrouridine synthase 1-like (S. cerevisiae)-like;
K05542 tRNA-dihydrouridine synthase 1 [EC:1.-.-.-]
Length=496
Score = 42.7 bits (99), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 35/142 (24%), Positives = 68/142 (47%), Gaps = 15/142 (10%)
Query 22 ALLAVAPMIAVSNTHFRNFMRLITRR---STVYTEMVVDSTILHNTHQLENHLGFDDV-- 76
A VAPM+ S +R L++RR YT M+ + + + +L + +V
Sbjct 34 ARFVVAPMVDQSELAWR----LLSRRHGAELCYTPMLHAQVFVRDANYRRENL-YSEVNQ 88
Query 77 -EHPIVCQLGGSNPQNVAEAATWAELAGY-DEVNLNVGCPSCRVVDKGSFGAALMRSPTK 134
+ P++ Q ++P+ +AA A+ Y D ++LN+GCP + +G +G L
Sbjct 89 EDRPLITQFCANDPEVFIQAALLAQ--DYCDAIDLNLGCPQ-MIAKRGHYGVFLQDEWDL 145
Query 135 VRDIVYEMKRRVQIPVTVKCRL 156
+ ++ ++ +P+T K R+
Sbjct 146 LEKMIKLANEKLSVPITCKIRV 167
> dre:393647 MGC63779; zgc:63779; K05544 tRNA-dihydrouridine synthase
3 [EC:1.-.-.-]
Length=660
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 41/145 (28%), Positives = 58/145 (40%), Gaps = 3/145 (2%)
Query 15 ETEISSRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFD 74
+ ++ R L +AP+ N FR + T EM + + +L L
Sbjct 308 KKQVDFRDKLYLAPLTTCGNLPFRRVCKRFGADITC-GEMAMCTNLLQGQASEWALLKRH 366
Query 75 DVEHPIVCQLGGSNPQNVAEAATW-AELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPT 133
E QL G P + A + D V++N GCP V KG G LM +
Sbjct 367 ASEDLFGVQLEGCFPDTMTRCAELLNQNIDVDFVDINSGCPIDLVYKKGG-GCGLMTRTS 425
Query 134 KVRDIVYEMKRRVQIPVTVKCRLGV 158
K IV M + +P+TVK R GV
Sbjct 426 KFEQIVRGMNSVLDVPLTVKIRTGV 450
> cel:Y37E11B.5 hypothetical protein; K05544 tRNA-dihydrouridine
synthase 3 [EC:1.-.-.-]
Length=554
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 43/151 (28%), Positives = 69/151 (45%), Gaps = 11/151 (7%)
Query 26 VAPMIAVSNTHFRNFMRLITRRSTVYT--EMVVDSTILHNTHQLENHLGFDDVEHPIVCQ 83
+AP+ V N FR R+ T EM + ++IL T + L E Q
Sbjct 208 LAPLTTVGNLPFR---RICVDYGADITCGEMALATSILSGTASEYSLLKRHPCEKIFGVQ 264
Query 84 LGGSNPQNVAEAATWA-ELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIVYEM 142
L G +A+A+ E D +++N+GCP VV++ G AL P K+ +++
Sbjct 265 LAGGFADTMAKASQIVVENFDVDFIDINMGCP-IDVVNQKGGGCALPSRPQKLFEVLAAT 323
Query 143 KRRVQ--IPVTVKCRLGVDN--LDSPEFTKQ 169
K + P+TVK R G+ L +PE+ +
Sbjct 324 KSVLGGCCPLTVKIRTGMKEGVLKAPEYVEH 354
> sce:YLR405W DUS4; Dihydrouridine synthase, member of a widespread
family of conserved proteins including Smm1p, Dus1p, and
Dus3p (EC:1.-.-.-); K05545 tRNA-dihydrouridine synthase 4
[EC:1.-.-.-]
Length=367
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 33/128 (25%), Positives = 61/128 (47%), Gaps = 7/128 (5%)
Query 20 SRALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHP 79
R + PM+ S FR R VY+ M++ + N H + L ++ + P
Sbjct 37 GRPVTIAGPMVRYSKLPFRQLCREYNV-DIVYSPMILAREYVRNEHARISDLSTNNEDTP 95
Query 80 IVCQLGGSNPQNVAEAATWAEL-AGY-DEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRD 137
++ Q+G + NVA+ + E+ A Y D + +N GCP + +G G AL+ + +
Sbjct 96 LIVQVGVN---NVADLLKFVEMVAPYCDGIGINCGCPIKEQIREG-IGCALIYNSDLLCS 151
Query 138 IVYEMKRR 145
+V+ +K +
Sbjct 152 MVHAVKDK 159
> sce:YNR015W SMM1, DUS2; Dihydrouridine synthase, member of a
family of dihydrouridine synthases including Dus1p, Smm1p,
Dus3p, and Dus4p; modifies uridine residues at position 20 of
cytoplasmic tRNAs (EC:1.-.-.-); K05543 tRNA-dihydrouridine
synthase 2 [EC:1.-.-.-]
Length=384
Score = 38.9 bits (89), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 53/99 (53%), Gaps = 14/99 (14%)
Query 80 IVCQLGGSNPQNVAEAATWA--ELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRD 137
++ Q+G ++P +AA +++G D +N GCP + G G+AL+R+P +
Sbjct 85 LIFQIGSASPALATQAALKVINDVSGID---INAGCPKHFSIHSG-MGSALLRTPDTLCL 140
Query 138 IVYEMKRRV----QIPVTVKCRLGVDNLDSPEFTKQFVE 172
I+ E+ + V P++VK RL LD+ + T Q V+
Sbjct 141 ILKELVKNVGNPHSKPISVKIRL----LDTKQDTLQLVK 175
> cpv:cgd5_2440 Ylr401cp-like protein with 2 CCCH domains plus
Dus1p tRNA dihydrouridine synthase Tim barrel ; K05544 tRNA-dihydrouridine
synthase 3 [EC:1.-.-.-]
Length=707
Score = 37.4 bits (85), Expect = 0.027, Method: Composition-based stats.
Identities = 38/138 (27%), Positives = 62/138 (44%), Gaps = 4/138 (2%)
Query 21 RALLAVAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHPI 80
R L +AP+ V N FR TV +EMV+ + IL L E
Sbjct 315 RNKLILAPLTTVGNLPFRRLCLKFGADVTV-SEMVLCNEILSGKASELALLKKSPEEKYF 373
Query 81 VCQLGGSNPQNVAEAATWA-ELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIV 139
QL G N + + + E +D +++N CP + DKG+ G+ L+ +++ +V
Sbjct 374 GIQLAGGNKSTIIKTGEFINEHCEFDFIDINAACPLKSLHDKGA-GSILVDRVSELETMV 432
Query 140 YEMKRRVQIP-VTVKCRL 156
+K+ + VTVK R+
Sbjct 433 KGLKKVMDGKLVTVKVRM 450
> sce:YLR401C DUS3; Dihydrouridine synthase, member of a widespread
family of conserved proteins including Smm1p, Dus1p, and
Dus4p; contains a consensus oleate response element (ORE)
in its promoter region (EC:1.-.-.-); K05544 tRNA-dihydrouridine
synthase 3 [EC:1.-.-.-]
Length=668
Score = 37.0 bits (84), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 37/137 (27%), Positives = 60/137 (43%), Gaps = 6/137 (4%)
Query 26 VAPMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGFDDVEHP---IVC 82
V+P+ V N +R MR + T Y+EM + ++ T+ E P +
Sbjct 297 VSPLTTVGNLPYRRLMRKLGADVT-YSEMALAVPLIQGTNSEWALPKAHTSEFPGFGVQV 355
Query 83 QLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIVYEM 142
+ A A ++ E+NLN GCP + +GS G+AL+ +P ++ + M
Sbjct 356 ACSKAWQAAKAAEALANSVSEISEINLNSGCPIDLLYRQGS-GSALLDNPARMIRCLNAM 414
Query 143 KR-RVQIPVTVKCRLGV 158
IP+TVK R G
Sbjct 415 NYVSKDIPITVKIRTGT 431
> hsa:1806 DPYD, DHP, DHPDHASE, DPD, MGC132008, MGC70799; dihydropyrimidine
dehydrogenase (EC:1.3.1.2); K00207 dihydropyrimidine
dehydrogenase (NADP+) [EC:1.3.1.2]
Length=1025
Score = 36.6 bits (83), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 35/140 (25%), Positives = 59/140 (42%), Gaps = 16/140 (11%)
Query 16 TEISSRALLAV--APMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGF 73
T +S R + PM + F N + LI+ ++ Y + +L+
Sbjct 584 TNVSPRIIRGTTSGPMYGPGQSSFLN-IELISEKTAAY--------WCQSVTELKADFPD 634
Query 74 DDVEHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPT 133
+ V I+C N + E A +E +G D + LN+ CP + + G A + P
Sbjct 635 NIVIASIMCSY---NKNDWTELAKKSEDSGADALELNLSCP--HGMGERGMGLACGQDPE 689
Query 134 KVRDIVYEMKRRVQIPVTVK 153
VR+I +++ VQIP K
Sbjct 690 LVRNICRWVRQAVQIPFFAK 709
> mmu:99586 Dpyd, AI315208, DPD, E330028L06Rik, MGC37940; dihydropyrimidine
dehydrogenase (EC:1.3.1.2); K00207 dihydropyrimidine
dehydrogenase (NADP+) [EC:1.3.1.2]
Length=1025
Score = 35.4 bits (80), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 32/140 (22%), Positives = 60/140 (42%), Gaps = 16/140 (11%)
Query 16 TEISSRALLAV--APMIAVSNTHFRNFMRLITRRSTVYTEMVVDSTILHNTHQLENHLGF 73
T +S R + P+ + F N + LI+ ++ Y H+ +L+
Sbjct 584 TNVSPRIIRGTTSGPLYGPGQSSFLN-IELISEKTAAY--------WCHSVTELKADFPD 634
Query 74 DDVEHPIVCQLGGSNPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPT 133
+ + I+C N + E + AE +G D + LN+ CP + + G A + P
Sbjct 635 NILIASIMCSY---NKSDWMELSKMAEASGADALELNLSCP--HGMGERGMGLACGQDPE 689
Query 134 KVRDIVYEMKRRVQIPVTVK 153
VR+I +++ V++P K
Sbjct 690 LVRNICRWVRQAVRVPFFAK 709
> cpv:cgd8_1140 Sec14d domain containing protein
Length=289
Score = 32.7 bits (73), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 40/74 (54%), Gaps = 2/74 (2%)
Query 2 AVIWLVNEIDTMGETEISSRAL-LAVAPM-IAVSNTHFRNFMRLITRRSTVYTEMVVDST 59
VI+ +EI + IS +L L + M I F++FMRLI+ Y+ +++++
Sbjct 80 KVIYSFSEISMKEKNFISHISLVLDFSEMGIYTCKVEFQSFMRLISEILDFYSPLLINTI 139
Query 60 ILHNTHQLENHLGF 73
I+H + L+ +L +
Sbjct 140 IIHKSKSLKENLWY 153
> cel:C25F6.3 dpyd-1; DihydroPYrimidine Dehydrogenase family member
(dpyd-1); K00207 dihydropyrimidine dehydrogenase (NADP+)
[EC:1.3.1.2]
Length=1059
Score = 32.3 bits (72), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 36/66 (54%), Gaps = 2/66 (3%)
Query 88 NPQNVAEAATWAELAGYDEVNLNVGCPSCRVVDKGSFGAALMRSPTKVRDIVYEMKRRVQ 147
N + E AT +E AG D + LN+ CP + +KG G A +SP V++I ++ V+
Sbjct 660 NKADWIELATKSEEAGADILELNLSCPH-GMGEKG-MGLACGQSPEIVKEICRWVRACVK 717
Query 148 IPVTVK 153
IP K
Sbjct 718 IPFFPK 723
Lambda K H
0.320 0.134 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4535951560
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40