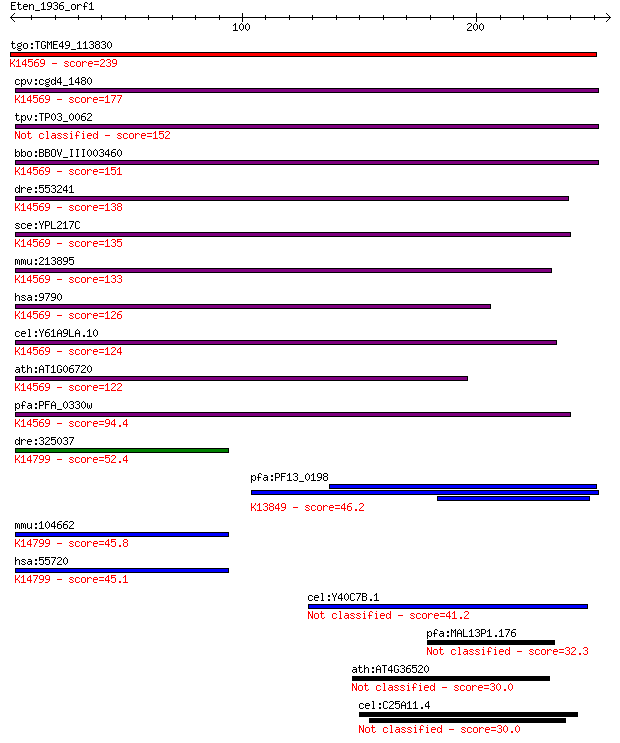
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1936_orf1
Length=256
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_113830 ribosome biogenesis protein BMS1, putative ;... 239 9e-63
cpv:cgd4_1480 BMS1 like GTpase involved in ribosome biogenesis... 177 3e-44
tpv:TP03_0062 hypothetical protein 152 1e-36
bbo:BBOV_III003460 17.m07327; hypothetical protein; K14569 rib... 151 2e-36
dre:553241 bms1l, im:7152071, wu:fb37d08; BMS1-like, ribosome ... 138 2e-32
sce:YPL217C BMS1; Bms1p; K14569 ribosome biogenesis protein BMS1 135 1e-31
mmu:213895 Bms1, AA408648, AU020092, AU043373, BB007109, BC030... 133 8e-31
hsa:9790 BMS1, BMS1L, KIAA0187; BMS1 homolog, ribosome assembl... 126 1e-28
cel:Y61A9LA.10 hypothetical protein; K14569 ribosome biogenesi... 124 3e-28
ath:AT1G06720 hypothetical protein; K14569 ribosome biogenesis... 122 1e-27
pfa:PFA_0330w AARP2; pfAARP2 protein; K14569 ribosome biogenes... 94.4 4e-19
dre:325037 tsr1, MGC123319, fc52b09, wu:fc52b09, zgc:123319; T... 52.4 2e-06
pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog A... 46.2 1e-04
mmu:104662 Tsr1, AU040765, AW550801, mKIAA1401; TSR1, 20S rRNA... 45.8 2e-04
hsa:55720 TSR1, FLJ10534, KIAA1401, MGC131829; TSR1, 20S rRNA ... 45.1 3e-04
cel:Y40C7B.1 hypothetical protein 41.2 0.003
pfa:MAL13P1.176 PfRh2b; reticulocyte binding protein 2, homolog B 32.3 1.6
ath:AT4G36520 heat shock protein binding 30.0 7.8
cel:C25A11.4 ajm-1; Apical Junction Molecule family member (aj... 30.0 8.2
> tgo:TGME49_113830 ribosome biogenesis protein BMS1, putative
; K14569 ribosome biogenesis protein BMS1
Length=1267
Score = 239 bits (609), Expect = 9e-63, Method: Compositional matrix adjust.
Identities = 129/258 (50%), Positives = 180/258 (69%), Gaps = 8/258 (3%)
Query 1 STKLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAIGTDGTFRA 60
S KLKL+GE K I + TAF+K MFNS LEVN C+GAKIQTVSGIRGQ+KKA+GTDGTFRA
Sbjct 998 SKKLKLLGEPKKIFKNTAFIKNMFNSDLEVNMCMGAKIQTVSGIRGQVKKALGTDGTFRA 1057
Query 61 SFEDKVLKSDLILCKTWVLVCPHAFYNPVLDLPGWRGLRLLSEIKKEKKIISKQKETLFH 120
+FEDK+L SDL++CKTW+ + P F NPVLD+ GW+ LR +EI++ ++ + K
Sbjct 1058 TFEDKILMSDLVVCKTWIKMQPRQFCNPVLDVEGWQRLRTQAEIRQALQLPTPTKPGSHP 1117
Query 121 SFG----KAPRKQHSFFPIRIPKQLAAKLPLHSRLKLQQG----RRVGKQQQQKERDMKK 172
G +A R+ F PIR+PKQL KLP H+R KLQ R++ + ++E D++K
Sbjct 1118 DGGLAALQAARRSKEFNPIRVPKQLMLKLPFHARTKLQHSTSKLRKLKGKALEEELDLRK 1177
Query 173 AFVSPNERRAAVLLQQLETLKQHRDKRRQQQQQQKARLKQQQTDKLQQQQQQQQKERQKR 232
VS +RR A LLQ+L+T+K R +RR++QQ++K + K ++++ ++Q E +KR
Sbjct 1178 PLVSAYDRRVAALLQRLQTIKNARVERRKEQQKEKRLKVAKAAAKKEEERARKQTEMRKR 1237
Query 233 RHSKQGKIEASMRKRLRL 250
R+ KQGKIE MRK++RL
Sbjct 1238 RYVKQGKIELGMRKKMRL 1255
> cpv:cgd4_1480 BMS1 like GTpase involved in ribosome biogenesis
; K14569 ribosome biogenesis protein BMS1
Length=1051
Score = 177 bits (450), Expect = 3e-44, Method: Composition-based stats.
Identities = 101/251 (40%), Positives = 161/251 (64%), Gaps = 10/251 (3%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAIGTDGTFRASF 62
KLKLVGE K I + TAF+ MFNS LEV++ +GAKIQTVSGIRGQ+KKAI T G FRA+F
Sbjct 809 KLKLVGEPKKIHKNTAFIHKMFNSDLEVSKFIGAKIQTVSGIRGQVKKAISTHGLFRATF 868
Query 63 EDKVLKSDLILCKTWVLVCPHAFYNPVLDLPGWRGLRLLSEIKKEKKI-ISKQKETLFHS 121
EDK+L SD++ CKTWV + P FYNPV+DLP WR +R +E+++E I ++ + ++ + +
Sbjct 869 EDKILLSDIVFCKTWVSMTPREFYNPVIDLPTWRRMRTQAELRRELNIPLAIKADSEYVT 928
Query 122 FGKAPRKQHSFFPIRIPKQLAAKLPLHSRLKLQQGRRVGKQQQQKERDMKKAFVSPNERR 181
P K+ F + +P +L +LP S+ K ++ K+++ S E+R
Sbjct 929 KQDRPEKKR-FNSVPVPSKLEKELPYASKTK-------NDSKKIKDKNQVAVIKSTFEKR 980
Query 182 AAVLLQQLETLKQHRDKRRQQQQQQKARLKQQQTDKLQQQQQQQQKERQKRRHSKQG-KI 240
A L Q+L T+++ + +R ++++ K + ++ L++ ++ + +ER+KRR++ QG KI
Sbjct 981 VANLFQRLSTIQKEKTAKRIEKKRIKREINIKRRQPLERIREAKNEERKKRRYALQGIKI 1040
Query 241 EASMRKRLRLD 251
+ RK + D
Sbjct 1041 DKQRRKLMMKD 1051
> tpv:TP03_0062 hypothetical protein
Length=295
Score = 152 bits (383), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 99/251 (39%), Positives = 147/251 (58%), Gaps = 15/251 (5%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAIGTDGTFRASF 62
KL L GE I + T F+K MFNS LEV +C+G+KIQT SGIRGQIKK I +G FRA+F
Sbjct 58 KLNLEGEAYKIMKNTCFIKNMFNSELEVIKCIGSKIQTSSGIRGQIKKPIEKNGAFRATF 117
Query 63 EDKVLKSDLILCKTWVLVCPHAFYNPVLDLPGWRGLRLLSEIKKEKKIISKQKETLFHSF 122
EDK+L SD+++ K+WV V FYN +LD G+R ++ ++E+KKE+ I + K
Sbjct 118 EDKILLSDIVVLKSWVNVETKRFYNLILDSEGFRRVKSIAELKKEEPIKADSKYERKELL 177
Query 123 GKAPRKQHSFFPIRIPKQLAAKLPLHSRLKLQQGRRVGKQQQQKERDMKKAFVSPN--ER 180
+ R+ F I+IPK++ KL SR K+ + +D V P+ E+
Sbjct 178 KRPVRR---FNEIKIPKKVIEKLAFSSRPKV----------IEHPKDDITVTVDPSEHEK 224
Query 181 RAAVLLQQLETLKQHRDKRRQQQQQQKARLKQQQTDKLQQQQQQQQKERQKRRHSKQGKI 240
R A LQ+L T+++ R ++R + +Q K KLQ+ ++ + KE +K +++KQ K
Sbjct 225 RIARTLQKLHTIRKDRLEKRLESIKQYKIKKDLDEGKLQEVRKTKLKEIKKAKYAKQTKK 284
Query 241 EASMRKRLRLD 251
E R +L LD
Sbjct 285 ERIKRSKLLLD 295
> bbo:BBOV_III003460 17.m07327; hypothetical protein; K14569 ribosome
biogenesis protein BMS1
Length=924
Score = 151 bits (382), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 92/254 (36%), Positives = 148/254 (58%), Gaps = 23/254 (9%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAIGTDGTFRASF 62
KLK+ G I + TAF+K MF S LEV +CLG+++ T SGIRG+IKK +G +G FRA+F
Sbjct 689 KLKVQGYPYKILKHTAFIKDMFTSELEVIKCLGSRLVTASGIRGEIKKPVGKNGAFRATF 748
Query 63 EDKVLKSDLILCKTWVLVCPHAFYNPVLDLPGWRGLRLLSEIKKEKKIISKQKETLFHSF 122
EDK+L SD++L K +V V F+NP++D +R +R ++E++ + I ++++
Sbjct 749 EDKILMSDIVLLKAFVSVPTRQFFNPMVDWSSFRRVRTIAELRTD---IGINPDSVYEPK 805
Query 123 GKAPRKQHSFFPIRIPKQLAAKLPLHSRLKLQQGRRVGKQQQQKERDMKKAFVSPNERRA 182
R + F PI++PK++ LP SR K+ + G E ER
Sbjct 806 ELLKRPERKFNPIKVPKKIIDNLPFSSRPKVYEQEDSGITLGHSEY----------ERSV 855
Query 183 AVLLQQLETLKQHR-DKRRQQQQQQKARLKQQQTDKLQQQQQQQQKER----QKRRHSKQ 237
A ++Q+L T+++ R +KR ++Q KA+L+ + +++Q KER +K R+ KQ
Sbjct 856 ANVMQRLLTIRKSRLEKRASERQLHKAKLRLE-----EERQAAASKERTKGIRKLRYIKQ 910
Query 238 GKIEASMRKRLRLD 251
GK EA+ R ++ LD
Sbjct 911 GKTEAAKRAKMCLD 924
> dre:553241 bms1l, im:7152071, wu:fb37d08; BMS1-like, ribosome
assembly protein (yeast); K14569 ribosome biogenesis protein
BMS1
Length=1221
Score = 138 bits (347), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 91/246 (36%), Positives = 142/246 (57%), Gaps = 18/246 (7%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAIGT-DGTFRAS 61
KLKL+G I + T F++GMFN+ LEV + GA I+TVSGI+GQIKKA+ T G FRA+
Sbjct 966 KLKLIGYPYKIFKNTCFIQGMFNTILEVAKFEGASIRTVSGIKGQIKKALRTPPGAFRAT 1025
Query 62 FEDKVLKSDLILCKTWVLVCPHAFYNPV--LDLP-----GWRGLRLLSEIKKEKKIISKQ 114
FED++L SD++ ++W V YNPV L +P W G+R L ++K + I +K
Sbjct 1026 FEDRLLMSDIVFLRSWYPVSVPRLYNPVTSLLMPVGQKHTWSGMRTLGQLKHDLGIHNKP 1085
Query 115 KETLFHSFGKAPRKQHSFFPIRIPKQLAAKLPLHSRLKLQQGRRVGKQQQQKERDMKKAF 174
K+ + R+ F P+RIPK+L LP S+ K Q + GK RD+++
Sbjct 1086 KQDSLYK--PIERQVRHFNPLRIPKELQKALPFKSKPKYMQSK--GKTP----RDLQRPA 1137
Query 175 V--SPNERRAAVLLQQLETLKQHRDKRRQQQQQQKARLKQQQTDKLQQQQQQQQKERQKR 232
V P+E++ A LL L T+ ++ K+ +Q+ K + +Q +K + ++QQ+ K +K+
Sbjct 1138 VIREPHEKKVAALLDALTTVYSYKTKKAIAEQRTKHKEFLKQKEKQEAERQQRLKVEKKK 1197
Query 233 RHSKQG 238
+ G
Sbjct 1198 TYRAMG 1203
> sce:YPL217C BMS1; Bms1p; K14569 ribosome biogenesis protein
BMS1
Length=1183
Score = 135 bits (340), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 88/247 (35%), Positives = 149/247 (60%), Gaps = 21/247 (8%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAIG-TDGTFRAS 61
KLKLVG I + TAF+K MF+S +EV R GA+I+TVSGIRG+IK+A+ +G +RA+
Sbjct 931 KLKLVGFPYKIFKNTAFIKDMFSSAMEVARFEGAQIKTVSGIRGEIKRALSKPEGHYRAA 990
Query 62 FEDKVLKSDLILCKTWVLVCPHAFYNPVLDL-----PGWRGLRLLSEIKKEKKI-ISKQK 115
FEDK+L SD+++ ++W V FYNPV L W+GLRL +I+ +
Sbjct 991 FEDKILMSDIVILRSWYPVRVKKFYNPVTSLLLKEKTEWKGLRLTGQIRAAMNLETPSNP 1050
Query 116 ETLFHSFGKAPRKQHSFFPIRIPKQLAAKLPLHSRLKLQQGRRVGKQQQQKERDMKKAFV 175
++ +H K R + F +++PK + +LP S++ K Q++K K+A V
Sbjct 1051 DSAYH---KIERVERHFNGLKVPKAVQKELPFKSQIHQM------KPQKKKTYMAKRAVV 1101
Query 176 -SPNERRAAVLLQQLETLKQHRDKRRQQQQ--QQKARLKQQQTDKLQQQQQQQQKERQKR 232
+E++A +Q++ T+ + +D +R++Q+ Q+K RLK+ K+++++ Q+ KE++K
Sbjct 1102 LGGDEKKARSFIQKVLTISKAKDSKRKEQKASQRKERLKKLA--KMEEEKSQRDKEKKKE 1159
Query 233 RHSKQGK 239
++ GK
Sbjct 1160 YFAQNGK 1166
> mmu:213895 Bms1, AA408648, AU020092, AU043373, BB007109, BC030906,
Bms1l, KIAA0187, mKIAA0187; BMS1 homolog, ribosome assembly
protein (yeast); K14569 ribosome biogenesis protein BMS1
Length=1284
Score = 133 bits (334), Expect = 8e-31, Method: Compositional matrix adjust.
Identities = 89/238 (37%), Positives = 134/238 (56%), Gaps = 20/238 (8%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAI-GTDGTFRAS 61
KLKL G I + T+F+KGMFNS LEV + GA I+TVSGIRGQIKKA+ +G FRAS
Sbjct 1030 KLKLTGFPFKIFKNTSFIKGMFNSALEVAKFEGAVIRTVSGIRGQIKKALRAPEGAFRAS 1089
Query 62 FEDKVLKSDLILCKTWVLVCPHAFYNPVLDL-------PGWRGLRLLSEIKKEKKI-ISK 113
FEDK+L SD++ +TW V AFYNPV L W G+R +++ I +
Sbjct 1090 FEDKLLMSDIVFMRTWYPVSIPAFYNPVTSLLKPVGEKDTWSGMRTTHQLRLAHGIKLKA 1149
Query 114 QKETLFHSFGKAPRKQHSFFPIRIPKQLAAKLPLHSRLKLQQGRRVGKQQQQKERDMKKA 173
K++L+ R++ F + IPK L LP ++ K Q + GK + ++R
Sbjct 1150 NKDSLYKPI---LRQKKHFNSLHIPKALQKALPFKNKPKTQA--KAGKVPRDRQR--PAV 1202
Query 174 FVSPNERRAAVLLQQLETLKQHRDKRRQQQQQQKARLKQQQTDKLQQQQQQQQKERQK 231
P+ER+ LL L T+ + K+ ++Q+ RL ++ K++Q++++ + RQK
Sbjct 1203 IREPHERKVLALLDALSTIHSQKMKKAKEQR----RLHNKEHVKMKQKEEEDKLRRQK 1256
> hsa:9790 BMS1, BMS1L, KIAA0187; BMS1 homolog, ribosome assembly
protein (yeast); K14569 ribosome biogenesis protein BMS1
Length=1282
Score = 126 bits (316), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 81/212 (38%), Positives = 117/212 (55%), Gaps = 16/212 (7%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAI-GTDGTFRAS 61
KLKL G I + T+F+KGMFNS LEV + GA I+TVSGIRGQIKKA+ +G FRAS
Sbjct 1026 KLKLTGFPYKIFKNTSFIKGMFNSALEVAKFEGAVIRTVSGIRGQIKKALRAPEGAFRAS 1085
Query 62 FEDKVLKSDLILCKTWVLVCPHAFYNPVLDL-------PGWRGLRLLSEIKKEKKI-ISK 113
FEDK+L SD++ +TW V AFYNPV L W G+R +++ + +
Sbjct 1086 FEDKLLMSDIVFMRTWYPVSIPAFYNPVTSLLKPVGEKDTWSGMRTTGQLRLAHGVRLKA 1145
Query 114 QKETLFHSFGKAPRKQHSFFPIRIPKQLAAKLPLHSRLKLQQGRRVGKQQQQKERDMKKA 173
K++L+ R++ F + IPK L LP ++ K Q + GK + + R
Sbjct 1146 NKDSLYKPI---LRQKKHFNSLHIPKALQKALPFKNKPKTQA--KAGKVPKDRRR--PAV 1198
Query 174 FVSPNERRAAVLLQQLETLKQHRDKRRQQQQQ 205
P+ER+ LL L T+ + K+ ++Q+
Sbjct 1199 IREPHERKILALLDALSTVHSQKMKKAKEQRH 1230
> cel:Y61A9LA.10 hypothetical protein; K14569 ribosome biogenesis
protein BMS1
Length=1055
Score = 124 bits (311), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 81/242 (33%), Positives = 131/242 (54%), Gaps = 23/242 (9%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAI-GTDGTFRAS 61
KLKL+G + I +KTAFVKGMFNS LEV + GA I+TV+GIRGQIKKAI +G FRA+
Sbjct 800 KLKLIGHPEKIFKKTAFVKGMFNSALEVAKFEGATIRTVAGIRGQIKKAIKAPEGAFRAT 859
Query 62 FEDKVLKSDLILCKTWVLVCPHAFYNPVLD-----LPGWRGLRLLSEIKKEKKIISKQKE 116
FEDK+L D++ ++WV V FY P+ D W G+R + +++ E + + Q +
Sbjct 860 FEDKILMRDIVFLRSWVTVPIPRFYTPISDHLQASAAAWIGMRTVGKMRSELGMGTPQNK 919
Query 117 TLFHSFGKAPRKQHSFFPIRIPKQLAAKLPLHSRLKLQQGRRVGKQQQQKERDMKKA--- 173
+ RK+ PI +P +L LP + Q +++KE+D A
Sbjct 920 D--SDYKPIVRKEFESAPIHLPPKLQKSLPFKMKPTYQ-------AREEKEKDSLVARHT 970
Query 174 --FVSPNERRAAVLLQQLETLKQHRDKRRQQQQQQKARLKQQQTDKLQQQQQQQQKERQK 231
+ P E + + L TL D ++Q+ K K+++ +++ + + +++K +
Sbjct 971 AVVLEPEEAKRERFMDMLRTL---NDVDLKKQENIKEGYKKRKAEEMAESEAKREKSIKS 1027
Query 232 RR 233
R+
Sbjct 1028 RK 1029
> ath:AT1G06720 hypothetical protein; K14569 ribosome biogenesis
protein BMS1
Length=1147
Score = 122 bits (306), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 73/206 (35%), Positives = 113/206 (54%), Gaps = 21/206 (10%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAI-------GTD 55
K+KLVG IK+KTAF+K MF S LE+ R G+ ++TVSGIRGQ+KKA +
Sbjct 896 KIKLVGTPCKIKKKTAFIKDMFTSDLEIARFEGSSVRTVSGIRGQVKKAGKNMLDNKAEE 955
Query 56 GTFRASFEDKVLKSDLILCKTWVLVCPHAFYNPVLDL-----PGWRGLRLLSEIKKEKKI 110
G R +FED++ SD++ + W V FYNP+ W G++ E+++E I
Sbjct 956 GIARCTFEDQIHMSDMVFLRAWTTVEVPQFYNPLTTALQPRDKTWNGMKTFGELRRELNI 1015
Query 111 -ISKQKETLFHSFGKAPRKQHSFFPIRIPKQLAAKLPLHSRLKLQQGRRVGKQQQQKERD 169
I K++L+ + RKQ F P++IPK+L LP S+ K + K+++ D
Sbjct 1016 PIPVNKDSLYKAI---ERKQKKFNPLQIPKRLEKDLPFMSKPK-----NIPKRKRPSLED 1067
Query 170 MKKAFVSPNERRAAVLLQQLETLKQH 195
+ + P ER+ ++QQ + L+ H
Sbjct 1068 KRAVIMEPKERKEHTIIQQFQLLQHH 1093
> pfa:PFA_0330w AARP2; pfAARP2 protein; K14569 ribosome biogenesis
protein BMS1
Length=1434
Score = 94.4 bits (233), Expect = 4e-19, Method: Composition-based stats.
Identities = 76/248 (30%), Positives = 120/248 (48%), Gaps = 19/248 (7%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAIGTDGTFRASF 62
KLKL+GE I + TAFVK MFNS LEV + L + T SGI+G IK I +G FR +F
Sbjct 1195 KLKLIGEPYKIFKNTAFVKNMFNSDLEVCKFLNCPVVTPSGIKGLIKNKINNNGDFRCTF 1254
Query 63 EDKVLKSDLILCKTWVLVCPHAFYNPVLDLPGWRGLRLLSEIKKEKKIISKQKETLFHSF 122
DK+ SD+++ K +V V FY D+ ++ ++E++ I +++
Sbjct 1255 ADKIRMSDIVILKLYVNVKIKKFY--YFDIEN--KIKSINELRYLYNIYVNHN----NNY 1306
Query 123 GKAPRKQHSFFPIRIPKQLAAKLPLHSR----LKLQQGRRVGKQQQQKERDMKKAFVS-- 176
P + I+I +L LP S+ KLQ + +Q + KK
Sbjct 1307 RSIPFRHFYHTKIQIKSKLLKDLPFKSKPKLFKKLQNHANLNNKQLTNNINNKKTKTDQI 1366
Query 177 -----PNERRAAVLLQQLETLKQHRDKRRQQQQQQKARLKQQQTDKLQQQQQQQQKERQK 231
PN + AA Q L T+K++ +++ + Q K ++ K+QQ + Q K+R+K
Sbjct 1367 NFNALPNPKLAAKWYQMLHTIKKNILDQKKAKSQLSYHKKLKEKLKIQQAKTQVVKQRKK 1426
Query 232 RRHSKQGK 239
+ K K
Sbjct 1427 ISYKKGRK 1434
> dre:325037 tsr1, MGC123319, fc52b09, wu:fc52b09, zgc:123319;
TSR1, 20S rRNA accumulation, homolog (yeast); K14799 pre-rRNA-processing
protein TSR1
Length=822
Score = 52.4 bits (124), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/91 (29%), Positives = 49/91 (53%), Gaps = 0/91 (0%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAIGTDGTFRASF 62
++ L G I R++A V+ MF + ++ +++T G RG IK+A+GT G + F
Sbjct 714 RIVLSGHPFKINRRSAVVRYMFFNRDDILWFKPVELRTKWGRRGHIKEALGTHGHMKCVF 773
Query 63 EDKVLKSDLILCKTWVLVCPHAFYNPVLDLP 93
+ ++ D +L + V PH Y+P + +P
Sbjct 774 DSQLRSQDTVLMNLYKRVYPHWTYDPYVPVP 804
> pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog
A; K13849 reticulocyte-binding protein
Length=3130
Score = 46.2 bits (108), Expect = 1e-04, Method: Composition-based stats.
Identities = 33/130 (25%), Positives = 70/130 (53%), Gaps = 16/130 (12%)
Query 137 IPKQLAAKLPLHSRLKLQQGRRVGKQQQ---QKERDMKKAFVSPNERRAAVLLQQLETLK 193
+ +Q +L LK Q+ R+ +++Q QKE ++K+ ++ A+ Q+ E L+
Sbjct 2739 LKRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQ 2798
Query 194 QHRDKRRQQQQ----------QQKARLKQQQTDKLQQQQ---QQQQKERQKRRHSKQGKI 240
+ + +RQ+Q+ Q++ LK+Q+ ++LQ+++ +Q+Q+ QK K+ +
Sbjct 2799 KEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELKRQEQ 2858
Query 241 EASMRKRLRL 250
E RK++ L
Sbjct 2859 ERLERKKIEL 2868
Score = 43.5 bits (101), Expect = 7e-04, Method: Composition-based stats.
Identities = 34/150 (22%), Positives = 81/150 (54%), Gaps = 16/150 (10%)
Query 104 IKKEKKIISKQKETLFHSFGKAPRKQHSFFPIRIPKQLAAKLPLHSRLKLQQGRRVGKQQ 163
++KE+++ +++E L + +K+ + +Q +L LK Q+ R+ K++
Sbjct 2747 LQKEEELKRQEQERLEREKQEQLQKEE-----ELKRQEQERLQKEEALKRQEQERLQKEE 2801
Query 164 QQKERDMKKAFVSPNERRAAVLLQQLETLKQHRDKRRQQQQQQKARLKQQQTDKLQQQQQ 223
+ K ++ ++ ER LQ+ E LK+ +R Q+++ LK+Q+ ++LQ++++
Sbjct 2802 ELKRQEQERL-----EREKQEQLQKEEELKRQEQERLQKEEA----LKRQEQERLQKEEE 2852
Query 224 --QQQKERQKRRHSKQGKIEASMRKRLRLD 251
+Q++ER +R+ + + E ++ +L D
Sbjct 2853 LKRQEQERLERKKIELAEREQHIKSKLESD 2882
Score = 32.3 bits (72), Expect = 1.6, Method: Composition-based stats.
Identities = 16/69 (23%), Positives = 45/69 (65%), Gaps = 4/69 (5%)
Query 183 AVLLQQLETLKQHRDKRRQQQQ----QQKARLKQQQTDKLQQQQQQQQKERQKRRHSKQG 238
A+ Q+ E L++ + +RQ+Q+ +++ +L++++ K Q+Q++ Q++E KR+ ++
Sbjct 2738 ALKRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERL 2797
Query 239 KIEASMRKR 247
+ E ++++
Sbjct 2798 QKEEELKRQ 2806
> mmu:104662 Tsr1, AU040765, AW550801, mKIAA1401; TSR1, 20S rRNA
accumulation, homolog (yeast); K14799 pre-rRNA-processing
protein TSR1
Length=803
Score = 45.8 bits (107), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/91 (30%), Positives = 45/91 (49%), Gaps = 0/91 (0%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAIGTDGTFRASF 62
++ L G I K A V+ MF + +V +++T G RG IK+ +GT G + SF
Sbjct 695 RVVLSGHPFKIFTKMAVVRYMFFNREDVMWFKPVELRTKWGRRGHIKEPLGTHGHMKCSF 754
Query 63 EDKVLKSDLILCKTWVLVCPHAFYNPVLDLP 93
+ K+ D +L + V P Y+P + P
Sbjct 755 DGKLKSQDTVLMNLYKRVFPKWTYDPYVPEP 785
> hsa:55720 TSR1, FLJ10534, KIAA1401, MGC131829; TSR1, 20S rRNA
accumulation, homolog (S. cerevisiae); K14799 pre-rRNA-processing
protein TSR1
Length=804
Score = 45.1 bits (105), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 28/91 (30%), Positives = 45/91 (49%), Gaps = 0/91 (0%)
Query 3 KLKLVGETKSIKRKTAFVKGMFNSHLEVNRCLGAKIQTVSGIRGQIKKAIGTDGTFRASF 62
++ L G I K A V+ MF + +V +++T G RG IK+ +GT G + SF
Sbjct 696 RVVLSGHPFKIFTKMAVVRYMFFNREDVLWFKPVELRTKWGRRGHIKEPLGTHGHMKCSF 755
Query 63 EDKVLKSDLILCKTWVLVCPHAFYNPVLDLP 93
+ K+ D +L + V P Y+P + P
Sbjct 756 DGKLKSQDTVLMNLYKRVFPKWTYDPYVPEP 786
> cel:Y40C7B.1 hypothetical protein
Length=916
Score = 41.2 bits (95), Expect = 0.003, Method: Composition-based stats.
Identities = 33/135 (24%), Positives = 68/135 (50%), Gaps = 20/135 (14%)
Query 128 KQHSFFPIRIPKQLAAKLPLHSRLKLQQ-------------GRRVGKQQQQKERDMKKAF 174
K H+ FP +IP ++ + L + QQ + +Q Q++E+ + + F
Sbjct 181 KNHNRFPKQIPVAISTREELQQQRHKQQLEEYEKRKEILRREEEIKQQNQEREKQINEDF 240
Query 175 VSPNERRAAVLLQQLETLKQHRDKRRQQQQQ--QKARLKQQQTDKLQQQQQQQQKERQK- 231
E+R L++ E +KQ +R +Q +K +L++ Q +KL ++ +Q + +R+K
Sbjct 241 ----EKRNQEKLKREEEIKQQNQEREKQINDDFEKRKLEKLQREKLFEKNKQAKLQREKA 296
Query 232 RRHSKQGKIEASMRK 246
+ S++ K+E R+
Sbjct 297 HQESERRKLEKLKRE 311
> pfa:MAL13P1.176 PfRh2b; reticulocyte binding protein 2, homolog
B
Length=3179
Score = 32.3 bits (72), Expect = 1.6, Method: Composition-based stats.
Identities = 20/57 (35%), Positives = 33/57 (57%), Gaps = 3/57 (5%)
Query 179 ERRAAVLLQQLETLK---QHRDKRRQQQQQQKARLKQQQTDKLQQQQQQQQKERQKR 232
+R+ LQ+ E LK Q R +R +Q+Q QK +++ + QQQ+ Q+ E QK+
Sbjct 2664 KRQEQERLQKEEELKRQEQERLEREKQEQLQKEEELRKKEQEKQQQRNIQELEEQKK 2720
> ath:AT4G36520 heat shock protein binding
Length=1422
Score = 30.0 bits (66), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 53/89 (59%), Gaps = 6/89 (6%)
Query 147 LHSRLKLQQGRRVGKQQQQKERDMKKAFVSPNERR----AAVLLQQLE-TLKQHRDKRRQ 201
+ +R K +Q R++ K+QQ+ E +K+AF E R A L Q+ E +K+ R+K
Sbjct 691 VEAREKAEQERKM-KEQQELELQLKEAFEKEEENRRMREAFALEQEKERRIKEAREKEEN 749
Query 202 QQQQQKARLKQQQTDKLQQQQQQQQKERQ 230
+++ ++AR K + +L+ +Q++KERQ
Sbjct 750 ERRIKEAREKAELEQRLKATLEQEEKERQ 778
> cel:C25A11.4 ajm-1; Apical Junction Molecule family member (ajm-1)
Length=1480
Score = 30.0 bits (66), Expect = 8.2, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 53/94 (56%), Gaps = 12/94 (12%)
Query 150 RLKLQQGRRVGKQQQQKERDMKKAFVSPNERRAAVLLQQLETLKQHRDKRRQQQQQQKAR 209
R++ Q RR ++Q +++R+ N RR +L Q E + R R QQ ++A
Sbjct 270 RIREDQARRQQEEQDRRDRE-------DNARR---ILAQREHQEMER-LREQQNLSERAL 318
Query 210 LKQQQTDKLQQQQQQQ-QKERQKRRHSKQGKIEA 242
++++ DK + QQ++ +++R+K+R + ++E+
Sbjct 319 AERERADKERLQQERLLRQQREKKRREEWDRLES 352
Score = 29.6 bits (65), Expect = 9.9, Method: Composition-based stats.
Identities = 20/84 (23%), Positives = 44/84 (52%), Gaps = 5/84 (5%)
Query 154 QQGRRVGKQQQQKERDMKKAFVSPNERRAAVLLQQLETLKQHRDKRRQQQQQQKARLKQQ 213
Q+ R+ +QQ ER + + + ER LQQ L+Q R+K+R+++ + ++
Sbjct 302 QEMERLREQQNLSERALAERERADKER-----LQQERLLRQQREKKRREEWDRLESIRLA 356
Query 214 QTDKLQQQQQQQQKERQKRRHSKQ 237
+ + +++ +KER R +++
Sbjct 357 EEEAELARRRALEKERIDREKAEE 380
Lambda K H
0.319 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9340616888
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40