bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1942_orf1
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
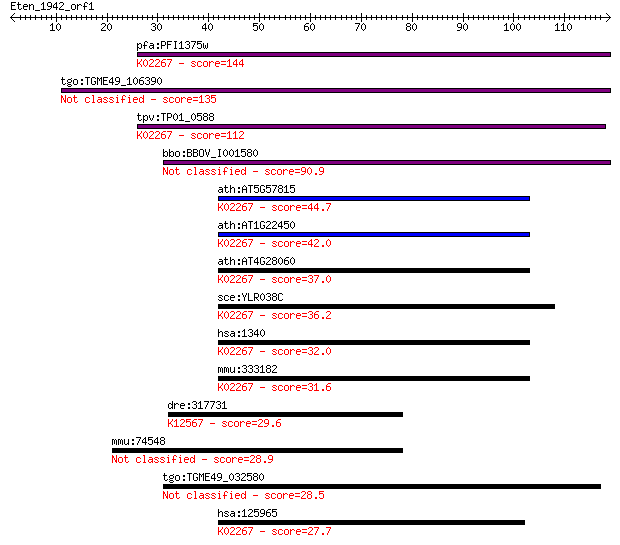
pfa:PFI1375w cytochrome c oxidase, putative (EC:1.10.2.2); K02... 144 7e-35
tgo:TGME49_106390 hypothetical protein 135 3e-32
tpv:TP01_0588 hypothetical protein; K02267 cytochrome c oxidas... 112 3e-25
bbo:BBOV_I001580 19.m02260; hypothetical protein 90.9 8e-19
ath:AT5G57815 cytochrome c oxidase subunit 6b, putative; K0226... 44.7 7e-05
ath:AT1G22450 COX6B; COX6B (CYTOCHROME C OXIDASE 6B); cytochro... 42.0 5e-04
ath:AT4G28060 cytochrome c oxidase subunit 6b, putative; K0226... 37.0 0.013
sce:YLR038C COX12; Cox12p (EC:1.9.3.1); K02267 cytochrome c ox... 36.2 0.024
hsa:1340 COX6B1, COX6B, COXG, COXVIb1; cytochrome c oxidase su... 32.0 0.50
mmu:333182 Cox6b2, 1700067P11Rik, BC048670, COXVIB2, MGC130249... 31.6 0.72
dre:317731 ttna, fi20g08, si:busm1-258d18.1, si:dz258d18.1, tt... 29.6 2.7
mmu:74548 Gsdmc4, 9030605I04Rik; gasdermin C4 28.9 3.8
tgo:TGME49_032580 acetyl-coenzyme A synthetase, putative (EC:6... 28.5 5.7
hsa:125965 COX6B2, COXVIB2, CT59, FLJ46422, MGC119094; cytochr... 27.7 9.6
> pfa:PFI1375w cytochrome c oxidase, putative (EC:1.10.2.2); K02267
cytochrome c oxidase subunit VIb [EC:1.9.3.1]
Length=103
Score = 144 bits (363), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 61/93 (65%), Positives = 79/93 (84%), Gaps = 0/93 (0%)
Query 26 YQRNIEELTAQNPHAEDPRLLQANQGSACALRYTMFCRCARELGEENPRCKYQYYRAQIA 85
+ +N +ELTAQNPHA DPR LQ NQ + CA RYT+FCRCARELGE++PRCK+QYYR+QIA
Sbjct 11 FIKNYDELTAQNPHASDPRFLQVNQFNHCAYRYTLFCRCARELGEDHPRCKFQYYRSQIA 70
Query 86 CTAEQMDDWNEHRSRGTCMFDIMPERSTKHLRQ 118
CTAEQ++DW+++R +GTC D +P++ T HLRQ
Sbjct 71 CTAEQLEDWDDNRQKGTCAMDTLPDKLTAHLRQ 103
> tgo:TGME49_106390 hypothetical protein
Length=191
Score = 135 bits (340), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 59/110 (53%), Positives = 83/110 (75%), Gaps = 2/110 (1%)
Query 11 PHCVLKMTKQEM--VQMYQRNIEELTAQNPHAEDPRLLQANQGSACALRYTMFCRCAREL 68
P V+KMT +QR++EELT+QNPH +DPR +Q NQ + C+LRY MF RCA+EL
Sbjct 82 PFFVVKMTDSNTNPALKFQRSVEELTSQNPHTDDPRSMQFNQMNTCSLRYAMFARCAKEL 141
Query 69 GEENPRCKYQYYRAQIACTAEQMDDWNEHRSRGTCMFDIMPERSTKHLRQ 118
G+++ RCKYQ+YRAQ+AC EQ++ W ++R +GTC FD++PE ST+H+ Q
Sbjct 142 GDDHTRCKYQFYRAQVACPIEQIELWEDYRQKGTCHFDLLPEFSTRHMWQ 191
> tpv:TP01_0588 hypothetical protein; K02267 cytochrome c oxidase
subunit VIb [EC:1.9.3.1]
Length=108
Score = 112 bits (280), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 48/92 (52%), Positives = 66/92 (71%), Gaps = 0/92 (0%)
Query 26 YQRNIEELTAQNPHAEDPRLLQANQGSACALRYTMFCRCARELGEENPRCKYQYYRAQIA 85
+ + ++ T NPHA DPR LQAN + C LRYT++CRC RELGE++PRCKYQYYR +++
Sbjct 16 FWESYDDYTLDNPHAGDPRCLQANNFNYCKLRYTLYCRCCRELGEDDPRCKYQYYRTELS 75
Query 86 CTAEQMDDWNEHRSRGTCMFDIMPERSTKHLR 117
CT + +D N+HR GTC DI+P R ++R
Sbjct 76 CTQDFLDLVNKHREEGTCSMDILPSRQLHNMR 107
> bbo:BBOV_I001580 19.m02260; hypothetical protein
Length=120
Score = 90.9 bits (224), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 46/100 (46%), Positives = 59/100 (59%), Gaps = 12/100 (12%)
Query 31 EELTAQNPHAEDPRLLQANQGSACALRYTMFCRCARELGEENPRCKYQYYRAQIACTAEQ 90
+ NPH DPR LQ Q + C LRY +FCRC REL E+NPRC+YQYYRA++ACT +
Sbjct 21 DHYNIDNPHVGDPRNLQFTQWNYCRLRYALFCRCCRELSEDNPRCRYQYYRAEMACTQDF 80
Query 91 MDDWNEHR------------SRGTCMFDIMPERSTKHLRQ 118
+D N++R S GT DI+P +LRQ
Sbjct 81 LDIANKYREQGNVGLLLRPPSLGTNTADILPTSQVFNLRQ 120
> ath:AT5G57815 cytochrome c oxidase subunit 6b, putative; K02267
cytochrome c oxidase subunit VIb [EC:1.9.3.1]
Length=78
Score = 44.7 bits (104), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 27/63 (42%), Positives = 31/63 (49%), Gaps = 4/63 (6%)
Query 42 DPRLLQANQGSACALRYTMFCRCARELGEENPRCK--YQYYRAQIACTAEQMDDWNEHRS 99
D R NQ C RY F RC GEE+ C+ +YYRA C E +D WNE R
Sbjct 13 DFRFPTTNQTRHCFTRYIEFHRCTTAKGEESNDCERFAKYYRA--LCPGEWVDKWNEQRE 70
Query 100 RGT 102
GT
Sbjct 71 SGT 73
> ath:AT1G22450 COX6B; COX6B (CYTOCHROME C OXIDASE 6B); cytochrome-c
oxidase; K02267 cytochrome c oxidase subunit VIb [EC:1.9.3.1]
Length=191
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 31/63 (49%), Gaps = 4/63 (6%)
Query 42 DPRLLQANQGSACALRYTMFCRCARELGEENPRCK--YQYYRAQIACTAEQMDDWNEHRS 99
D R NQ C RY + RC G++ P C ++YR+ C +E +D WNE R
Sbjct 125 DFRFPTTNQTRHCFTRYVEYHRCVAAKGDDAPECDKFAKFYRS--LCPSEWVDRWNEQRE 182
Query 100 RGT 102
GT
Sbjct 183 NGT 185
> ath:AT4G28060 cytochrome c oxidase subunit 6b, putative; K02267
cytochrome c oxidase subunit VIb [EC:1.9.3.1]
Length=164
Score = 37.0 bits (84), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 30/69 (43%), Gaps = 10/69 (14%)
Query 42 DPRLLQANQGSACALRYTMFCR------CARELGEENPRCK--YQYYRAQIACTAEQMDD 93
D R NQ C RY F R C GE+ C+ +YYRA C E +D
Sbjct 93 DFRFPTTNQTRHCFTRYIEFHRYSCVIECTTAKGEDANECERFAKYYRA--LCPGEWVDK 150
Query 94 WNEHRSRGT 102
WNE R GT
Sbjct 151 WNEQRETGT 159
> sce:YLR038C COX12; Cox12p (EC:1.9.3.1); K02267 cytochrome c
oxidase subunit VIb [EC:1.9.3.1]
Length=83
Score = 36.2 bits (82), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 28/66 (42%), Gaps = 0/66 (0%)
Query 42 DPRLLQANQGSACALRYTMFCRCARELGEENPRCKYQYYRAQIACTAEQMDDWNEHRSRG 101
D R Q NQ C Y + +C GE+ CK + C + ++ W++ R +G
Sbjct 15 DARFPQQNQTKHCWQSYVDYHKCVNMKGEDFAPCKVFWKTYNALCPLDWIEKWDDQREKG 74
Query 102 TCMFDI 107
DI
Sbjct 75 IFAGDI 80
> hsa:1340 COX6B1, COX6B, COXG, COXVIb1; cytochrome c oxidase
subunit VIb polypeptide 1 (ubiquitous) (EC:1.9.3.1); K02267
cytochrome c oxidase subunit VIb [EC:1.9.3.1]
Length=86
Score = 32.0 bits (71), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 27/64 (42%), Gaps = 3/64 (4%)
Query 42 DPRLLQANQGSACALRYTMFCRCAREL---GEENPRCKYQYYRAQIACTAEQMDDWNEHR 98
D R NQ C Y F RC + + G + C++ Q C + DW+E R
Sbjct 18 DSRFPNQNQTRNCWQNYLDFHRCQKAMTAKGGDISVCEWYQRVYQSLCPTSWVTDWDEQR 77
Query 99 SRGT 102
+ GT
Sbjct 78 AEGT 81
> mmu:333182 Cox6b2, 1700067P11Rik, BC048670, COXVIB2, MGC130249,
MGC130250; cytochrome c oxidase subunit VIb polypeptide
2; K02267 cytochrome c oxidase subunit VIb [EC:1.9.3.1]
Length=88
Score = 31.6 bits (70), Expect = 0.72, Method: Compositional matrix adjust.
Identities = 17/64 (26%), Positives = 26/64 (40%), Gaps = 3/64 (4%)
Query 42 DPRLLQANQGSACALRYTMFCRCAREL---GEENPRCKYQYYRAQIACTAEQMDDWNEHR 98
DPR NQ C + + RC + + G+ C+Y + C + WNE
Sbjct 20 DPRFPNQNQTRNCYQNFLDYHRCVKTMNRRGKSTQPCEYYFRVFHSLCPISWVQRWNEQI 79
Query 99 SRGT 102
+GT
Sbjct 80 KQGT 83
> dre:317731 ttna, fi20g08, si:busm1-258d18.1, si:dz258d18.1,
ttn, wu:fi04b11, wu:fi20g08; titin a; K12567 titin [EC:2.7.11.1]
Length=32757
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 25/46 (54%), Gaps = 1/46 (2%)
Query 32 ELTAQNPHAEDPRLLQANQGSACALRYTMFCRCARELGEENPRCKY 77
E A +P E P++L+ Q AL + RC R +G+ +P C++
Sbjct 1840 EKVALSPSMEAPKILERIQSQTVALADEVRFRC-RVIGKPDPECQW 1884
> mmu:74548 Gsdmc4, 9030605I04Rik; gasdermin C4
Length=480
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 14/57 (24%), Positives = 26/57 (45%), Gaps = 0/57 (0%)
Query 21 EMVQMYQRNIEELTAQNPHAEDPRLLQANQGSACALRYTMFCRCARELGEENPRCKY 77
E+V+ + + ++ P P+LL QG A+ Y + C ++ NPR +
Sbjct 387 ELVKSILQTNFKYSSNTPFTLQPQLLAPLQGEGLAITYELLEECGLKMELNNPRSTW 443
> tgo:TGME49_032580 acetyl-coenzyme A synthetase, putative (EC:6.2.1.1)
Length=821
Score = 28.5 bits (62), Expect = 5.7, Method: Composition-based stats.
Identities = 25/89 (28%), Positives = 38/89 (42%), Gaps = 3/89 (3%)
Query 31 EELTAQNPHAEDPRLLQANQG--SACALRYTMFCRCARELGEENPRCKYQYYRAQIACTA 88
+EL+ + HA+ ++ A+ G + Y A +L P K R+Q CT
Sbjct 288 QELSTRIDHAKPKAIIAASCGIEPRRIVPYKEGLEHALQLSSHKPEFKVILQRSQHECTL 347
Query 89 EQMD-DWNEHRSRGTCMFDIMPERSTKHL 116
E D DW+ G+ DI+P S L
Sbjct 348 ESSDIDWDCFIKSGSGQCDILPVDSNHPL 376
> hsa:125965 COX6B2, COXVIB2, CT59, FLJ46422, MGC119094; cytochrome
c oxidase subunit VIb polypeptide 2 (testis); K02267 cytochrome
c oxidase subunit VIb [EC:1.9.3.1]
Length=88
Score = 27.7 bits (60), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 16/63 (25%), Positives = 24/63 (38%), Gaps = 3/63 (4%)
Query 42 DPRLLQANQGSACALRYTMFCRCAR---ELGEENPRCKYQYYRAQIACTAEQMDDWNEHR 98
DPR NQ C + + RC + G+ C+Y + C ++ WNE
Sbjct 20 DPRFPSQNQIRNCYQNFLDYHRCLKTRTRRGKSTQPCEYYFRVYHSLCPISWVESWNEQI 79
Query 99 SRG 101
G
Sbjct 80 KNG 82
Lambda K H
0.323 0.132 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2027872200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40