bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1961_orf1
Length=169
Score E
Sequences producing significant alignments: (Bits) Value
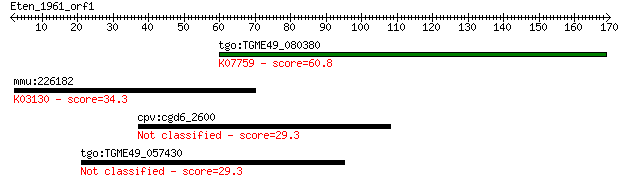
tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143); K0... 60.8 2e-09
mmu:226182 Taf5, 6330528C20Rik, AV117817; TAF5 RNA polymerase ... 34.3 0.19
cpv:cgd6_2600 hypothetical protein 29.3 6.2
tgo:TGME49_057430 hypothetical protein 29.3 6.4
> tgo:TGME49_080380 non-transmembrane antigen (EC:3.2.1.143);
K07759 poly(ADP-ribose) glycohydrolase [EC:3.2.1.143]
Length=553
Score = 60.8 bits (146), Expect = 2e-09, Method: Composition-based stats.
Identities = 37/112 (33%), Positives = 57/112 (50%), Gaps = 3/112 (2%)
Query 60 AILLPQNPQGWQQVQQILAEVEAGKVPKTAGEYKQLFTSILTAAGNWRVDKPLRI---GP 116
AIL + W + + IL V +G P + L +L +GN R +K RI G
Sbjct 47 AILRTPVEEDWGKTKYILNFVASGFGPADPQQLASLQKRLLKLSGNIRTEKNKRILQRGL 106
Query 117 LTHHLATHKNLSKSLLPFLASLARRLPDFFPSGIRHLGRDNPAVHLRRIEVL 168
L + LPF+A+L R+ + FPSG++++ +NP VHLR+I+V
Sbjct 107 LNYLEENPGKFFSHDLPFMATLVMRIDELFPSGLQYITPENPQVHLRKIQVF 158
> mmu:226182 Taf5, 6330528C20Rik, AV117817; TAF5 RNA polymerase
II, TATA box binding protein (TBP)-associated factor; K03130
transcription initiation factor TFIID subunit 5
Length=801
Score = 34.3 bits (77), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 27/72 (37%), Positives = 36/72 (50%), Gaps = 4/72 (5%)
Query 2 GAAAAVAAALLLLLFVSPPGFAAAAPPGDDDGAVAGTSAAAAAAAA----AAAAAQQQQG 57
GA A A+ALL + S PG AA PPG + S +A+ AA A+ A + Q
Sbjct 139 GAGAEAASALLSRVTASVPGSAAPEPPGTGASVTSVFSGSASGPAAPGKVASVAVEDQPD 198
Query 58 LRAILLPQNPQG 69
+ A+L N QG
Sbjct 199 VSAVLSAYNQQG 210
> cpv:cgd6_2600 hypothetical protein
Length=692
Score = 29.3 bits (64), Expect = 6.2, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 32/73 (43%), Gaps = 4/73 (5%)
Query 37 GTSAAAAAAAAAAAAAQQQQGLRAILLPQNPQGWQQVQQILA--EVEAGKVPKTAGEYKQ 94
+S + A A ++ G+ + Q PQGW+ V E E KVP A E +
Sbjct 300 ASSQSTATPQPQVRGAVRKSGVPGVYWSQKPQGWRVVYYTGKDREFEYFKVPANASE--E 357
Query 95 LFTSILTAAGNWR 107
+ + IL A +R
Sbjct 358 IISEILEVAKRFR 370
> tgo:TGME49_057430 hypothetical protein
Length=2250
Score = 29.3 bits (64), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 20/74 (27%), Positives = 32/74 (43%), Gaps = 13/74 (17%)
Query 21 GFAAAAPPGDDDGAVAGTSAAAAAAAAAAAAAQQQQGLRAILLPQNPQGWQQVQQILAEV 80
G AAAP GD DG +G+S A+ A++ A W +V +A
Sbjct 898 GVCAAAPEGDSDGRGSGSSTYRRASCASSVGAASSD-------------WAEVPGPVAST 944
Query 81 EAGKVPKTAGEYKQ 94
A ++P T + ++
Sbjct 945 RASRLPATQAQARR 958
Lambda K H
0.319 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4222647260
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40